Abstract
Mitogen-activated protein kinases (MAPKs) are evolutionarily conserved proteins that function as key signal transduction components in fungi, plants, and mammals. During interaction between phytopathogenic fungi and plants, fungal MAPKs help to promote mechanical and/or enzymatic penetration of host tissues, while plant MAPKs are required for activation of plant immunity. However, new insights suggest that MAPK cascades in both organisms do not operate independently but that they mutually contribute to a highly interconnected molecular dialogue between the plant and the fungus. As a result, some pathogenesis-related processes controlled by fungal MAPKs lead to the activation of plant signaling, including the recruitment of plant MAPK cascades. Conversely, plant MAPKs promote defense mechanisms that threaten the survival of fungal cells, leading to a stress response mediated in part by fungal MAPK cascades. In this review, we make use of the genomic data available following completion of whole-genome sequencing projects to analyze the structure of MAPK protein families in 24 fungal taxa, including both plant pathogens and mycorrhizal symbionts. Based on conserved patterns of sequence diversification, we also propose the adoption of a unified fungal MAPK nomenclature derived from that established for the model species Saccharomyces cerevisiae. Finally, we summarize current knowledge of the functions of MAPK cascades in phytopathogenic fungi and highlight the central role played by MAPK signaling during the molecular dialogue between plants and invading fungal pathogens.
INTRODUCTION
The development of all organisms relies on the ability of cells to sense and respond rapidly to changes in the surrounding environment. To coordinate appropriate cellular actions, eukaryotes use diverse receptors that perceive specific cues and relay information to intracellular signaling pathways. The interface created during the interaction between a phytopathogenic fungus and its host is a rich source of signals that feed into this molecular dialogue, resulting in rapid and highly structured responses in both protagonists.
Despite completely different lifestyles, fungi and plants rely on many analogous signaling pathways to coordinate their respective cellular actions. Among evolutionarily conserved pathways, mitogen-activated protein kinase (MAPK) cascades function as key signal transducers that use protein phosphorylation/dephosphorylation cycles to channel information. MAPK cascades in all eukaryotes generally consist of three interlinked protein kinases (PKs) that are sequentially activated (Widmann et al., 1999). Activated MAPK kinase kinases (MAP3Ks) first phosphorylate two Ser and/or Thr residues located within the activation loop of MAPK kinases (MAP2Ks). Activated MAP2Ks in turn trigger MAPK activation through dual phosphorylation of a highly conserved activation loop that possesses the hallmark motif -TXY-. Activated MAPKs can then phosphorylate downstream substrates, affecting their biochemical properties and leading to specific output responses.
Arabidopsis thaliana has been widely employed as a model to examine MAPK functions in the plant kingdom. Plant MAPKs have been shown to regulate numerous cellular processes, including biotic stress relief (Pitzschke et al., 2009; Andreasson and Ellis, 2010). In fungi, the study of MAPKs from Saccharomyces cerevisiae has provided pivotal insights that have contributed greatly to our understanding of MAPK signaling in all eukaryotes (Chen and Thorner, 2007). This has been particularly important for studies of other fungi, including both human and plant pathogens, where orthologous MAPK signaling modules have been found to be involved in the control of infection-related morphogenesis (IRM), virulence, cell wall biogenesis, and stress responses (Xu, 2000; Zhao et al., 2007; Rispail et al., 2009).
In this review, we aimed at identifying the full complement of MAPK signaling components in a range of taxonomically diverse fungi that yet all interact with plants. Based on the analysis of recovered protein sequences, we could resolve the fungal MAPK family into four separate clades, while three distinct subgroups were defined for each of the fungal MAP2K and MAP3K protein families. As clustering of these signaling components is recapitulated in the model fungal species S. cerevisiae, we propose a unified nomenclature that relies on gene/protein names from yeast to identify homologous candidates from the investigated fungal species. To avoid ambiguous annotation of orthologous candidates, we also make use of three-letter species acronyms that indicate the origin of each gene or protein (Table 1). Finally and as a complement to this, we summarize current knowledge of the functions of MAPK cascades in phytopathogenic fungi and highlight the central role played by MAPK signaling during the molecular dialogue between plants and fungal pathogens.
Table 1. Plant-Interacting Fungi and Three-Letter Species Prefixes.
| Phylum | Organism | Prefix | Plant Disease | Infection Strategy |
| Ascomycota | A. brassicicola | Abr | Brassica dark leaf spot disease | Necrotroph |
| Ascomycota | B. cinerea | Bci | Polyphage (e.g., grape noble rot) | Necrotroph |
| Ascomycota | C. heterostrophus | Che | Southern corn leaf blight disease | Nectrotroph |
| Ascomycota | C. parasitica | Cpa | Chestnut blight disease | Necrotroph |
| Ascomycota | F. graminearum | Fgr | Wheat head-blight disease | Necrotroph |
| Ascomycota | F. oxysporum | Fox | Polyphage (several formae speciales) | Hemibiotroph |
| Ascomycota | Fusarium verticillioides | Fve | Rice bakanae disease | Necrotroph |
| Basidiomycota | H. annosum sensu lato | Han | Conifer root rot disease | Necrotroph |
| Basidiomycota | L. bicolor | Lbi | None: mutualist | Ectomycorrhizae |
| Ascomycota | M. oryzae | Mor | Rice blast disease | Hemibiotroph |
| Basidiomycota | M. larici-populina | Mlp | Poplar leaf rust disease | Obligate biotroph |
| Ascomycota | Mycosphaerella fijiensis | Mfi | Banana (Musa spp) black leaf spot disease | Necrotroph |
| Ascomycota | M. graminicola | Mgr | Septoria leaf blotch of wheat | Necrotroph |
| Ascomycota | Mycosphaerella pini | Mpi | Pine needle blight disease | Hemibiotroph |
| Ascomycota | Mycosphaerella populorum | Mpo | Septoria leaf spot and canker of poplar | Hemibiotroph |
| Basidiomycota | P. graminis-tritici | Pgr | Wheat and barley stem rust disease | Obligate biotroph |
| Basidiomycota | P. triticina | Put | Wheat leaf rust disease | Obligate biotroph |
| Ascomycota | Pyrenophora tritici-repentis | Ptr | Wheat yellow leaf spot disease | Necrotroph |
| Ascomycota | S. cerevisiae | Sce | None: does not interact with plants | None |
| Ascomycota | S. nodorum | Sno | S. nodorum blotch of wheat | Necrotroph |
| Ascomycota | Sclerotinia sclerotiorum | Ssc | White mold disease | Necrotroph |
| Ascomycota | Trichoderma virens | Tvi | None: biocontrol agent | Mycoparasitism |
| Ascomycota | T. melanosporum | Tme | None: mutualist | Ectomycorrhizae |
| Basidiomycota | U. maydis | Uma | Maize smut disease | Biotroph |
| Ascomycota | V. dahliae | Vda | Wilt disease | Necrotroph |
| Not included in phylogenetic analysis | ||||
| Ascomycota | C. purpurea | Cpu | Ergot of rye and other grains | Biotroph |
| Ascomycota | C. orbiculare | Cor | Cucumber (Cucumis sativus) anthracnose disease | Hemibiotroph |
| Basidiomycota | P. striiformis | Pst | Wheat stripe rust | Obligate biotroph |
| Ascomycota | P. teres | Pte | Barley net blotch | Necrotroph |
MAPKs FROM BUDDING YEAST
In S. cerevisiae, MAPK cascades constitute essential signaling pathways that are involved in key aspects of the yeast life cycle (Chen and Thorner, 2007). As a result, these protein modules as well as both their upstream regulators and downstream substrates are highly conserved even in distantly related fungi, including plant pathogens (Rispail et al., 2009). S. cerevisiae thus represents an excellent model for the study of MAPK signaling in fungi. To allow better understanding of MAPK signaling in phytopathogenic fungi, we first provide a brief description of prototypical MAPK cascades from yeast.
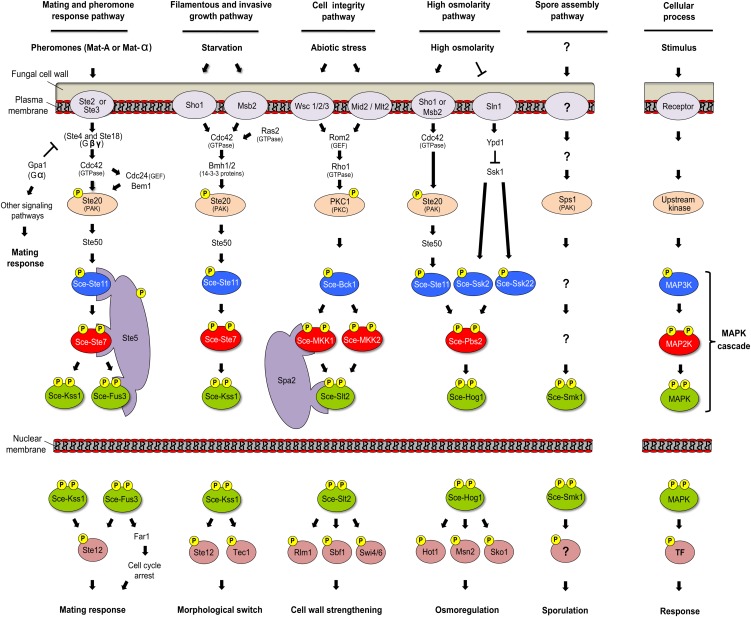
In the budding yeast, five MAPK pathways regulate mating, invasive growth, cell wall integrity, hyperosmolarity responses, and ascospore formation (Chen and Thorner, 2007). The yeast mating response depends on G protein–coupled receptors (Ste2 and Ste3) that bind cognate peptidic pheromones (Figure 1). This binding results in dissociation of the inhibitory Gα subunit Gpa1, from Ste4 and Ste18, which respectively function as stimulatory Gβ and Gγ subunits. Released Gβγ subunits associate with the scaffolding protein Ste5, and the p21-activated kinase (PAK) Ste20, to create a protein complex that activates the MAP3K Ste11. Ste11 is the entry point for a MAPK cascade that also includes MAP2K Ste7 and two partially redundant MAPKs, Kss1 and Fus3. The cyclin-dependent kinase (CDK) inhibitor Far1 and the transcription factor (TF) Ste12, which respectively control cell cycle arrest and expression of mating-responsive genes, are primary targets of the pheromone response pathway (Figure 1).
Figure 1.
MAPK Pathways in S. cerevisiae.
Yeast cells rely on four MAPK cascades to regulate mating, invasive growth, cell integrity, and high osmolarity. Smk1 may be part of a fifth, yet undefined, pathway that regulates ascospore formation. Activation of MAPK cascades depends on transmembrane receptors that perceive extracellular cues and translate information to intracellular signaling components, including GTPases, GEFs, and PKs. Upstream regulators may be shared between distinct signaling pathways, and timely activation of a specific cascade must be tightly regulated. In some cases, signal specificity is achieved through the use of scaffolding proteins promoting interaction between suitable MAPK signaling components. Following activation, MAPKs phosphorylate an array of substrates, including TFs that induce a transcriptional shift controlling output responses. See text for more details on each signaling pathway. In light of this study, the acronym “Sce” (for S. cerevisiae) was added to the standard nomenclature of yeast MAPKs.
Many components of the pheromone pathway, including Ste20, Ste50, Ste11, Ste7, Kss1, and Ste12, are also involved in regulating filamentous growth (Figure 1). This response is stimulated upon starvation and leads to invasive growth in haploid cells or pseudohyphal development in diploid cells. Under nutrient-rich conditions, inactive Kss1 localizes to the nucleus, where it sequesters TFs such as Ste12 and Tec1. Phosphorylation of Kss1 relieves its negative regulation and results in TF release. However, upstream components of the pheromone pathway, such as the pheromone receptors, heterotrimeric G proteins, and the Ste5 scaffold protein, are not required for filamentation. In this developmental context, activation of the Ste11-Ste7-Kss1 module instead depends on osmosensors Sho1 and Msb2 (Figure 1).
A third yeast MAPK cascade forms part of the cell integrity sensing pathway and consists of the MAP3K Bck1, two redundant MAP2Ks, MKK1 and MKK2, as well as the Slt2 MAPK (Figure 1). This cascade is required to maintain cell wall integrity and is involved in compensatory responses against environmental stresses like high temperature or exposition to heavy metals. Surface sensors perceive external cues and relay information to Rom2, a guanine exchange factor (GEF) that regulates the activity of the Rho1 GTPase. Active Rho1 then activates PKC1, a homolog of the α, β, and γ isoforms of mammalian PK C. PKC1 then phosphorylates Bck1 to initiate MAPK signaling through the Slt2 cascade. Associated with this module is the Spa2 adaptor protein, which directs appropriate MAPK signaling components to cell growth sites. Activated Slt2 then modifies downstream targets, such as the TFs Rlm1, Sbf1, and Swi4/6, which are involved in regulating the transcriptional programming required for cell wall biogenesis (Figure 1).
In yeast cells grown under hypertonic conditions, the high osmolarity glycerol (HOG) pathway (Figure 1) is required for accumulation of osmoprotectant molecules and maintenance of an osmotic gradient across the stressed plasma membrane. This pathway leads to the activation of Hog1, a stress-related MAPK. The Pbs2 MAP2K and three MAP3Ks (Ste11, Ssk2, and Ssk22) complete this MAPK module, which can be activated by two signaling input branches converging at the level of Pbs2. While activation of Ssk2 and Ssk22 depends on a two-component His kinase phosphorelay system (Sln1, Ypd1, and Ssk1), activation of Ste11 depends on a signaling pathway shared with the filamentous growth pathway and comprising proteins Sho1, Msb2, Cdc42, Ste20, and Ste50. Several TFs, including Hot1, Msn2, and Sko1, lie downstream of Hog1 and control expression of genes involved in osmotic as well as oxidative stress responses (Figure 1).
Smk1, the fifth MAPK from S. cerevisiae, is unique because it is only expressed during late stages of meiosis, prior to ascospore enclosure (Krisak et al., 1994). Smk1, which has no homolog in other fungi except in some ascomycetous yeasts, regulates spore morphogenesis by controlling assembly of the ascospore wall. The Smk1 pathway is the least well defined, with neither the upstream MAP2K nor MAP3K so far identified (Figure 1). Since PAK Sps1 is also involved in ascospore wall assembly, it is believed to work upstream of the Smk1 MAPK (Friesen et al., 1994).
PHYLOGENY OF MAPK FAMILIES IN PLANT-INTERACTING FUNGI
Considering the importance of MAPK cascades in fungal biology and virulence against plants (see the MAPKs from phytopathogenic fungi section), we took advantage of genomic databases that are available following completion of genome sequencing projects (U.S. Department of Energy Joint Genome Institute, Broad Institute of MIT and Harvard, and Genoscope Sequencing Center, France; see Supplemental Table 1 online) to identify the full complement of associated MAPK signaling components. Overall, the genomes of 24 taxonomically diverse fungi were analyzed in our survey. A total of 260 genes and corresponding proteins were identified, including 112 MAPKs, 74 MAP2Ks, and 74 MAP3Ks (see Supplemental Data Sets 1 to 3, respectively, and Supplemental References 1 online). Importantly, more than 50% of all identified candidates correspond to PKs that have never been described in the literature, suggesting that the assembled information will provide a useful resource for researchers wishing to compare and investigate MAPK modules in plant-interacting fungi. In the case of model plant pathogens Magnaporthe oryzae, Ustilago maydis, Fusarium spp, and Botrytis cinerea, our screen was accurate enough to retrieve all the previously identified MAPK signaling components (Dean et al., 2005; García-Pedrajas et al., 2008; Rispail et al., 2009; Ma et al., 2010; Amselem et al., 2011; see Supplemental Data Sets 1 to 3 online). We therefore presume that remaining gene models and associated proteins identified here correspond to the full complement of MAPK signaling components in the investigated fungi.
MAPKs
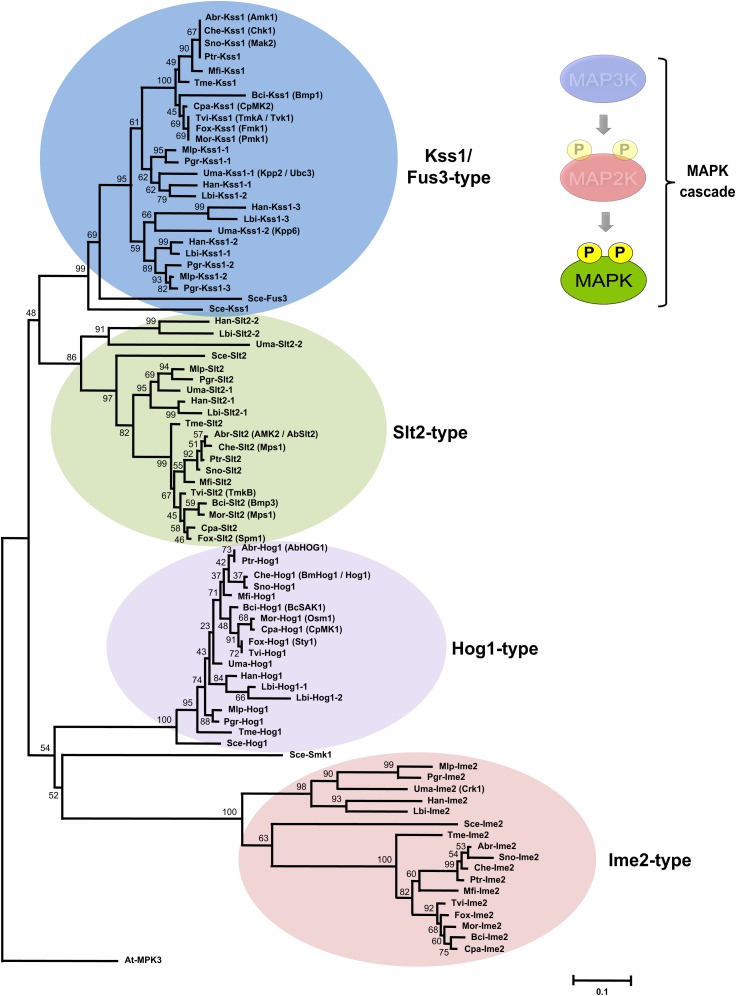
Full-length MAPK protein sequences were aligned (see Supplemental Data Set 4 online), and a phylogenetic analysis was conducted (Figure 2). This showed that the fungal MAPK family can be resolved into four separate clades, each of which contained at least one representative member from each of the investigated fungal species (Figure 2). Certain MAPK homologs display a strikingly high level of similarity, even though they belong to different fungal species (e.g., Kss1/Fus3-type homologs from Dothideomycetes; Figure 2; see Supplemental Data Set 1 online). With the exception of Verticillium dahliae, which has two Hog1 homologs, all the ascomycetes examined have four conserved MAPK candidates, including a single homolog for each of the yeast Kss1 (or Fus3), Slt2, and Hog1 prototypical MAPKs (Figure 2; see Supplemental Data Set 1 online). On the other hand, basidiomycete species generally possess a larger number of MAPKs, with up to seven or eight candidates in the Agaricomycetes (Figure 2; see Supplemental Data Set 1 online). The increased number of MAPKs in these species most likely results from duplication events that created extra gene copies retained during diversification of the various basidiomycete species. Gene duplication is observed for some Slt2 and Hog1 homologs, although this feature is particularly prominent for MAPKs homologous to yeast Kss1 and Fus3 (Figure 2; see Supplemental Data Set 1 online). Since most of the duplicated MAPKs are found on scaffolds that have not yet been assigned to a specific chromosome, it is not clear which kind of duplication events might be involved in the expansion of the MAPK family in basidiomycetes. However, since no obvious MAPK gene cluster could be identified, this suggests that family expansion is not associated primarily with tandem gene duplication, but rather with whole-genome or large segmental duplication events. Interestingly, the dichotomy existing between ascomycetes and basidomycetes is recapitulated in the clustering of MAPKs (Figure 2; this is also true for MAP2Ks and MAP3Ks). For basidiomycetous species, the segregation of MAPK candidates also reflects the established phylogeny associated with fungal classes (Ustilaginomycetes, Pucciniomycetes, and Agaricomycetes; Figure 2). All the investigated species also contain a single MAPK-related protein that is homologous to yeast Ime2 (Figure 2; see Supplemental Data Set 1 online). When compared with classical MAPKs, Ime2 homolog sequences have diverged more extensively, yet these proteins have been sufficiently preserved to define a fourth clade of MAPKs (Figure 2).
Figure 2.
Phylogenetic Relationships of MAPKs from Plant-Interacting Fungi.
Genome assembly from various fungi was searched using amino acid sequence of yeast MAPKs as queries. Retrieved gene models were accepted only if corresponding protein displayed consensus sequences of Ser/Thr PKs, including conserved Asp and Lys residues within the active site (D[L/I/V]K motif), and an appropriately positioned activation loop comprising the conserved -TXY- phosphorylation motif. Full-length PKs were next aligned with ClustalW (see Supplemental Data Set 4 online) using plant MAPK At-MPK3 as an outgroup. The following alignment parameters were used: for pairwise alignment, gap opening, 10.0, and gap extension, 0.1; for multiple alignment, gap opening, 10.0, and gap extension, 0.20. Resulting alignments were submitted to Molecular Evolutionary Genetics Analysis 4 (MEGA4) software (Tamura et al., 2007) to generate a neighbor-joining tree derived from 5000 replicates. Bootstrap values are indicated on the nodes of each branch. A colored circle depicts each type of MAPKs, and a species acronym indicates the origin of each protein (Table 1; see Supplemental Data Set 1 online). In relevant cases, previous MAPK nomenclature is indicated in parenthesis.
MAP2Ks
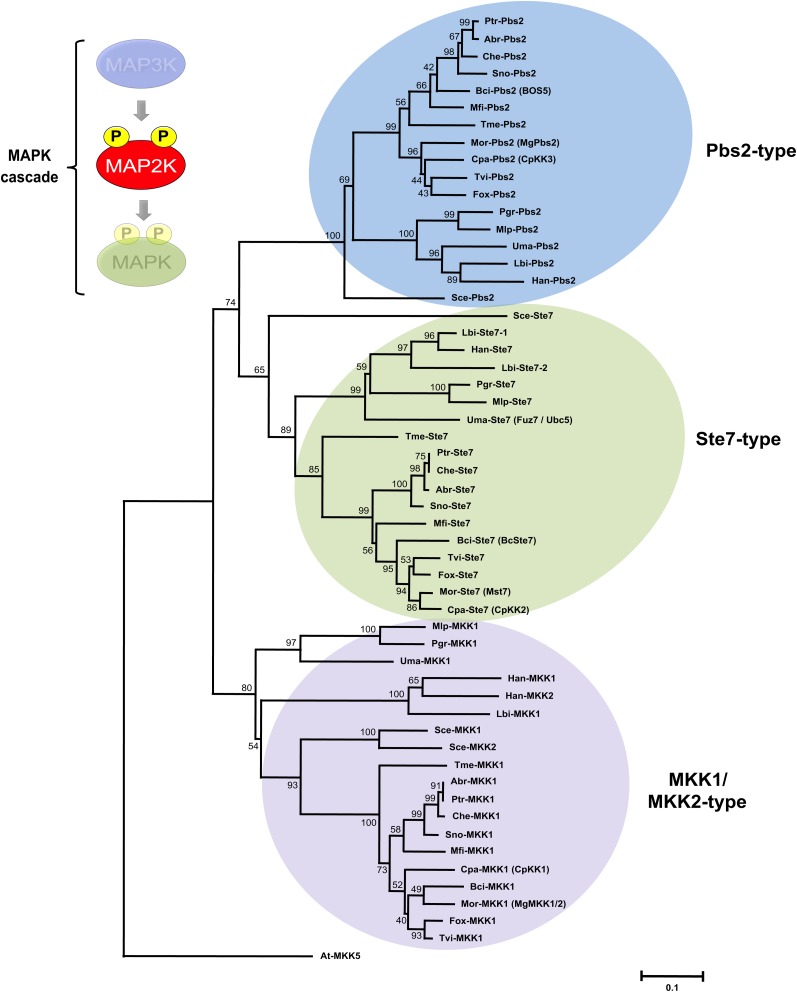
The results of phylogenetic analysis of MAP2K sequences indicate that this protein family can be divided into three clades, each of which contains at least one representative from each of the examined fungi (Figure 3; see Supplemental Data Set 5 online). With the exception of the two Agaricomycetes, all surveyed fungal species have three MAP2K candidates, which appear to be the homologs for the yeast Ste7, MKK1 (or MKK2), and Pbs2 prototypical MAP2Ks (Figure 3; see Supplemental Data Set 2 online). In contrast with the pattern observed among the basidiomycetous MAPKs, it seems that gene duplication has not led to expansion of the fungal MAP2K family. The only obvious duplication events, including the presence of two MKK1/2 homologs in Heterobasidion annosum sensu lato (also referred to as H. irregulare; see Olson et al., 2012) and two Ste7 homologs in Laccaria bicolor (Martin et al., 2008), seem to be lineage-specific events since they are not recapitulated in the other Agaricomycetes examined, namely, L. bicolor and H. annosum sensu lato (Figure 3; see Supplemental Data Set 2 online). The relatively small number of MAP2Ks encoded in these genomes also suggests that each of these proteins likely activates multiple MAPKs, a scenario that may be more prominent in basidiomycetes that possess recently duplicated gene pairs in several clades of MAPKs. Experimental support for this prediction comes from the study of U. maydis, where the MAP2K Fuz7 (here renamed Uma-Ste7 according to our unified nomenclature; see below) has been shown to operate upstream of several MAPKs (Garrido et al., 2004; Di Stasio et al., 2009).
Figure 3.
Phylogenetic Relationships of MAP2Ks from Plant-Interacting Fungi.
Genome assembly from various fungi was searched using amino acid sequence of yeast MAP2Ks as queries. Retrieved gene models were accepted only if corresponding protein contained consensus sequences of dual-specificity PKs, including conserved Asp and Lys residues within the active site (D[L/I/V]K motif), and an appropriately positioned activation loop comprising the conserved [S/T]xxx[S/T] phosphorylation motif. Full-length PKs were next aligned with ClustalW (see Supplemental Data Set 5 online) using plant MAP2K At-MKK5 as an outgroup. The following alignment parameters were used: for pairwise alignment, gap opening, 10.0, and gap extension, 0.1; for multiple alignment, gap opening, 10.0, and gap extension, 0.20. Resulting alignments were submitted to MEGA4 software to generate a neighbor-joining tree derived from 5000 replicates. Bootstrap values are indicated on the nodes of each branch. A colored circle depicts each type of MAP2K and a species acronym indicates the origin of each protein (Table 1; see Supplemental Data Set 2 online). In relevant cases, previous MAP2K nomenclature is indicated in parenthesis.
MAP3Ks
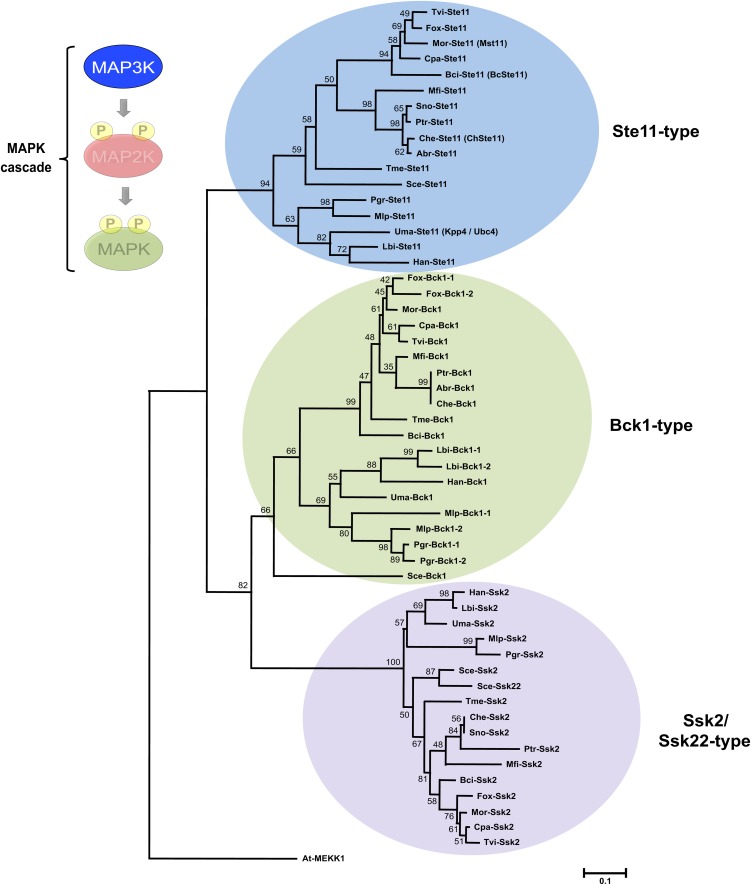
Phylogenetic analysis of fungal MAP3K sequences demonstrates that this family also consists of three well-resolved clades, defined by homologs of the yeast Ste11, Bck1, and Ssk2/Ssk22 prototypical MAP3Ks (Figure 4; see Supplemental Data Set 6 online). Surprisingly, the ascomycetes Alternaria brassicicola and Stagonospora nodorum possess only two predicted MAP3Ks and lack Ssk2/Ssk22 or Bck1 homologs, respectively (Figure 4; see Supplemental Data Set 3 online). This situation may be due to sequencing artifacts, since MAPKs and MAP2Ks that presumably function downstream of the apparently missing MAP3Ks can be identified in the genome of both fungi. With the exception of Fusarium oxysporum, all examined ascomycetes have a single candidate for each of yeast Ste11, Bck1, and Ssk2/Ssk22 MAP3Ks (Figure 4; see Supplemental Data Set 3 online). In F. oxysporum, duplication of Bck1 homologs seems lineage specific, as this pattern is not conserved even in closely related Fusarium species (Figure 4; see Supplemental Data Set 3 online; Ma et al., 2010). Interestingly, lineage-specific duplication of Bck1 homologs can also be identified in basidiomycetes such as the rust fungi Melampsora larici-populina and Puccinia graminis-tritici as well as within the ectomycorrhizal fungus L. bicolor (Figure 4; see Supplemental Data Set 3 online; Martin et al., 2008; Duplessis et al., 2011). Even though refined functional analysis of each paralog has yet to be reported, the duplication of Bck1 homologs suggests that the cell integrity pathway operating in several plant-interacting fungi may be more complex than that in yeast and that new specialized functions may have arisen following recent duplication of specific signaling components.
Figure 4.
Phylogenetic Relationships of MAP3Ks from Plant-Interacting Fungi.
Genome assembly from various fungi was searched using amino acid sequence of yeast MAP3Ks as queries. Retrieved gene models were accepted only if corresponding protein contained consensus sequences for Ser/Thr PKs, including conserved Asp and Lys residues within the active site (D[L/I/V]K motif).
Comparisons with other eukaryotic MAP3Ks were also conducted to confirm protein identification (data not shown). Full-length PKs were aligned with ClustalW (see Supplemental Data Set 6 online) using plant MAP3K At-MEKK1 as an outgroup. The following alignment parameters were used: for pairwise alignment, gap opening, 10.0, and gap extension, 0.1; for multiple alignment, gap opening, 10.0, and gap extension, 0.20. Resulting alignments were submitted to MEGA4 software to generate a neighbor-joining tree derived from 5000 replicates. Bootstrap values are indicated on the nodes of each branch. A colored circle depicts each type of MAP3K and a species acronym indicates the origin of each protein (Table 1; see Supplemental Data Set 3 online). In relevant cases, previous MAP3K nomenclature is indicated in parentheses.
MAPK Signaling Components from Symbiotic Fungi
The development of long-lasting compatible interactions between mutualistic telluric fungi and the roots of forest trees requires highly controlled reprogramming of cellular activities, and consistent with this, signaling genes are overrepresented in collections of ESTs isolated from symbiotically associated partners (Martin et al., 2001; Sundaram et al., 2001; Voiblet et al., 2001; Peter et al., 2003). Earlier attempts to investigate gene expression during mycorrhiza development reported the presence of MAPKs expressed in the free-living mycelium of the basidiomycetes Pisolithus microcarpus and L. bicolor (Peter et al., 2003) and the induction of several signaling-related transcripts, including those encoding MAPKs, during mycorrhiza development (Voiblet et al., 2001; Duplessis et al., 2005; Le Quéré et al., 2005). The recent availability of genome sequences from ectomycorrhizal fungi (L. bicolor and Tuber melanosporum) has now made it possible to identify the full complement of MAPK signaling components in plant-beneficial fungi (Martin et al., 2008, 2010). Although they belong to distinct taxa (Basidiomycota for L. bicolor and Ascomycota for T. melanosporum), both of these fungi establish ectomycorrhizal symbioses with forest trees. Genome sequence analysis shows that MAPKs are relatively well conserved in T. melanosporum compared with other ascomycetes, whereas L. bicolor presents a few gene family expansions compared with other fungi (for more details, see phylogenetic classification of MAPK families in the preceding sections). Genome-scale transcriptome studies show that MAPK genes are relatively highly expressed in the free-living mycelium, fruiting bodies, and ectomycorrhiza compared with other conserved genes (Martin et al., 2008, 2010), indicating that these signaling pathways likely play a role in the development and functioning of the symbiotic association. Although no genome sequence is yet available from endomycorrhizal fungi in the Glomeromycota, the recent report of nonredundant virtual transcripts from the fungus Glomus intraradices confirms the presence and expression of several MAPK signaling components (Tisserant et al., 2012).
At the functional level, only limited information is currently available on MAPK signaling in symbiotic fungi. In the ectomycorrhizal fungus Tuber borchii, the Kss1/Fus3-type MAPK TBMK (here renamed Tbo-Kss1 according to our unified nomenclature; see below) becomes phosphorylated during interaction of the fungus with its host tree, Tilia americana (Menotta et al., 2006). Interestingly, ectopic expression of this MAPK in F. oxysporum strains lacking Fmk1 (here renamed Fox-Kss1; see below) partially restores their ability to grow invasively (Menotta et al., 2006). This suggests that MAPK signaling could play an important role during the presymbiotic colonization phase, perhaps by modulating expression of target genes necessary to allow host infection and establishment of functional ectomycorrhizae. The induction of fungal MAPK transcript expression at later stages of ectomycorrhizal development (Voiblet et al., 2001; Duplessis et al., 2005; Martin et al., 2008, 2010) also suggests that MAPK signaling may be involved in communication events after the symbiosis is established.
UNIFIED NOMENCLATURE FOR FUNGAL MAPK FAMILIES
In the past years, MAPK signaling has been studied in a wide array of fungi, including several plant pathogens and symbionts (reviewed in Xu, 2000; Zhao et al., 2007; Rispail et al., 2009; this article). During this time, research reports have generated disparate and confusing gene/protein designations that do not necessarily reflect putative orthologous relationships between homologs from distinct fungal species (see Supplemental Data Sets 1 to 3 online). Trivial naming also marked the plant MAPK family, before establishment of a unified nomenclature based on the model species, Arabidopsis (Ichimura, 2002; Hamel et al., 2006).
In light of the strong evolutionary conservation between fungal MAPK signaling components (Figures 2 to 4), we propose the adoption of a systematic nomenclature derived from that established for the model species S. cerevisiae. MAPK cascades from yeast are viewed as prototypical signaling modules, and the ancient patterns of diversification that gave rise to the modern fungal MAPK families are well conserved in this ascomycete (Zhao et al., 2007; Rispail et al., 2009; this article). Based on these principles, we therefore suggest that fungal MAPKs should be referred to as either belonging to the Kss1/Fus3, Slt2, or Hog1 type (Figure 2; see Supplemental Data Set 1 online). We also propose the designation of a fourth clade that includes conserved homologs of the MAPK-related protein Ime2 from yeast (Figure 2; see Supplemental Data Set 1 online). For each of the MAP2K and MAP3K families, we propose the designation of three subgroups, reflecting the phylogenetic architecture of each family. MAP2Ks should now belong to the Ste7, MKK1/MKK2, or Pbs2 type (Figure 3; see Supplemental Data Set 2 online), whereas MAP3Ks should now belong to the Ste11, Bck1, or Ssk2/Ssk22 type (Figure 4; see Supplemental Data Set 3 online).
To discriminate between signaling components from various fungal species, we recommend use of a three-letter acronym preceding each protein type (e.g., Abr for A. brassicicola; Table 1). For basidiomycetes that possess clearly paralogous forms of a given kinase, additional numbering should be included (e.g., Han-Slt2-1 and Han-Slt2-2 versus Abr-Slt2). We recognize that such a naming and numbering protocol is driven essentially by predicted evolutionary relationships and that conservation of biological function within most of these relationships remains to be confirmed. Indeed, disruption of a particular MAPK type may well have unequal phenotypic effects in various fungal species, since those effects will be modulated by interactions of the kinase within the larger genetic and cellular context of each species. Nonetheless, the systematic nomenclature proposed here should facilitate species-to-species comparisons and limit the expansion of trivial naming that already marks the MAPK family from phytopathogenic fungi (see Supplemental Data Set 1 online).
MAPKs FROM PHYTOPATHOGENIC FUNGI
Following characterization of the first MAPK from a fungal plant pathogen (Xu and Hamer, 1996), similar strategies have been employed to examine the role of MAPKs in other phytopathogenic fungi. Generally, degenerate primers allow PCR-based amplification of MAPK genes, and associated functions are assessed through loss-of-function studies. Knockout strains can, for instance, be examined for phenotypes involving altered vegetative growth and virulence. More recently, comparative genomics has facilitated study of fungal signaling pathways (Rispail et al., 2009), and complete MAPK cascades have been functionally defined in model plant pathogens.
The Kss1-1/Kss1-2 MAPK Cascade in U. maydis
U. maydis is the causal agent of maize (Zea mays) smut disease, an infection characterized by the development of tumor-like structures called galls (Kahmann and Kämper, 2004; Brefort et al., 2009). This basiodiomycete has been widely used as a model species to study dimorphism, a process that refers to the ability of certain fungi to switch between unicellular and multicellular growth forms to maintain resource uptake under changing conditions (Nadal et al., 2008). For some phytopathogenic fungi, this morphology switch is a prerequisite to plant infection, and in most cases, an encounter with the host is responsible for triggering the dimorphic shift.
In the absence of host plants, U. maydis develops as a saprobic, haploid, unicellular organism that is incapable of infecting maize (Kahmann and Kämper, 2004; Brefort et al., 2009). Upon contact with the host, compatible haploid cells mate and initiate formation of a dikaryotic filament. This transition marks the initiation of a parasitic growth stage typified by appressorium formation, penetration, and the formation of filamentous hyphae inside plant tissues. Mating of U. maydis cells depends on a pheromone-receptor system that is coupled to MAPK signaling (Figure 5; Kahmann and Kämper, 2004; Brefort et al., 2009). Continued parasitic growth depends on the same MAPK cascade, as well as on another signaling pathway that is activated by modulation of the intracellular level of cAMP (Figure 5).
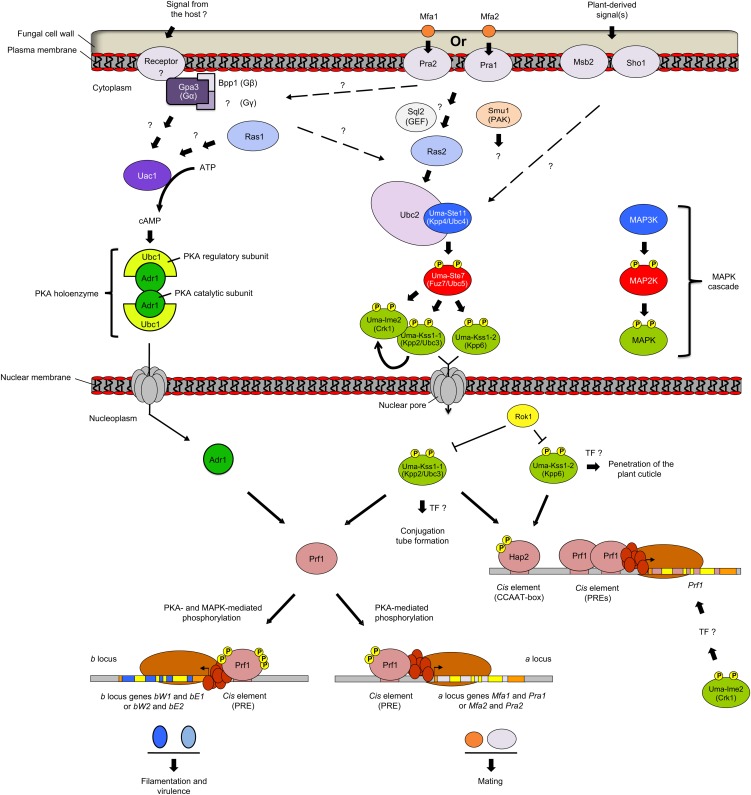
Figure 5.
Mating, Filamentous Growth, and Virulence in U. maydis.
In Ustilago, several MAPKs regulate mating by acting downstream of the pheromone receptor (see text for details). Along with cAMP signaling, MAPKs are also involved in filamentous growth and virulence. The cAMP pathway comprises heterotrimeric G proteins that function upstream of the adenylate cyclase Uac1. cAMP-dependent PKA holoenzyme works as a tetramer comprising two catalytic subunits (Adr1) and two regulatory subunits (Ubc1). When cAMP level is low, binding of regulatory subunits prevents catalytic activity. At higher cAMP levels, conformational changes allow release of catalytic subunits that enter in the nucleus. MAPK and cAMP signaling converge toward TF Prf1, which controls expression of pheromone- and virulence-induced genes. Hap2 is another MAPK substrate controlling expression of Prf1. In light of this study, previous MAPK names are depicted in parentheses. This figure has been adapted from Nadal et al. (2008).
The U. maydis mating apparatus includes a seven-transmembrane receptor (Pra1 or Pra2), which recognizes cognate lipopeptide pheromones (Mfa2 or Mfa1, respectively; Figure 5; Kahmann and Kämper, 2004; Brefort et al., 2009). Downstream of the receptor lies a MAPK cascade consisting of the MAP3K Uma-Ste11 (previously named Kpp4/Ubc4), the MAP2K Uma-Ste7 (Fuz7/Ubc5), and the MAPK Uma-Kss1-1 (Kpp2/Ubc3; Figure 5; Banuett and Herskowitz, 1994; Mayorga and Gold, 1999; Andrews et al., 2000). Ste7 is essential for mating, filamentous growth, and virulence and was the first component of the cascade to be characterized (Banuett and Herskowitz, 1994). It was later found that deletion of Kss1-1 partially compromises mating, formation of filamentous dikaryons, and virulence (Mayorga and Gold, 1999; Müller et al., 2003b). Partial phenotypes are correlated with the existence of Uma-Kss1-2 (also named Kpp6; Figure 5), a closely related MAPK that possesses an unusually long N-terminal extension (Brachmann et al., 2003). Kss1-2 participates in regulation of virulence since deletion mutants are unable to penetrate the plant cuticule (Brachmann et al., 2003). Most importantly, Kss1-1/Kss1-2 double knockouts fail to mate and are nonpathogenic, suggesting that these MAPKs possess at least partially overlapping functions during mating and virulence (Figure 5; Zhao et al., 2007; Brefort et al., 2009; Rispail et al., 2009). Consistent with this, Kss1-1 and Kss1-2 share Ste7 as an upstream activator, as well as Rok1, a member of the dual-specificity protein phosphatase family, as a negative regulator (Figure 5; Di Stasio et al., 2009).
A loss-of-function study also indicates that Ustilago MAP3K Ste11 lies upstream of the Ste7-Kss1-1 module, since Δste11 mutants are defective in both mating and virulence (Figure 5; Andrews et al., 2000; Müller et al., 2003b). Ste11 also interacts with Ubc2 (Figure 5), an adaptor protein with homology to Ste50 from S. cerevisiae (Figure 1; Klosterman et al., 2008). Ubc2 had previously been associated with regulation of the Kss1-1 cascade, since its deletion impairs pheromone response and virulence (Mayorga and Gold, 2001). Ubc2 harbors sterile α motif (SAM) and Ras association domains that are typical of Ste50-like adaptors from ascomycetous fungi, as well as Src homology 3 domains that are conserved only in Ubc2-like homologs from basidiomycetous fungi (Klosterman et al., 2008). Yeast two-hybrid (Y2H) assays indicate that Ubc2 and Ste11 interact via their respective SAM domains, and deletion experiments have shown that the SAM and Ras association domains are indispensable for mating and filamentous growth, while Src homology 3 domains specifically regulate virulence (Klosterman et al., 2008).
One of the Ras proteins from U. maydis has also been positioned upstream of Ubc2 and its cognate MAPK cascade (Figure 5; Lee and Kronstad, 2002). Thus, expression of a dominant active ras2 allele promotes pseudohyphal growth in a manner dependent on the Kss1-1 MAPK cascade. One possible activator of Ras2 is the GEF protein Sql2 (Figure 5; Müller et al., 2003a). However, unlike ras2 mutants, sql2 knockouts are defective in plant infection but not in mating. It has been suggested that Sql2 may activate Ras2 only under certain circumstances, for example, following perception of host-derived signals (Klose et al., 2004). Other signaling components acting between the Ustilago pheromone receptors and the Kss1-1 MAPK cascade remain unknown and may differ from those operating in the yeast pheromone pathway (Figure 1). For instance, none of the four Gα subunits from U. maydis could be linked with pheromone receptors (Zhao et al., 2007). In addition, the PAK Smu1 is dispensable for mating and plant infection (Figure 5; Smith et al., 2004), even though it shares high homology with PAK Ste20 from yeast (Figure 1).
In U. maydis, pheromone perception induces transcription of several target genes, including those located within the loci “a” and “b” (Kahmann and Kämper, 2004; Brefort et al., 2009). The “a” locus contains two genes encoding mating pheromones and receptors, while the “b” locus contains two homeodomain genes named bE and bW (Figure 5). Regulation of these genes requires the Kss1-1/Kss1-2 cascade as well as cAMP signaling. These pathways converge toward Prf1, a high mobility group domain TF that recognizes pheromone response elements in regulatory regions of target genes (Figure 5). The promoter of Prf1 itself contains two pheromone response elements, which most likely allows self-regulation of the TF (Figure 5; Kahmann and Kämper, 2004; Brefort et al., 2009).
Regulation of Prf1 also depends on its phosphorylation status (Figure 5), and this TF has six putative MAPK phosphorylation sites (Müller et al., 1999). Mutation of these sites creates an allele that retains its ability to drive basal expression of “a” genes but loses its capacity to induce “b” gene expression following pheromone stimulation. Refined delineation of Prf1 phosphorylation patterns demonstrated that only three MAPK phosphorylation sites are required for Prf1 function (Figure 5; Kaffarnik et al., 2003). Prf1 also contains two PKA phosphorylation sites that are sufficient and essential for “a” gene induction (Figure 5; Kaffarnik et al., 2003). Kss1-1 and PKA both interact with Prf1 in vivo, and kinase assays confirm their ability to phosphorylate this TF. Taken together, these results imply that regulation of “a” genes depends solely on Prf1 phosphorylation by PKA, while expression of “b” genes requires phosphorylation by both PKA and MAPKs (Figure 5). Prf1 therefore constitutes a key crosstalk point at which signals derived from both MAPK and cAMP signaling pathways are integrated, with differential phosphorylation status controlling the ability of Prf1 to discriminate between target gene promoters (Figure 5; Kaffarnik et al., 2003).
Hap2 is another TF that was shown to operate downstream of MAPKs in U. maydis. This protein was identified as a Kss1-2–interacting protein but was later found to associate equally well with Kss1-1 (Mendoza-Mendoza et al., 2009). Hap2 regulates Prf1 gene expression by binding to a cis-element called the CCAAT-box (Figure 5). Deletion of Hap2 abolishes mating and results in defective expression of pheromone-responsive genes. Mutation of predicted Hap2 MAPK phosphorylation sites attenuates the pheromone response (Mendoza-Mendoza et al., 2009), suggesting that phosphorylation of this TF is required to fine-tune the expression of Prf1 (Figure 5). Epistasis studies also suggest that more unidentified TFs likely operate downstream of Kss1-1 and Kss1-2 to regulate conjugation tube formation and penetration of the plant cuticle (Figure 5).
The Kss1 MAPK Cascade in M. oryzae
Upon host sensing, many phytopathogenic fungi differentiate specialized structures allowing hyphal penetration of plant tissues. MAPKs have been associated with this process, also referred to as IRM. Typically, IRM is initiated shortly after germination of asexual spores called conidia. Emerging germ tubes grow and differentiate into a dome-shaped appressorium that adheres tightly to the hydrophobic plant surface. Following formation of a penetration peg, the appressorium generates strong turgor pressure that ultimately breaches underlying plant tissues (Figure 6; Talbot, 2003; Ebbole, 2007; Wilson and Talbot, 2009).
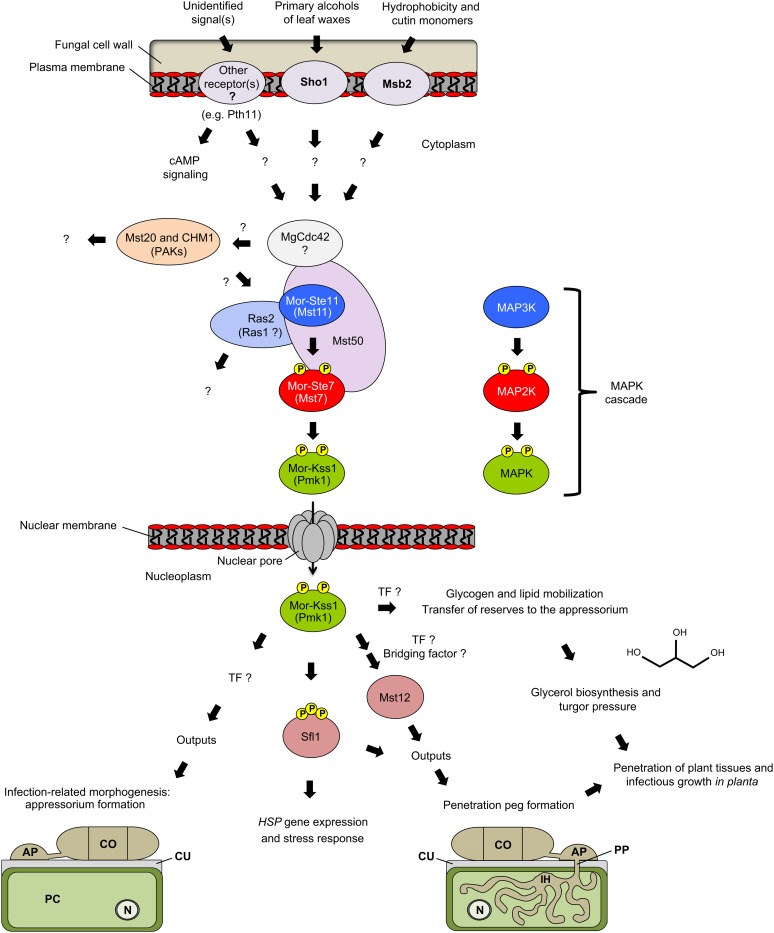
Figure 6.
The Pathogenicity Pathway from M. oryzae.
Following sensing of host signals, the pathogenicity pathway is activated. Components mediating early steps of signal transduction converge toward a protein complex comprising Mst50 and MAPK signaling components. Following activation by Ste7, the Kss1 MAPK stimulates differentiation of appressoria and controls formation of turgor pressure. Nuclear localization of Kss1 is consistent with the fact that TFs accomplish MAPK function. Study of Mst12 indicates that IRM and infectious growth are separated processes, both required for full pathogenicity. Mst12 is not involved in appressorium formation but functions downstream of Kss1 to control penetration peg formation and proliferation inside host tissues. Sfl1 is another MAPK substrate regulating expression of stress-related genes. In light of this study, previous MAPK names are depicted in parentheses. This figure has been adapted from Park et al. (2006). AP, appressorium; CO, conidia; CU, cuticule; IH, infection hyphae; N, nucleus; PC, plant cell; PP, penetration peg.
One of the most well-studied appressorium-forming fungi is the ascomycete M. oryzae (formerly named Magnaporthe grisea), a hemibiotroph that causes a devastating rice (Oryza sativa) blast disease (Talbot, 2003; Ebbole, 2007; Wilson and Talbot, 2009). In this pathogen, cAMP regulates recognition of the plant surface, a prerequisite for the initiation of IRM. This process also depends on Mor-Kss1 (previously named Pmk1), a MAPK that stimulates appressorium formation (Figure 6). Kss1 knockouts still recognize hydrophobic surfaces and respond to cAMP but fail to form appressoria or produce lesions on susceptible rice plants (Xu and Hamer, 1996). Δkss1 strains also lack the ability to mobilize glycogen and lipid reserves that allow glycerol accumulation and generation of high hydrostatic turgor (Figure 6; Thines et al., 2000). In line with this, Kss1 expression is induced during appressorium formation, and the corresponding protein predominantly accumulates in the appressorium nucleus (Bruno et al., 2004). Mutant strains are also unable to colonize wounded rice tissue, indicating that this MAPK is not only required for early penetration steps but also for infectious growth once inside the plant (Xu and Hamer, 1996).
Based on sequence comparisons with yeast and epistasis studies, the MAP2K Mor-Ste7 (Mst7) and the MAP3K Mor-Ste11 (Mst11) have been positioned upstream of Mor-Kss1 (Figure 6; Zhao et al., 2005). M. oryzae strains lacking ste7 and ste11 fail to form appressoria and are nonpathogenic, while ectopic expression of an active Ste7 allele restores appressorium formation in ste7 and ste11 genetic backgrounds. Moreover, expression of an active Ste7 allele results in phosphorylation of the downstream Kss1 MAPK. On the other hand, appressoria formed by such complemented lines fail to penetrate rice leaves, indicating that active Ste7 can only partially restore defects of ste7 and ste11 mutants (Zhao et al., 2005). Coimmunoprecipitation and bimolecular fluorescence complementation assays also indicate that Ste7 and Kss1 physically interact during appressorium formation and that this interaction depends on a MAPK-docking site located at the N terminus of the MAP2K (Zhao and Xu, 2007).
Y2H assays also reveal that Ste7 and Ste11 interact with Mst50 (Figure 6; Zhao et al., 2005), a homolog of the yeast adaptor protein Ste50 (Figure 1). This ternary association most likely results in the formation of a complex that stabilizes the otherwise weak interaction between Mor-Ste7 and Mor-Ste11 (Zhao et al., 2005). Strains lacking Mst50 are defective in appressorium formation and fail to infect rice (Park et al., 2006). Expression of active Ste7 complements appressorium formation, but not lesion development, in mst50 knockout lines, and coimmunoprecipitation experiments confirms the physical interaction between Ste7, Ste11, and Mst50 (Figure 6; Park et al., 2006).
Adaptor protein Mst50 also interacts with the GTPase Cdc42 (Figure 6; Park et al., 2006), a homolog of Cdc42 from yeast (Figure 1). However, Mor-Cdc42 is unlikely to participate in the activation of the Mor-Kss1 cascade since it is dispensable for appressorium formation and plant infection (Zhao et al., 2007). Two M. oryzae PAKs, Mst20 and CHM1, have also been characterized in search of other candidate working as upstream regulators (Li et al., 2004). Mst20 is dispensable for appressorium formation and plant infection, whereas Δchm1 mutants are only reduced in appressorium formation and still grow invasively on wounded rice leaves. This indicates that pak mutants are phenotypically dissimilar from Δkss1 strains and suggests that Mst20 and CHM1 individually play no critical role in activating the Kss1 pathway from M. oryzae (Figure 6; Li et al., 2004).
Mst50 and Ste11 also interact with GTPases Ras1 and Ras2 (Figure 6; Park et al., 2006). Ras1 knockouts have no defect in appressorium formation or plant infection, whereas lethality hinders characterization of Ras2 knockouts. Nonetheless, expression of an active Ras2 allele stimulates appressorium formation in a wild-type M. oryzae strain, but not in mst50, ste11, ste7, or kss1 mutants, indicating that at least one function of Ras2 is mediated through the Kss1 pathway (Figure 6; Park et al., 2006).
Examination of the Magnaporthe infection process also uncovered Kss1-dependent responses involving the TF Mst12 (Figure 6; Park et al., 2002). Mst12 belongs to a family of proteins that are homologous to Ste12, a well-known substrate of yeast MAPKs (Figure 1). Mutant strains lacking Mst12 are nonpathogenic in rice (Park et al., 2002) and produce classic dome-shaped appressoria that, however, fail to penetrate plant surface. It is thought that Mst12 controls cytoskeleton reorganization associated with penetration peg formation (Park et al., 2004a) and that mutants lacking this TF fail to orient the physical forces exerted by appressorium turgor. mst12 mutants are also compromised in infectious growth since inoculation through wound sites does not allow fungal spreading (Park et al., 2002). The Kss1 cascade from M. oryzae thus regulates appressorium differentiation and infectious growth by relying on output responses that are uncoupled, with yet unidentified TF(s) mediating appressorium formation and Mst12 promoting penetration peg formation and invasive development (Figure 6).
In light of the direct phosphorylation of Ste12 by yeast MAPKs (Figure 1), and because corresponding Magnaporthe mutants display partially overlapping phenotypes, it is tempting to hypothesize that there is a direct functional relationship between Mor-Kss1 and Mst12. Interestingly, however, Y2H assays detected only a weak interaction between the two proteins (Park et al., 2002). Mst12 homologs have also been characterized in other phytopathogenic fungi (Rispail and Di Pietro, 2010), and again no direct evidence supports a physical interaction between these TFs and upstream MAPKs. In line with this, all Ste12-like TFs from filamentous fungi lack the protein motif that mediates interaction between Ste12 and yeast MAPKs (Wong Sak Hoi and Dumas, 2010). These observations argue for a conserved function of Ste12-like TFs downstream of MAPKs but also for an indirect relationship involving yet unidentified players that may function as bridging factors for these key signaling components (Figure 6).
In an attempt to identify further Mor-Kss1 substrates, a protein named Sfl1 was recently identified out of a screen for phosphorylated TFs (Figure 6; Li et al., 2011). Sfl1 belongs to the heat shock factor family and contains a putative MAPK docking site as well as three predicted MAPK phosphorylation sites. Coimmunoprecipitation experiments indicate that Kss1 and Sfl1 interact in vivo, and deletion of Sfl1 results in significant reduction of virulence against rice and barley (Hordeum vulgare; Li et al., 2011). Mutant strains differentiate appressoria but appear to be defective for invasive growth. sfl1 mutants also show increased sensitivity to elevated temperatures, a finding most likely related to the fact that this TF controls expression of genes encoding heat shock proteins (Figure 6; Li et al., 2011). Screening of a cDNA subtraction library also identified GAS1 and GAS2, two Kss1-regulated genes encoding small proteins required for full appressorium function and virulence (Xue et al., 2002). Comparative analysis of ESTs obtained from wild-type appressoria and kss1 mutant germlings also revealed other potential genetic targets of this key MAPK (Ebbole et al., 2004).
Kss1/Fus3-Type MAPKs in Other Phytopathogenic Fungi
Several MAPKs regulate morphogenetic transitions associated with plant infection by U. maydis and M. oryzae. Importantly, these infection-related MAPKs are evolutionarily related to Kss1 and Fus3 (Figure 2), which regulate mating and filamentous growth in S. cerevisiae (Figure 1). Homologous Kss1/Fus3-type MAPKs have also been characterized in other fungal plant pathogens, and functional analyses confirm that this class of PKs plays important functions in establishment of various infection strategies.
In the case of investigated pathogens that form well-defined appressoria, including Cochliobolus heterostrophus (Lev et al., 1999), Colletotrichum orbiculare (formerly known as C. lagenarium; Takano et al., 2000), and Pyrenophora teres (Ruiz-Roldán et al., 2001), Kss1/Fus3-type MAPKs are required for appressorium formation. More recently, the MAP3K MEKK1 (Cor-Ste11 according to our proposed nomenclature) was found to act upstream of the MAPK Cmk1 (here renamed Cor-Kss1) in C. orbiculare (Sakaguchi et al., 2010). C. orbiculare strains lacking Ste11 and Kss1 both fail to form an appressorium, while nuclear localization of a Kss1–green fluorescent protein fusion is diminished in the ste11 deletion mutants. In C. heterostrophus, the MAP3K Ste11 was proposed to work upstream of the MAPK CHK1 (here renamed Che-Kss1), since loss-of-function study revealed that Ste11 and Kss1 are both required for appressorium formation (Izumitsu et al., 2009). In addition, the Che-Kss1 MAPK was previously shown to be required for timely induction of genes encoding cellulolytic enzymes and controlling host tissue penetration (Lev and Horwitz, 2003).
Other phytopathogenic fungi do not form appressoria and thus rely on distinct strategies to colonize host tissues. For instance, penetration of plant aerial parts can occur through stomata that provide direct access to underlying photosynthetic mesophyll tissues. In the wheat (Triticum aestivum) pathogen Mycosphaerella graminicola, the MAPK gene Fus3 (here renamed Mgr-Kss1) is essential for colonization of the host, and knockout mutants fail to penetrate stomata (Cousin et al., 2006). Based on homology with putative orthologs from M. oryzae, additional components of the Mgr-Kss1 pathway have been identified, and complementation assays have emphasized the evolutionary conservation of this important signaling pathway (Kramer et al., 2009). In the cereal pathogen S. nodorum, deletion of the MAPK gene Mak2 (here renamed Sno-Kss1) produces strains that achieve penetration of stomata but are still nonpathogenic because of their inability to produce infection structures within the host mesophyll (Solomon et al., 2005).
The activity of Kss1/Fus3-type MAPKs is also associated with virulence of several soil-borne pathogens that cause wilt disease symptoms in a variety of crops. In F. oxysporum, deletion of the MAPK gene Fmk1 (Fox-Kss1 in our analysis) results in strains that fail to differentiate penetration hyphae and that display reduced transcript level of the pectate lyase gene pl1 (Di Pietro et al., 2001). In V. dahliae, disruption of the MAPK gene VMK1 (here renamed Vda-Kss1) also results in strains that have reduced virulence against a variety of host plants (Rauyaree et al., 2005). In the necrotrophic fungus A. brassicicola, disruption of the MAPK gene Amk1 (here renamed Abr-Kss1) results in strains that are nonpathogenic on intact plants but that still colonize damaged host tissues (Cho et al., 2007). Unlike Mor-Kss1, this suggests that Abr-Kss1 specifically controls host penetration, whereas subsequent proliferation within the host does not predominantly rely on this MAPK. Accordingly, Alternaria mutants lacking this Kss1/Fus3-type MAPK produce lower amounts of hydrolytic enzymes that are usually required for penetration of host tissues (Cho et al., 2007). In B. cinerea, inactivation of the MAPK gene BMP1 (here renamed Bci-Kss1) results in strains that are essentially nonpathogenic, since infection hyphae fail to penetrate and macerate plant tissues (Zheng et al., 2000; Doehlemann et al., 2006). Independent studies also revealed that deletion of the MAPK gene MAP1/Gpmk1 (here renamed Fgr-Kss1) hinders pathogenicity of Fusarium graminearum, the causal agent of wheat head-blight disease (Jenczmionka et al., 2003; Urban et al., 2003). Again, apathogenicity of Fgr-kss1 mutants could be associated with diminished or delayed induction of enzymatic activity normally associated with degradation of the plant cell wall (Jenczmionka and Schäfer, 2005).
The function of Kss1/Fus3-type MAPKs was also assessed in biotrophic fungi, including the hemibiotroph pathogen Claviceps purpurea (Mey et al., 2002b). C. purpurea mutants lacking the MAPK gene mk1 (here renamed Cpu-Kss1) are nonpathogenic, while complementation assays reveal that Cpu-Kss1 can rescue M. oryzae strains lacking their corresponding Kss1 gene. Kss1/Fus3-type MAPKs from obligate biotrophic fungi have also been studied, even though lack of genetic transformation and gene disruption strategies hinders epistasis study. Pharmacological inhibitors nevertheless demonstrated the importance of MAPK signaling in appressorial development of the powdery mildew fungus Blumeria graminis (Kinane and Oliver, 2003). Complementation assays in the surrogate basidiomycete U. maydis also indicate that MAPK1 (here renamed Put-Kss1-2), from the rust fungus Puccinia triticina, can overcome mating and pathogenicity defects of U. maydis kss1-1 and kss1-2 single mutants or kss1-1 kss1-2 double mutants (Hu et al., 2007). It was also shown that MAPK1 (here renamed Pst-Kss1), from the rust fungus Puccinia striiformis, partially complements the corresponding kss1 mutants from F. graminearum and M. oryzae (Guo et al., 2011). Interestingly, the expression of Pst-Kss1 is induced at early infection stages and peaks during formation of the haustorium, suggesting that Kss1/Fus3-type MAPKs are involved in the virulence of rust fungi, perhaps by controlling IRM associated with the differentiation of haustoria.
In light of more than 30 published studies, it can generally be assumed that Kss1/Fus3-type MAPKs are multifunctional pathogenicity factors required for virulence of biologically and taxonomically diverse phytopathogenic fungi. Interestingly, Δkss1 mutants from the multihost pathogen F. oxysporum are nonpathogenic in tomato (Solanum lycopersicum; Di Pietro et al., 2001) but remain fully pathogenic in a murine model system (Ortoneda et al., 2004). This indicates that MAPK signaling plays functionally distinct roles during infection of plants and infection of other organisms and that Kss1/Fus3-type MAPKs share an ancient function in pathogenicity against plants.
Slt2-Type MAPKs in Phytopathogenic Fungi
Homologs of the yeast Kss1 and Fus3 are not the only MAPKs whose function is critical for virulence of pathogenic fungi (Zhao et al., 2007; Rispail et al., 2009). Many pathogenesis-related MAPKs are indeed more closely related to Slt2, the yeast cell wall integrity-associated MAPK (Figure 1). In fungal plant pathogens, the first characterized Slt2-type MAPK was Mps1 from M. oryzae (Xu et al., 1998), which we therefore suggest renaming Mor-Slt2 (see Supplemental Data Set 2 online). Mor-Slt2 mutants are nonpathogenic since, while they differentiate appressoria, these fail to penetrate plant tissues. This MAPK also controls cell wall integrity as reflected in the observation that M. oryzae mutants are hypersensitive to cell wall–degrading enzymes and that aging colonies grown on plates undergo autolysis in the absence of osmotic stabilization. Taken together, these results suggest that Mor-Slt2 controls overall cell wall strength as well as remodeling of the appressorium wall during host infection (Xu et al., 1998). Recently, it was shown that, upon plant infection, α-1,3-glucans accumulate in the fungal cell wall. Interestingly, these polysaccharides tend to be localized close to the outer layer of the cell wall, farther from the fungal cell membrane (Fujikawa et al., 2009). Such uneven distribution could favor masking of chitin and β-1,3-glucans, which are highly sensitive to plant hydrolases. Interestingly, accumulation of reinforcing α-1,3-glucans does not occur in Mor-slt2 knockout strains, a situation possibly reflecting one way in which this MAPK regulates strengthening of the fungal cell wall during plant colonization (Fujikawa et al., 2009).
Our understanding of the cell wall integrity pathway from M. oryzae was also expanded by the characterization of MCK1, a MAP3K that we suggest renaming Mor-Bck1 as it displays high homology to Bck1 from yeast. Following epistasis study, Mor-Bck1 was suggested to function upstream of Mor-Slt2 to ensure proper appressorium function (Jeon et al., 2008). Two nuclear localized proteins from M. oryzae were also found to interact with Slt2 and are believed to act as key downstream substrates mediating MAPK function (Mehrabi et al., 2008; Qi et al., 2012). These are the MADS box TF Mig1 and the APSES TF Swi6, the respective homologs of Slt2 substrates Rlm1 and Swi6 in yeast (Figure 1). Disruption of Mig1 results in the differentiation of normal appressoria, penetration pegs, and primary infectious hyphae. However, mig1 mutants fail to infect rice tissues, since they are blocked in the differentiation of secondary infection structures (Mehrabi et al., 2008). In the case of Mor-Swi6, deletion mutants display reduced hyphal growth, abnormal formation of conidia and appressoria, as well as altered appressorium function and pathogenicity (Qi et al., 2012). Although it remains to be established whether Mor-Slt2 can phosphorylate Mig1 and Mor-Swi6, these results suggest that several TFs function downstream of this MAPK to promote pathogenicity and sustain infectious growth in planta.
The cell wall integrity signaling pathway from U. maydis was also recently resolved and shown to involve MAP3K Bck1, MAP2K MKK1, and MAPK Mpk1 (here renamed Uma-Slt2-1; Carbó and Pérez-Martín, 2010). In contrast with yeast, where activation of the cell wall integrity pathway arrests the cell cycle in the G2 phase, activation of the corresponding pathway in U. maydis forces cells to escape the G2 phase. This effect is correlated with a decrease in CDK inhibitory phosphorylation, which itself depends on the nuclear accumulation of the mitotic protein phosphatase Cdc25 following activation of the Slt2-1 MAPK (Carbó and Pérez-Martín, 2010). The authors proposed that cell cycle adaptation to stress most likely evolved differently in divergent fungi, so that each species can move toward a cell cycle phase that is most appropriate for responding to the environmental signals encountered.
While the virulence function of the Ustilago cell wall integrity signaling pathway still remains to be established, Slt2 homologs from other plant pathogenic fungi have been linked to pathogenicity. In C. orbiculare, the MAPK MAF1 (here renamed Cor-Slt2) regulates early penetration steps, and deletion mutants produce elongated germ tubes that lack an appressorium (Kojima et al., 2002). In C. purpurea, the Slt2 homolog (previously known as MK2) is necessary for host penetration and corresponding deletion mutants retain only a limited ability to colonize plant tissues (Mey et al., 2002a). In B. cinerea, bmp3 (here renamed Bci-slt2) mutants are defective in surface sensing, plant penetration, and induction of necrotic lesions (Rui and Hahn, 2007). In M. graminicola, Slt2 mutants show normal penetration of wheat stomata but are highly reduced in virulence because infectious hyphae fail to branch out (Mehrabi et al., 2006a). Finally, MGV1 (here renamed Fgr-Slt2) was shown to control heterokaryon formation and accumulation of trichothecene mycotoxins in F. graminearum (Hou et al., 2002). In light of these various findings, the study of Slt2-type MAPKs has defined a second fungal MAPK cascade that plays an evolutionarily conserved role in virulence against plants.
While the role of Slt2-type MAPKs in fungal pathogenesis seems to be broadly conserved, their function in preserving cell wall integrity seems to vary among fungal pathogens. As seen for the Mor-slt2 mutants, Cpu-slt2, Fgr-slt2, Mgr-slt2, and Uma-slt2 mutants all have weakened cell walls and show increased sensitivity to cell wall–degrading enzymes and to compounds interfering with cell wall biosynthesis (Hou et al., 2002; Mey et al., 2002a; Mehrabi et al., 2006a; Carbó and Pérez-Martín, 2010). By contrast, deletion of Slt2 homologs from C. orbiculare and B. cinerea has no obvious effect on cell wall sensitivity to lytic enzymes and inhibitors (Kojima et al., 2002; Rui and Hahn, 2007). The reasons for this discrepancy may be related to the possibility that not all Slt2 homologs have retained their function in cell wall integrity or that a functionally redundant pathway controls this response in some, but not all, phytopathogenic fungi.
Hog1-Type MAPKs in Phytopathogenic Fungi
In S. cerevisiae, Hog1 lies downstream of a branched signaling pathway that responds to high osmolarity conditions (Figure 1). However, other deleterious environmental conditions also activate the HOG pathway (Hayashi and Maeda, 2006; Marques et al., 2006; Panadero et al., 2006). Consistent with this, Hog1 homologs carry a -TGY- phosphorylation motif, which is a hallmark of stress-activated MAPKs in fungal and mammalian systems.
The M. oryzae MAPK OSM1 (here renamed Mor-Hog1) was the first Hog1-type MAPK to be characterized from phytopathogenic fungi (Dixon et al., 1999). Deletion mutants are highly sensitive to osmotic stress and show severe morphological defects when grown under hyperosmotic conditions. These phenotypes are correlated with reduced ability of mutant strains to accumulate osmoprotectant molecules in the stressed mycelium. On the other hand, strains lacking Mor-Hog1 generate normal appressorial turgor since they can still accumulate glycerol in their appressoria. At the time, this suggested that Hog1 signaling is not required for appressorium function and that an independent pathway controls IRM in M. oryzae (Dixon et al., 1999).
In phytopathogenic fungi, two other Hog1 homologs are dispensable for virulence. These are C. orbiculare OSC1 (here renamed Cor-Hog1; Kojima et al., 2004) and Bipolaris oryzae SRM1 (here renamed Bor-Hog1; Moriwaki et al., 2006). On the other hand, deletion of Mgr-Hog1 produces nonpathogenic M. graminicola strains that are impaired in mating and thus unable to switch from a yeast-like form to filamentous growth (Mehrabi et al., 2006b). In B. cinerea, deletion of SAK1 (here renamed Bci-Hog1) also creates nonpathogenic strains that are unable to penetrate unwounded plant tissues (Segmüller et al., 2007). Taken as a whole, these results indicate that Hog1-type MAPKs are required for virulence of some, but not all, plant pathogens. The reasons for this difference remain unclear but could be related to the disparity of fungal invasion strategies and/or ability to cope with host counterdefenses (Zhao et al., 2007).
While deletion of Hog1-type MAPKs has variable and species-specific consequences for fungal virulence, the effect on resistance to stress appears more universal. For instance, hypersensitivity to osmotic stress is not only reported for the Mor-hog1 mutants (Dixon et al., 1999) but also for mutants of Cryphonectria parasitica, C. orbiculare, B. oryzae, M. graminicola, and B. cinerea lacking their respective Hog1 homologs (Kojima et al., 2004; Park et al., 2004b; Mehrabi et al., 2006b; Moriwaki et al., 2006; Segmüller et al., 2007). In filamentous fungi, reports have also linked the activity of group III His kinases to the phosphorylation status of Hog1-type MAPKs. For instance, the C. heterostrophus His kinase Dic1p promotes stress-mediated activation of Che-Hog1 (Yoshimi et al., 2005), while deletion of His kinase gene HSK1 compromises Aal-Hog1 activation in response to stress in Alternaria alternata (Lin and Chung, 2010). These studies provide the first hints of how upstream regulation of the HOG pathway might be organized in phytopathogenic fungi.
In several fungal species, it has also been reported that mutants blocked in components of the HOG pathway are more resistant to a variety of fungicides (Kojima et al., 2004; Motoyama et al., 2005, 2008; Yoshimi et al., 2005; Mehrabi et al., 2006b; Moriwaki et al., 2006; Viaud et al., 2006). In at least three cases, treatment with fungicides also results in the activation of Hog1-type MAPKs (Kojima et al., 2004; Yoshimi et al., 2005; Segmüller et al., 2007). This suggests that the fungicidal properties of several compounds may be due to an overstimulation of the HOG pathway, leading to uncontrolled accumulation of osmoprotectant molecules and concomitant swelling of fungal cells.
Ime2 Homologs: A Novel Subfamily of MAPKs Conserved in All Eukaryotes?
More than 20 years ago, S. cerevisiae Inducer of Meiosis2 (Ime2) was identified as a gene expressed exclusively during meiosis (Smith and Mitchell, 1989; Yoshida et al., 1990). It was later shown that Ime2 is required for entry and progression of the meiotic cell cycle via the destabilization of its transcriptional activator, Ime1 (Guttmann-Raviv et al., 2002). Following in-depth characterization of its function, Ime2 is now considered as a key node controlling various steps of meiosis, via the fine-tuned regulation of several substrates that are, in some cases, shared with yeast CDK1 (Irniger, 2011). However, recent reports focusing on Ime2 or closely related homologs from various fungal species suggest that this class of PKs is not only involved in the control of meiosis but also in the regulation of a variety of processes, including ascospore formation, pseudohyphal growth, and sexual reproduction in response to light and nutrient deprivation (Irniger, 2011).
To date, all identified Ime2 homologs share conserved topology, including an N-terminal region that contains a PK domain displaying high homology to the CDK proteins. The kinase domain of Ime2-type proteins also harbors an activation loop containing a -TXY- motif, the hallmark of the MAPK protein family. Until recently, Ime2 homologs had not been defined as true MAPKs, even though dual phosphorylation of the -TAY- motif found in yeast Ime2 is required for its activity (Schindler et al., 2003). In fact, there is still no evidence that MAP2Ks from yeast can activate Ime2, and modification of the Ime2 -TAY- motif is associated with both autophosphorylation and CDK-activating kinase1 activities (Schindler et al., 2003; Schindler and Winter, 2006). On the other hand, characterization of Cdk-related kinase1, an Ime2 homolog from U. maydis that was here renamed Uma-Ime2, now suggests that at least certain Ime2 homologs behave like true MAPKs and that Ime2-like proteins may indeed constitute a novel subfamily of MAPKs (Figure 2; Garrido et al., 2004).
In Ustilago, Ime2 promotes morphogenetic transition through the control of Prf1 expression (Figure 5; Garrido and Pérez-Martín, 2003; Garrido et al., 2004). As a result, strains lacking this PK are impaired in mating and formation of dikaryotic filaments because hyphal fusion between opposite mating types fails to occur (Garrido et al., 2004). Since mating of Ustilago haploid cells is a prerequisite for plant infection, Ime2 is also necessary for pathogenicity, and mutant strains cause drastically reduced numbers of tumors compared with wild-type strains. Interestingly, the MAP2K Ste7, which stimulates mating and virulence by operating upstream of MAPKs Kss1-1 and Kss1-2, also targets the -TXY- motif of Ime2 for phosphorylation (Figure 5; Garrido et al., 2004). In addition, Ime2 physically interacts with Kss1-1, and its full activity is dependent on this MAPK. This suggests that in U. maydis, activation of Ime2 is tightly linked with signaling that goes through the Kss1-1 MAPK module and that this PK exerts its function both in parallel to Kss1-1 and as a substrate for this MAPK (Figure 5; Garrido et al., 2004; Irniger, 2011).
Ime2 homologs are not only conserved in fungi but also in all eukaryotic taxa examined (Krylov et al., 2003). In mammals, for instance, the Ime2 homolog, male germ cell–associated kinase (Mak), has an expression pattern that correlates with sexual development (Jinno et al., 1993). In Arabidopsis, three Mak-homologous kinases show overall sequence similarity to MAPKs and possess a -TEY- phosphorylation motif that is typically associated with extracellular signal-regulated kinase–type MAPKs (Tena et al., 2001; Champion et al., 2004). In the original plant MAPKs survey, Arabidopsis Mak-homologous kinases were not classified as true MAPKs because they show sequence relatedness to both MAPKs and CDC2-like PKs (Ichimura, 2002). In view of the novel data now linking Ste7 and Ime2 from U. maydis, it will be important to investigate whether MAP2Ks can activate Ime2-type proteins in other eukaryotic taxa, including in plant and animal systems. If this were indeed the case, this exciting new feature would certainly argue for inclusion of the Ime2-like kinases as bona fide MAPKs.
MOLECULAR DIALOGUE BETWEEN PLANTS AND FUNGAL PATHOGENS
Upon host sensing, fungal MAPKs are important for the virulence of phytopathogens because they control mating, IRM, reinforcement of the fungal cell wall, and expression of virulence factors (Zhao et al., 2007; Rispail et al., 2009; this article). Using mechanical pressure and/or enzymatic activity, fungal pathogens breach the plant surface in order to access desired nutrients (Figure 7, top left). However, during fungal infection of plants, MAPK signaling is critical not only for the attacking fungi but also for activation of defense responses in plant cells.
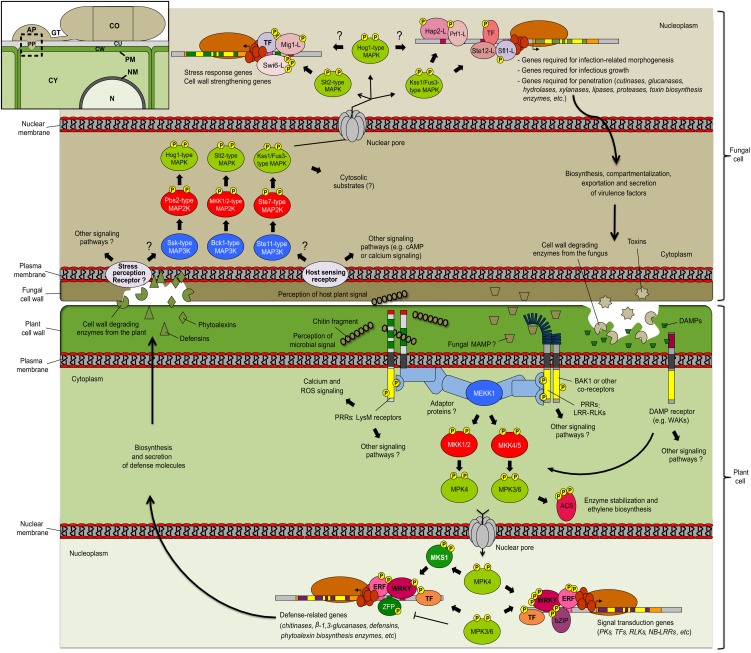
Figure 7.
MAPK Signaling during Interaction of Pathogenic Fungi and Plants.
Fungal pathogens perceive plant-derived signals using plasma membrane receptors. This leads to the activation of fungal MAPK cascades that modulate TF activity and promote expression of genetic targets. Among induced genes are those associated with mating, IRM, mycotoxin biosynthesis, and degradation of the plant cell wall. On the other hand, plant receptors perceive molecular signatures associated with fungal structures or activity. This results in the activation of plant signaling pathways, including modified Ca2+ homeostasis, oxidative burst, and changes in MAPK phosphorylation status. Plant MAPKs promote biosynthesis of stress hormone and modulate gene expression through the phosphorylation of TFs. Output responses, including cell wall–degrading enzymes and antimicrobial compounds, affect fungal cell integrity and threaten pathogen survival. Fungal MAPK cascades also participate in compensatory responses allowing protection of hyphae against plant defenses. Overall, MAPK cascades can be viewed as conserved signaling modules involved in the molecular dialogue between fungal pathogens and plants. AP, appressorium; CO, conidia; CU, cuticule; CW, cell wall; CY, cytoplasm; GT, germ tube; N, nucleus; NM, nuclear membrane; PM, plasma membrane; PP, penetration peg; ROS, reactive oxygen species.
Activation of MAPK Cascades in the Infected Plants
Plant MAPK cascades lie downstream of a sensitive surveillance system that monitors threats of infection (Figure 7). The plant surveillance apparatus includes pattern recognition receptors (PRRs) that allow detection of molecular signatures also known as microbe-associated molecular patterns (MAMPs; Boller and Felix, 2009). For instance, the extracellular portion of LysM receptors possesses chitin binding activity (Figure 7; Iizasa et al., 2010; Petutschnig et al., 2010), and LysM receptors are necessary for chitin-mediated activation of defense in Arabidopsis (Miya et al., 2007; Wan et al., 2008) and rice (Kaku et al., 2006). Receptor-like kinases harboring extracellular Leu-rich repeats have also been linked to the detection of microbes (Zipfel et al., 2004, 2006), and members from this large PK family likely participate in detection of several fungal MAMPs (Figure 7). Endogenous molecules referred to as damage-associated molecular patterns (DAMPs) can also induce plant defense following their release from damaged plant cell walls (Figure 7). Wall-associated kinases have recently been identified as DAMP receptors (Kohorn et al., 2009; Brutus et al., 2010), and these, together with signals from other PRRs, promote activation of several signaling pathways regulating plant basal defense (Figure 7).
In Arabidopsis, two complete MAPK cascades have been shown to function downstream of PRRs (Figure 7; Asai et al., 2002; Gao et al., 2008; Qiu et al., 2008b). Perception of elicitors thus leads to the activation of at least three stress-responsive MAPKs: MPK3, MPK4, and MPK6 (Asai et al., 2002; Wan et al., 2004; Mészáros et al., 2006; Denoux et al., 2008; Galletti et al., 2011). Further study of plant MAPKs has uncovered several output responses that contribute to plant defense activation. For example, At-MPK3 and At-MPK6 were shown to promote stabilization of certain isoforms of 1-aminocyclopropane-1-carboxylic acid synthase, a key step in Botrytis-induced production of ethylene (Figure 7; Liu and Zhang, 2004; Han et al., 2010). Large-scale in vitro analysis also indicates that TFs are well represented among the primary targets of stress-responsive MAPKs (Feilner et al., 2005; Popescu et al., 2009). In planta, MAPKs have been shown to alter the function of several TFs (Figure 7), including WRKYs in Arabidopsis and Nicotiana (Andreasson et al., 2005; Ishihama et al., 2011; Mao et al., 2011), the bZIP At-VIP1 (Djamei et al., 2007), ethylene-related proteins At-EIN3 (Yoo et al., 2008) and At-ERF104 (Bethke et al., 2009), as well as Pti-ZFP1, a poplar (Populus trichocarpa) Zn finger protein (Hamel et al., 2011).
Through posttranslational modification of TFs, plant MAPKs regulate expression of target genes, including several that are involved in signal transduction and defense (Figure 7). Notably, MPK3/6 were shown to promote synthesis of camalexin, the main antimicrobial phytoalexin in Arabidopsis (Ren et al., 2008). Epistasis analysis places both MAPKs upstream of Phytoalexin Deficient2/3 (PAD2/3), two camalexin biosynthesis genes. Moreover, ectopic expression of active upstream MAP2K or MAP3K induces camalexin biosynthesis in the absence of pathogen attack. In wild-type plants, Botrytis-induced production of camalexin is preceded by MPK3/6 activation, and the production of this secondary metabolite is compromised in mpk3 and mpk6 knockout mutants (Ren et al., 2008).
More recently, it was shown that WRKY33 is required for MPK3/6-mediated induction of camalexin biosynthesis in Arabidopsis (Mao et al., 2011). Following pathogen attack, wrky33 knockout plants are compromised in the induction of camalexin biosynthesis genes as well as in production of the metabolite. In vivo analysis indicates that WRKY33 is a direct substrate of MPK3/6, and complementation assays show that phosphorylation is involved in WRKY33-mediated induction of camalexin biosynthesis (Mao et al., 2011). Interestingly, WRKY33 was also identified as part of a multiprotein complex that includes MPK4 and one of its substrates, MKS1 (Andreasson et al., 2005). In the absence of pathogen, the MPK4-MKS1-WRKY33 ternary complex is stabilized, preventing WRKY33 from activating target gene promoters (Qiu et al., 2008a). Following pathogen perception, MPK4 is activated, resulting in MKS1 phosphorylation and dissolution of the protein complex (Figure 7). Released WRKY33 can then bind its genetic targets, including the promoter of PAD3 (Qiu et al., 2008a).
Fungal MAPK Cascades and Compensatory Response against Host Defenses
Camalexin produced by the challenged plant probably exerts its antifungal activity by causing cell membrane damage in the attacking fungi (Figure 7). In the necrotroph A. brassicicola, the cellular response elicited by such damage requires both the cell wall integrity and HOG pathways, leading to the activation of fungal MAPKs Abr-Slt2 and Abr-Hog1, respectively (Joubert et al., 2011). Alternaria strains lacking these MAPKs are not only hypersensitive to camalexin, but also to brassinin, a structurally related phytoalexin from Brassica species (Joubert et al., 2011). In F. graminearum, two MAPKs, Fgr-Kss1 and Fgr-Slt2, appear to play a major role in fungal sensitivity to certain plant defensins, a small family of pathogenesis-related proteins (Figure 7; Ramamoorthy et al., 2007). Taken as a whole, these findings suggest that fungal MAPKs are not only required for plant infection but also for regulation of compensatory responses that help to preserve fungal cell integrity during exposure to plant defenses. As a result, fungal strains lacking MAPK signaling components may exhibit reduced virulence because of their inability to cope with plant defenses.
Specificity of Mutually Activated MAPK Pathways
Many features associated with fungal pathogenicity and plant defense are dependent on signaling through MAPK cascades (Figure 7). As a result, these modules can be viewed as crucial players that promote the two-way molecular dialogue between pathogenic fungi and plants. On the other hand, it is not clear whether such patterns of mutual activation are necessarily specific to MAPK modules or if other shared eukaryotic signaling pathways exhibit such a bilateral contribution to the highly complex molecular dialogue between both protagonists.
Part of the answer may come from the comparison of conserved eukaryotic signaling pathways that have been characterized in both plants and fungi. In mammalian and fungal systems, for instance, cyclic nucleotides are viewed as central secondary messengers that promote activation of conserved PKA/PKG/PKC (AGC) signaling pathways. In phytopathogenic fungi like U. maydis, perception of host-derived signals leads to an increase in the cellular concentrations of cAMP, resulting in the activation of PKA signaling and associated promotion of virulence (Figure 5; Kahmann and Kämper, 2004; Brefort et al., 2009). Modulation of cAMP levels also results in altered plant cell signaling, although plant genomes do not appear to encode prototypical PKA enzymes (Robert and Offringa, 2008). Of the 37 AGC kinases identified in Arabidopsis (Galván-Ampudia and Offringa, 2007), none has so far been directly involved in MAMP-triggered immunity. In fact, the majority of plant AGC kinases belong to a plant-specific subgroup, called the AGCVIII kinases, members of which are mainly involved in auxin signaling (Galván-Ampudia and Offringa, 2007; Robert and Offringa, 2008) and other developmental processes (Zhang and McCormick, 2009). Therefore, despite their wide distribution in eukaryotes, including plants and fungi, it seems unlikely that AGC kinases contribute directly to the molecular dialogue between plants and phytopathogenic fungi, since the plant homologs do not actively participate in early-induced plant immunity.
Proteins allowing perception of calcium (Ca2+), another ubiquitous secondary messenger, are also likely candidates as crucial contributors to the two-way molecular dialogue between plants and interacting fungi. In animals and fungi, intracellular Ca2+ sensors include calmodulins (CaMs), which interact with calcineurin B (CNB) upon Ca2+ stimulation. These protein complexes then bind to calcineurin A, a highly conserved protein phosphatase (Luan, 2009). In phytopathogenic fungi, CaMs and CNBs have been involved in virulence (Lee and Lee, 1998; Warwar et al., 2000; Uhm et al., 2003; Harel et al., 2006; Ahn and Suh, 2007; Egan et al., 2009; Cervantes-Chávez et al., 2011), and comparative genomics of the Ca2+/CaM/CNB pathway suggest that it is highly conserved in fungi (Rispail et al., 2009).
In plants, Ca2+ signaling also depends on CaMs, as well as a group of CNB-related proteins, referred to as the CNB-like (CBL) proteins (Batistič and Kudla, 2009; Tena et al., 2011). Like their animals and fungi counterparts, CBLs were anticipated to act upstream of a protein phosphatase, but instead they were found to form a complex regulatory network with members of the CBL-interacting PK (CIPK) family (Luan, 2009). In plants, CBL–CIPK interactions regulate various stress responses and represent a fundamental paradigm shift that distinguishes signaling in animals and fungi from that in plants (Luan, 2009).
The Ca2+ signaling machinery of plants also possesses an exclusive feature related to the evolution and expansion of the calcium-dependent PK family, a group of plant-specific proteins that combine unique Ca2+ sensing and signaling capabilities within the same gene product (Batistič and Kudla, 2009). Several members of the calcium-dependent PK family are involved in stress responses, including defense gene upregulation in response to MAMPs (Tena et al., 2011). As a result, while the Ca2+ signaling pathways from plants and interacting fungi share some components, striking differences exist in pathway architecture, regulation, and complexity.
Although the full range of signaling strategies operating in plants and fungi has not been defined, comparison of other characterized pathways suggests that very few of these strategies display the striking level of architectural conservation (from membrane receptors to nuclear TFs) that typifies the MAPK pathways. Bilateral activation of MAPK modules from interacting plants and pathogenic fungi may therefore possess a certain level of specificity compared with other signaling pathways, possibly as a consequence of the outstanding MAPK versatility with regards to environmental responsiveness.
UPCOMING CHALLENGES AND CONCLUDING REMARKS
Completion of whole-genome sequencing projects marks the entry of fungal MAPK research in the postgenomic era. Many questions regarding fungal MAPK signaling remain unanswered, and upcoming studies should, for instance, focus on the understanding of upstream mechanisms leading to MAPK activation. Recently, homologs of the yeast osmosensors Sho1 and Msb2 (Figure 1) have been shown to act upstream of pathogenesis-related MAPK cascades in U. maydis (Figure 5; Lanver et al., 2010), M. oryzae (Figure 6; Liu et al., 2011), and F. oxysporum (Pérez-Nadales and Di Pietro, 2011). Along with the previously identified G protein–coupled receptor Pth11 from M. oryzae (Figure 6; DeZwaan et al., 1999; Kulkarni et al., 2003), these candidate receptors likely ensure MAPK cascade activation following recognition of specific ligands. However, identity of the full array of signaling components lying between surface receptors and MAPK modules is still missing. Comparative and functional genomics should also facilitate characterization of less-well-defined MAPK pathways, including those that engage the Slt2- and Hog1-type MAPKs (Rispail et al., 2009). While upstream regulation of Slt2-type MAPK cascades remains largely unexplored in phytopathogenic fungi, group III His kinases have been shown to operate upstream of Hog1-type MAPKs (Yoshimi et al., 2005; Lin and Chung, 2010). With regards to fungal pathogenicity against plants, complete characterization of all the MAPK signaling pathways is crucial, since genetic evidence points to extensive crosstalk between parallel MAPK cascades operating in C. orbiculare (Takano et al., 2000; Kojima et al., 2002), M. oryzae (Xu and Hamer, 1996; Dixon et al., 1999), and C. heterostrophus (Eliahu et al., 2007; Igbaria et al., 2008).
Comparison of genomic sequences also highlights new challenges regarding the study of MAPK signaling in basidiomycetes. For instance, duplication of several MAPK genes raises the question of the extent of functional redundancy between paralogous MAPKs. In U. maydis, Kss1-1 and Kss1-2 exhibit partially redundant functions during mating and virulence (Zhao et al., 2007; Brefort et al., 2009; Rispail et al., 2009), but careful study of single deletion mutants also shows that these MAPKs still display some degree of specialization. So far, Ustilago is the only species in which functional redundancy between paralogous MAPKs has been addressed experimentally, and the situation may be even more complex in other fungi, where up to three paralogs can be identified in a single MAPK clade. Fine-grained approaches, including promoter-reporter fusions and functional complementation of double and triple knockout mutants, will be needed to decipher discrete functions of such closely related MAPKs. Downstream of the cascade, paralogous MAPKs may also possess unique and shared substrates, and identification of these targets will be a prerequisite to fully understand the output responses controlled by each MAPK. Currently, only a few TFs have been linked to MAPK signaling, and characterization of deletion mutants suggests that more MAPK substrates await discovery. In addition to TFs, MAPKs from other eukaryotic taxa are well known for their propensity to phosphorylate cytosolic as well as microtubule-associated proteins. This pattern has yet to be confirmed in phytopathogenic fungi, even though new potential interacting partners have recently been identified (Zhang et al., 2011).
It is paradoxical that, while tripartite MAPK modules have been conserved throughout evolution of all eukaryotes, these control an extraordinary array of responses that are specific to each species. How such an ancient and evolutionarily conserved mechanism can regulate so many output responses is a fascinating, yet unanswered, question. Study of the interactions between plants and fungal pathogens exemplifies the plasticity of MAPK cascades operating simultaneously in both protagonists. MAPK phosphorelays are not only conserved in phytopathogenic fungi but also in other eukaryotes that interact with plants, including mycorrhizal fungi, oomycetes, insects, and nematodes. Relying on different strategies, these varied organisms positively or negatively affect plant fitness, employing mechanisms that may often rely on well-conserved MAPK signaling pathways. In line with this, it was recently shown that an oomycete MAPK is required for virulence of Phytophthora sojae against soybean (Glycine max; Li et al., 2010). Activation of plant MAPKs also occurs following interactions with other eukaryotes, including following perception of oomycete elicitors (Kroj et al., 2003) and feeding by herbivorous insects (Kandoth et al., 2007; Wu et al., 2007). Taken together, these reports strongly suggest that MAPK cascades are not only required for the molecular dialogue between plants and pathogenic fungi but also for the exchanges occurring between plants and eukaryotes from other taxa.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. List of the Genomic Databases Used in This Study.
Supplemental Data Set 1. Origin and Gene Model Number of MAPKs from Plant-Interacting Fungi.
Supplemental Data Set 2. Origin and Gene Model Number of MAP2Ks from Plant-Interacting Fungi.
Supplemental Data Set 3. Origin and Gene Model Number of MAP3Ks from Plant-Interacting Fungi.
Supplemental Data Set 4. Amino Acid Alignment Used to Produce Phylogeny of Fungal MAPKs Shown in Figure 2.
Supplemental Data Set 5. Amino Acid Alignment Used to Produce Phylogeny of Fungal MAP2Ks shown in Figure 3.
Supplemental Data Set 6. Amino Acid Alignment Used to Produce Phylogeny of Fungal MAP3Ks shown in Figure 4.
Supplemental References 1. List of the Literature Cited in the Supplemental Data Sets.
Supplementary Material
Acknowledgments
We thank Jim Kronstad for helpful discussions and critical reading of the manuscript. L.-P.H. and M.-C.N. are the recipients of postdoctoral fellowships from Fonds Québécois de la Recherche sur la Nature et les Technologies. We apologize to our colleagues whose work could not be cited here due to space limitations.
AUTHOR CONTRIBUTIONS
L.-P.H. conducted in silico search of gene models and conceived the figures and tables. L.-P.H. and M.-C.N. drafted the article. S.D. wrote the section describing MAPK signaling components from symbiotic fungi. S.D. and B.E.E. critically revised the article and provided important intellectual content. All authors read, helped to edit, and approved the final version of the article.
References
- Ahn I.P., Suh S.C. (2007). Calcium/calmodulin-dependent signaling for pre-penetration development in Cochliobolus miyabeanus infecting rice. J. Gen. Plant Pathol. 73: 113–120 [Google Scholar]
- Amselem J., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7: e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E., Ellis B. (2010). Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 15: 106–113 [DOI] [PubMed] [Google Scholar]
- Andreasson E., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24: 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews D.L., Egan J.D., Mayorga M.E., Gold S.E. (2000). The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant Microbe Interact. 13: 781–786 [DOI] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Banuett F., Herskowitz I. (1994). Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8: 1367–1378 [DOI] [PubMed] [Google Scholar]
- Batistič O., Kudla J. (2009). Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 1793: 985–992 [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Brachmann A., Schirawski J., Müller P., Kahmann R. (2003). An unusual MAP kinase is required for efficient penetration of the plant surface by Ustilago maydis. EMBO J. 22: 2199–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brefort T., Doehlemann G., Mendoza-Mendoza A., Reissmann S., Djamei A., Kahmann R. (2009). Ustilago maydis as a pathogen. Annu. Rev. Phytopathol. 47: 423–445 [DOI] [PubMed] [Google Scholar]
- Bruno K.S., Tenjo F., Li L., Hamer J.E., Xu J.R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3: 1525–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbó N., Pérez-Martín J. (2010). Activation of the cell wall integrity pathway promotes escape from G2 in the fungus Ustilago maydis. PLoS Genet. 6: e1001009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Chávez J.A., Ali S., Bakkeren G. (2011). Response to environmental stresses, cell-wall integrity, and virulence are orchestrated through the calcineurin pathway in Ustilago hordei. Mol. Plant Microbe Interact. 24: 219–232 [DOI] [PubMed] [Google Scholar]
- Champion A., Kreis M., Mockaitis K., Picaud A., Henry Y. (2004). Arabidopsis kinome: After the casting. Funct. Integr. Genomics 4: 163–187 [DOI] [PubMed] [Google Scholar]
- Chen R.E., Thorner J. (2007). Function and regulation in MAPK signaling pathways: Lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773: 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y., Cramer R.A., Jr, Kim K.H., Davis J., Mitchell T.K., Figuli P., Pryor B.M., Lemasters E., Lawrence C.B. (2007). The Fus3/Kss1 MAP kinase homolog Amk1 regulates the expression of genes encoding hydrolytic enzymes in Alternaria brassicicola. Fungal Genet. Biol. 44: 543–553 [DOI] [PubMed] [Google Scholar]
- Cousin A., Mehrabi R., Guilleroux M., Dufresne M., Van Der Lee T., Waalwijk C., Langin T., Kema G.H. (2006). The MAP kinase-encoding gene MgFus3 of the non-appressorium phytopathogen Mycosphaerella graminicola is required for penetration and in vitro pycnidia formation. Mol. Plant Pathol. 7: 269–278 [DOI] [PubMed] [Google Scholar]
- Dean R.A., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- Denoux C., Galletti R., Mammarella N., Gopalan S., Werck D., De Lorenzo G., Ferrari S., Ausubel F.M., Dewdney J. (2008). Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1: 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan T.M., Carroll A.M., Valent B., Sweigard J.A. (1999). Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11: 2013–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A., García-MacEira F.I., Méglecz E., Roncero M.I. (2001). A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39: 1140–1152 [PubMed] [Google Scholar]
- Di Stasio M., Brefort T., Mendoza-Mendoza A., Münch K., Kahmann R. (2009). The dual specificity phosphatase Rok1 negatively regulates mating and pathogenicity in Ustilago maydis. Mol. Microbiol. 73: 73–88 [DOI] [PubMed] [Google Scholar]
- Dixon K.P., Xu J.R., Smirnoff N., Talbot N.J. (1999). Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11: 2045–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A., Pitzschke A., Nakagami H., Rajh I., Hirt H. (2007). Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 318: 453–456 [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Berndt P., Hahn M. (2006). Different signalling pathways involving a Galpha protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol. Microbiol. 59: 821–835 [DOI] [PubMed] [Google Scholar]
- Duplessis S., Courty P.E., Tagu D., Martin F. (2005). Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus. New Phytol. 165: 599–611 [DOI] [PubMed] [Google Scholar]
- Duplessis S., et al. (2011). Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 108: 9166–9171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole D.J. (2007). Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45: 437–456 [DOI] [PubMed] [Google Scholar]
- Ebbole D.J., Jin Y., Thon M., Pan H., Bhattarai E., Thomas T., Dean R. (2004). Gene discovery and gene expression in the rice blast fungus, Magnaporthe grisea: Analysis of expressed sequence tags. Mol. Plant Microbe Interact. 17: 1337–1347 [DOI] [PubMed] [Google Scholar]
- Egan J.D., García-Pedrajas M.D., Andrews D.L., Gold S.E. (2009). Calcineurin is an antagonist to PKA protein phosphorylation required for postmating filamentation and virulence, while PP2A is required for viability in Ustilago maydis. Mol. Plant Microbe Interact. 22: 1293–1301 [DOI] [PubMed] [Google Scholar]
- Eliahu N., Igbaria A., Rose M.S., Horwitz B.A., Lev S. (2007). Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1. Eukaryot. Cell 6: 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilner T., et al. (2005). High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4: 1558–1568 [DOI] [PubMed] [Google Scholar]
- Friesen H., Lunz R., Doyle S., Segall J. (1994). Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 8: 2162–2175 [DOI] [PubMed] [Google Scholar]
- Fujikawa T., Kuga Y., Yano S., Yoshimi A., Tachiki T., Abe K., Nishimura M. (2009). Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 73: 553–570 [DOI] [PubMed] [Google Scholar]
- Galletti R., Ferrari S., De Lorenzo G. (2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157: 804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván-Ampudia C.S., Offringa R. (2007). Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 12: 541–547 [DOI] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- García-Pedrajas M.D., Nadal M., Bölker M., Gold S.E., Perlin M.H. (2008). Sending mixed signals: Redundancy vs. uniqueness of signaling components in the plant pathogen, Ustilago maydis. Fungal Genet. Biol. 45 (Suppl 1): S22–S30 [DOI] [PubMed] [Google Scholar]
- Garrido E., Pérez-Martín J. (2003). The crk1 gene encodes an Ime2-related protein that is required for morphogenesis in the plant pathogen Ustilago maydis. Mol. Microbiol. 47: 729–743 [DOI] [PubMed] [Google Scholar]
- Garrido E., Voss U., Müller P., Castillo-Lluva S., Kahmann R., Pérez-Martín J. (2004). The induction of sexual development and virulence in the smut fungus Ustilago maydis depends on Crk1, a novel MAPK protein. Genes Dev. 18: 3117–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Dai X., Xu J.R., Wang Y., Bai P., Liu F., Duan Y., Zhang H., Huang L., Kang Z. (2011). Molecular characterization of a Fus3/Kss1 type MAPK from Puccinia striiformis f. sp. tritici, PsMAPK1. PLoS ONE 6: e21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttmann-Raviv N., Martin S., Kassir Y. (2002). Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol. Cell. Biol. 22: 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L.P., Benchabane M., Nicole M.C., Major I.T., Morency M.J., Pelletier G., Beaudoin N., Sheen J., Séguin A. (2011). Stress-responsive mitogen-activated protein kinases interact with the EAR motif of a poplar zinc finger protein and mediate its degradation through the 26S proteasome. Plant Physiol. 157: 1379–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L.P., et al. (2006). Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11: 192–198 [DOI] [PubMed] [Google Scholar]
- Han L., Li G.J., Yang K.Y., Mao G., Wang R., Liu Y., Zhang S. (2010). Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Harel A., Bercovich S., Yarden O. (2006). Calcineurin is required for sclerotial development and pathogenicity of Sclerotinia sclerotiorum in an oxalic acid-independent manner. Mol. Plant Microbe Interact. 19: 682–693 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Maeda T. (2006). Activation of the HOG pathway upon cold stress in Saccharomyces cerevisiae. J. Biochem. 139: 797–803 [DOI] [PubMed] [Google Scholar]
- Hou Z., Xue C., Peng Y., Katan T., Kistler H.C., Xu J.R. (2002). A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 15: 1119–1127 [DOI] [PubMed] [Google Scholar]
- Hu G., Kamp A., Linning R., Naik S., Bakkeren G. (2007). Complementation of Ustilago maydis MAPK mutants by a wheat leaf rust, Puccinia triticina homolog: Potential for functional analyses of rust genes. Mol. Plant Microbe Interact. 20: 637–647 [DOI] [PubMed] [Google Scholar]
- Ichimura K. MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Igbaria A., Lev S., Rose M.S., Lee B.N., Hadar R., Degani O., Horwitz B.A. (2008). Distinct and combined roles of the MAP kinases of Cochliobolus heterostrophus in virulence and stress responses. Mol. Plant Microbe Interact. 21: 769–780 [DOI] [PubMed] [Google Scholar]
- Iizasa E., Mitsutomi M., Nagano Y. (2010). Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J. Biol. Chem. 285: 2996–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S. (2011). The Ime2 protein kinase family in fungi: more duties than just meiosis. Mol. Microbiol. 80: 1–13 [DOI] [PubMed] [Google Scholar]
- Ishihama N., Yamada R., Yoshioka M., Katou S., Yoshioka H. (2011). Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23: 1153–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumitsu K., Yoshimi A., Kubo D., Morita A., Saitoh Y., Tanaka C. (2009). The MAPKK kinase ChSte11 regulates sexual/asexual development, melanization, pathogenicity, and adaptation to oxidative stress in Cochliobolus heterostrophus. Curr. Genet. 55: 439–448 [DOI] [PubMed] [Google Scholar]
- Jenczmionka N.J., Maier F.J., Lösch A.P., Schäfer W. (2003). Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase gpmk1. Curr. Genet. 43: 87–95 [DOI] [PubMed] [Google Scholar]
- Jenczmionka N.J., Schäfer W. (2005). The Gpmk1 MAP kinase of Fusarium graminearum regulates the induction of specific secreted enzymes. Curr. Genet. 47: 29–36 [DOI] [PubMed] [Google Scholar]
- Jeon J., Goh J., Yoo S., Chi M.H., Choi J., Rho H.S., Park J., Han S.S., Kim B.R., Park S.Y., Kim S., Lee Y.H. (2008). A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol. Plant Microbe Interact. 21: 525–534 [DOI] [PubMed] [Google Scholar]
- Jinno A., Tanaka K., Matsushime H., Haneji T., Shibuya M. (1993). Testis-specific mak protein kinase is expressed specifically in the meiotic phase in spermatogenesis and is associated with a 210-kilodalton cellular phosphoprotein. Mol. Cell. Biol. 13: 4146–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert A., Bataille-Simoneau N., Campion C., Guillemette T., Hudhomme P., Iacomi-Vasilescu B., Leroy T., Pochon S., Poupard P., Simoneau P. (2011). Cell wall integrity and high osmolarity glycerol pathways are required for adaptation of Alternaria brassicicola to cell wall stress caused by brassicaceous indolic phytoalexins. Cell. Microbiol. 13: 62–80 [DOI] [PubMed] [Google Scholar]
- Kaffarnik F., Müller P., Leibundgut M., Kahmann R., Feldbrügge M. (2003). PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 22: 5817–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann R., Kämper J. (2004). Ustilago maydis: How its biology relates to pathogenic development. New Phytol. 164: 31–42 [DOI] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth P.K., Ranf S., Pancholi S.S., Jayanty S., Walla M.D., Miller W., Howe G.A., Lincoln D.E., Stratmann J.W. (2007). Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc. Natl. Acad. Sci. USA 104: 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinane J., Oliver R.P. (2003). Evidence that the appressorial development in barley powdery mildew is controlled by MAP kinase activity in conjunction with the cAMP pathway. Fungal Genet. Biol. 39: 94–102 [DOI] [PubMed] [Google Scholar]
- Klose J., de Sá M.M., Kronstad J.W. (2004). Lipid-induced filamentous growth in Ustilago maydis. Mol. Microbiol. 52: 823–835 [DOI] [PubMed] [Google Scholar]
- Klosterman S.J., Martinez-Espinoza A.D., Andrews D.L., Seay J.R., Gold S.E. (2008). Ubc2, an ortholog of the yeast Ste50p adaptor, possesses a basidiomycete-specific carboxy terminal extension essential for pathogenicity independent of pheromone response. Mol. Plant Microbe Interact. 21: 110–121 [DOI] [PubMed] [Google Scholar]
- Kohorn B.D., Johansen S., Shishido A., Todorova T., Martinez R., Defeo E., Obregon P. (2009). Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 60: 974–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K., Kikuchi T., Takano Y., Oshiro E., Okuno T. (2002). The mitogen-activated protein kinase gene MAF1 is essential for the early differentiation phase of appressorium formation in Colletotrichum lagenarium. Mol. Plant Microbe Interact. 15: 1268–1276 [DOI] [PubMed] [Google Scholar]
- Kojima K., Takano Y., Yoshimi A., Tanaka C., Kikuchi T., Okuno T. (2004). Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 53: 1785–1796 [DOI] [PubMed] [Google Scholar]
- Kramer B., Thines E., Foster A.J. (2009). MAP kinase signalling pathway components and targets conserved between the distantly related plant pathogenic fungi Mycosphaerella graminicola and Magnaporthe grisea. Fungal Genet. Biol. 46: 667–681 [DOI] [PubMed] [Google Scholar]
- Krisak L., Strich R., Winters R.S., Hall J.P., Mallory M.J., Kreitzer D., Tuan R.S., Winter E. (1994). SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8: 2151–2161 [DOI] [PubMed] [Google Scholar]
- Kroj T., Rudd J.J., Nürnberger T., Gäbler Y., Lee J., Scheel D. (2003). Mitogen-activated protein kinases play an essential role in oxidative burst-independent expression of pathogenesis-related genes in parsley. J. Biol. Chem. 278: 2256–2264 [DOI] [PubMed] [Google Scholar]
- Krylov D.M., Nasmyth K., Koonin E.V. (2003). Evolution of eukaryotic cell cycle regulation: stepwise addition of regulatory kinases and late advent of the CDKs. Curr. Biol. 13: 173–177 [DOI] [PubMed] [Google Scholar]
- Kulkarni R.D., Kelkar H.S., Dean R.A. (2003). An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28: 118–121 [DOI] [PubMed] [Google Scholar]
- Lanver D., Mendoza-Mendoza A., Brachmann A., Kahmann R. (2010). Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis. Plant Cell 22: 2085–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Kronstad J.W. (2002). ras2 Controls morphogenesis, pheromone response, and pathogenicity in the fungal pathogen Ustilago maydis. Eukaryot. Cell 1: 954–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Lee Y.H. (1998). Calcium/calmodulin-dependent signaling for appressorium formation in the plant pathogenic fungus Magnaporthe grisea. Mol. Cells 8: 698–704 [PubMed] [Google Scholar]
- Lev S., Horwitz B.A. (2003). A mitogen-activated protein kinase pathway modulates the expression of two cellulase genes in Cochliobolus heterostrophus during plant infection. Plant Cell 15: 835–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Quéré A., Wright D.P., Söderström B., Tunlid A., Johansson T. (2005). Global patterns of gene regulation associated with the development of ectomycorrhiza between birch (Betula pendula Roth.) and Paxillus involutus (Batsch) Fr. Mol. Plant Microbe Interact. 18: 659–673 [DOI] [PubMed] [Google Scholar]
- Lev S., Sharon A., Hadar R., Ma H., Horwitz B.A. (1999). A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: Diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 96: 13542–13547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Wang Y., Tao K., Dong S., Huang Q., Dai T., Zheng X., Wang Y. (2010). PsSAK1, a stress-activated MAP kinase of Phytophthora sojae, is required for zoospore viability and infection of soybean. Mol. Plant Microbe Interact. 23: 1022–1031 [DOI] [PubMed] [Google Scholar]
- Li G., Zhou X., Kong L., Wang Y., Zhang H., Zhu H., Mitchell T.K., Dean R.A., Xu J.R. (2011). MoSfl1 is important for virulence and heat tolerance in Magnaporthe oryzae. PLoS ONE 6: e19951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Xue C., Bruno K., Nishimura M., Xu J.R. (2004). Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant Microbe Interact. 17: 547–556 [DOI] [PubMed] [Google Scholar]
- Lin C.H., Chung K.R. (2010). Specialized and shared functions of the histidine kinase- and HOG1 MAP kinase-mediated signaling pathways in Alternaria alternata, a filamentous fungal pathogen of citrus. Fungal Genet. Biol. 47: 818–827 [DOI] [PubMed] [Google Scholar]
- Liu W., Zhou X., Li G., Li L., Kong L., Wang C., Zhang H., Xu J.R. (2011). Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 7: e1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Ma L.J., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z., Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques J.M., Rodrigues R.J., de Magalhães-Sant’ana A.C., Gonçalves T. (2006). Saccharomyces cerevisiae Hog1 protein phosphorylation upon exposure to bacterial endotoxin. J. Biol. Chem. 281: 24687–24694 [DOI] [PubMed] [Google Scholar]
- Martin F., et al. (2008). The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452: 88–92 [DOI] [PubMed] [Google Scholar]
- Martin F., Duplessis S., Ditengou F., Lagrange H., Voiblet C., Lapeyrie F. (2001). Developmental cross talking in the ectomycorrhizal symbiosis: Signals and communication genes. New Phytol. 151: 145–154 [DOI] [PubMed] [Google Scholar]
- Martin F., et al. (2010). Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464: 1033–1038 [DOI] [PubMed] [Google Scholar]
- Mayorga M.E., Gold S.E. (1999). A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34: 485–497 [DOI] [PubMed] [Google Scholar]
- Mayorga M.E., Gold S.E. (2001). The ubc2 gene of Ustilago maydis encodes a putative novel adaptor protein required for filamentous growth, pheromone response and virulence. Mol. Microbiol. 41: 1365–1379 [DOI] [PubMed] [Google Scholar]
- Mehrabi R., Ding S., Xu J.R. (2008). MADS-box transcription factor mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot. Cell 7: 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi R., Van der Lee T., Waalwijk C., Gert H.J. (2006a). MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant Microbe Interact. 19: 389–398 [DOI] [PubMed] [Google Scholar]
- Mehrabi R., Zwiers L.H., de Waard M.A., Kema G.H. (2006b). MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola. Mol. Plant Microbe Interact. 19: 1262–1269 [DOI] [PubMed] [Google Scholar]
- Mendoza-Mendoza A., Eskova A., Weise C., Czajkowski R., Kahmann R. (2009). Hap2 regulates the pheromone response transcription factor prf1 in Ustilago maydis. Mol. Microbiol. 72: 683–698 [DOI] [PubMed] [Google Scholar]
- Menotta M., Pierleoni R., Amicucci A., Sisti D., Cerasi A., Millo E., Chiarantini L., Stocchi V. (2006). Characterization and complementation of a Fus3/Kss1 type MAPK from Tuber borchii, TBMK. Mol. Genet. Genomics 276: 126–134 [DOI] [PubMed] [Google Scholar]
- Mészáros T., Helfer A., Hatzimasoura E., Magyar Z., Serazetdinova L., Rios G., Bardóczy V., Teige M., Koncz C., Peck S., Bögre L. (2006). The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 48: 485–498 [DOI] [PubMed] [Google Scholar]
- Mey G., Held K., Scheffer J., Tenberge K.B., Tudzynski P. (2002a). CPMK2, an SLT2-homologous mitogen-activated protein (MAP) kinase, is essential for pathogenesis of Claviceps purpurea on rye: evidence for a second conserved pathogenesis-related MAP kinase cascade in phytopathogenic fungi. Mol. Microbiol. 46: 305–318 [DOI] [PubMed] [Google Scholar]
- Mey G., Oeser B., Lebrun M.H., Tudzynski P. (2002b). The biotrophic, non-appressorium-forming grass pathogen Claviceps purpurea needs a Fus3/Pmk1 homologous mitogen-activated protein kinase for colonization of rye ovarian tissue. Mol. Plant Microbe Interact. 15: 303–312 [DOI] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki A., Kubo E., Arase S., Kihara J. (2006). Disruption of SRM1, a mitogen-activated protein kinase gene, affects sensitivity to osmotic and ultraviolet stressors in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol. Lett. 257: 253–261 [DOI] [PubMed] [Google Scholar]
- Motoyama T., Kadokura K., Ohira T., Ichiishi A., Fujimura M., Yamaguchi I., Kudo T. (2005). A two-component histidine kinase of the rice blast fungus is involved in osmotic stress response and fungicide action. Fungal Genet. Biol. 42: 200–212 [DOI] [PubMed] [Google Scholar]
- Motoyama T., Ochiai N., Morita M., Iida Y., Usami R., Kudo T. (2008). Involvement of putative response regulator genes of the rice blast fungus Magnaporthe oryzae in osmotic stress response, fungicide action, and pathogenicity. Curr. Genet. 54: 185–195 [DOI] [PubMed] [Google Scholar]
- Müller P., Aichinger C., Feldbrügge M., Kahmann R. (1999). The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34: 1007–1017 [DOI] [PubMed] [Google Scholar]
- Müller P., Katzenberger J.D., Loubradou G., Kahmann R. (2003a). Guanyl nucleotide exchange factor Sql2 and Ras2 regulate filamentous growth in Ustilago maydis. Eukaryot. Cell 2: 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P., Weinzierl G., Brachmann A., Feldbrügge M., Kahmann R. (2003b). Mating and pathogenic development of the Smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2: 1187–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal M., García-Pedrajas M.D., Gold S.E. (2008). Dimorphism in fungal plant pathogens. FEMS Microbiol. Lett. 284: 127–134 [DOI] [PubMed] [Google Scholar]
- Olson A., et al. (March 28, 2012). Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. New Phytol. http://dx.doi.org/10.1111/j.1469-8137.2012.04128.x [DOI] [PubMed] [Google Scholar]
- Ortoneda M., Guarro J., Madrid M.P., Caracuel Z., Roncero M.I., Mayayo E., Di Pietro A. (2004). Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect. Immun. 72: 1760–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panadero J., Pallotti C., Rodríguez-Vargas S., Randez-Gil F., Prieto J.A. (2006). A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 281: 4638–4645 [DOI] [PubMed] [Google Scholar]
- Park G., Bruno K.S., Staiger C.J., Talbot N.J., Xu J.R. (2004a). Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol. Microbiol. 53: 1695–1707 [DOI] [PubMed] [Google Scholar]
- Park G., Xue C., Zhao X., Kim Y., Orbach M., Xu J.R. (2006). Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell 18: 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G., Xue C., Zheng L., Lam S., Xu J.R. (2002). MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 15: 183–192 [DOI] [PubMed] [Google Scholar]
- Park S.M., Choi E.S., Kim M.J., Cha B.J., Yang M.S., Kim D.H. (2004b). Characterization of HOG1 homologue, CpMK1, from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol. Microbiol. 51: 1267–1277 [DOI] [PubMed] [Google Scholar]
- Pérez-Nadales E., Di Pietro A. (2011). The membrane mucin Msb2 regulates invasive growth and plant infection in Fusarium oxysporum. Plant Cell 23: 1171–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M., Courty P.E., Kohler A., Delaruelle C., Martin D., Tagu D., Frey-Klett P., Duplessis M., Chalot M., Podila G.K., Martin F. (2003). Analysis of expressed sequence tags from the ectomycorrhizal basidiomycetes Laccaria bicolor and Pisolithus microcarpus. New Phytol. 159: 117–129 [DOI] [PubMed] [Google Scholar]
- Petutschnig E.K., Jones A.M., Serazetdinova L., Lipka U., Lipka V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285: 28902–28911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23: 80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Wang Q., Dou X., Wang W., Zhao Q., Lv R., Zhang H., Zheng X., Wang P., Zhang Z. (February 9, 2012). MoSwi6, an APSES family transcription factor, interacts with MoMps1 and is required for hyphal and conidial morphogenesis, appressorial function and pathogenicity of Magnaporthe oryzae. Mol. Plant Pathol. http://dx.doi.org/10.1111/j.1364-3703.2011.00779.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.L., et al. (2008a). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., Mackinlay J., Loake G.J., Mundy J., Morris P.C. (2008b). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy V., Zhao X., Snyder A.K., Xu J.R., Shah D.M. (2007). Two mitogen-activated protein kinase signalling cascades mediate basal resistance to antifungal plant defensins in Fusarium graminearum. Cell. Microbiol. 9: 1491–1506 [DOI] [PubMed] [Google Scholar]
- Rauyaree P., Ospina-Giraldo M.D., Kang S., Bhat R.G., Subbarao K.V., Grant S.J., Dobinson K.F. (2005). Mutations in VMK1, a mitogen-activated protein kinase gene, affect microsclerotia formation and pathogenicity in Verticillium dahliae. Curr. Genet. 48: 109–116 [DOI] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.Y., Han L., Mao G., Glazebrook J., Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispail N., Di Pietro A. (2010). The homeodomain transcription factor Ste12: Connecting fungal MAPK signalling to plant pathogenicity. Commun. Integr. Biol. 3: 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispail N., et al. (2009). Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46: 287–298 [DOI] [PubMed] [Google Scholar]
- Robert H.S., Offringa R. (2008). Regulation of auxin transport polarity by AGC kinases. Curr. Opin. Plant Biol. 11: 495–502 [DOI] [PubMed] [Google Scholar]
- Rui O., Hahn M. (2007). The Slt2-type MAP kinase Bmp3 of Botrytis cinerea is required for normal saprotrophic growth, conidiation, plant surface sensing and host tissue colonization. Mol. Plant Pathol. 8: 173–184 [DOI] [PubMed] [Google Scholar]
- Ruiz-Roldán M.C., Maier F.J., Schäfer W. (2001). PTK1, a mitogen-activated-protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity of Pyrenophora teres on barley. Mol. Plant Microbe Interact. 14: 116–125 [DOI] [PubMed] [Google Scholar]
- Sakaguchi A., Tsuji G., Kubo Y. (2010). A yeast STE11 homologue CoMEKK1 is essential for pathogenesis-related morphogenesis in Colletotrichum orbiculare. Mol. Plant Microbe Interact. 23: 1563–1572 [DOI] [PubMed] [Google Scholar]
- Schindler K., Benjamin K.R., Martin A., Boglioli A., Herskowitz I., Winter E. (2003). The Cdk-activating kinase Cak1p promotes meiotic S phase through Ime2p. Mol. Cell. Biol. 23: 8718–8728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler K., Winter E. (2006). Phosphorylation of Ime2 regulates meiotic progression in Saccharomyces cerevisiae. J. Biol. Chem. 281: 18307–18316 [DOI] [PubMed] [Google Scholar]
- Segmüller N., Ellendorf U., Tudzynski B., Tudzynski P. (2007). BcSAK1, a stress-activated mitogen-activated protein kinase, is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryot. Cell 6: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.G., García-Pedrajas M.D., Hong W., Yu Z., Gold S.E., Perlin M.H. (2004). An ste20 homologue in Ustilago maydis plays a role in mating and pathogenicity. Eukaryot. Cell 3: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.E., Mitchell A.P. (1989). A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon P.S., Waters O.D., Simmonds J., Cooper R.M., Oliver R.P. (2005). The Mak2 MAP kinase signal transduction pathway is required for pathogenicity in Stagonospora nodorum. Curr. Genet. 48: 60–68 [DOI] [PubMed] [Google Scholar]
- Sundaram S., Kim S.J., Suzuki H., Mcquattie C.J., Hiremah S.T., Podila G.K. (2001). Isolation and characterization of a symbiosis-regulated ras from the ectomycorrhizal fungus Laccaria bicolor. Mol. Plant Microbe Interact. 14: 618–628 [DOI] [PubMed] [Google Scholar]
- Takano Y., Kikuchi T., Kubo Y., Hamer J.E., Mise K., Furusawa I. (2000). The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant Microbe Interact. 13: 374–383 [DOI] [PubMed] [Google Scholar]
- Talbot N.J. (2003). On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57: 177–202 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tena G., Asai T., Chiu W.L., Sheen J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4: 392–400 [DOI] [PubMed] [Google Scholar]
- Tena G., Boudsocq M., Sheen J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines E., Weber R.W., Talbot N.J. (2000). MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12: 1703–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserant E., et al. (2012). The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol. 193: 755–769 [DOI] [PubMed] [Google Scholar]
- Uhm K.H., Ahn I.P., Kim S., Lee Y.H. (2003). Calcium/calmodulin-dependent signaling for prepenetration development in Colletotrichum gloeosporioides. Phytopathology 93: 82–87 [DOI] [PubMed] [Google Scholar]
- Urban M., Mott E., Farley T., Hammond-Kosack K. (2003). The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 4: 347–359 [DOI] [PubMed] [Google Scholar]
- Viaud M., Fillinger S., Liu W., Polepalli J.S., Le Pêcheur P., Kunduru A.R., Leroux P., Legendre L. (2006). A class III histidine kinase acts as a novel virulence factor in Botrytis cinerea. Mol. Plant Microbe Interact. 19: 1042–1050 [DOI] [PubMed] [Google Scholar]
- Voiblet C., Duplessis S., Encelot N., Martin F. (2001). Identification of symbiosis-regulated genes in Eucalyptus globulus-Pisolithus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. Plant J. 25: 181–191 [DOI] [PubMed] [Google Scholar]
- Wan J., Zhang S., Stacey G. (2004). Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant Pathol. 5: 125–135 [DOI] [PubMed] [Google Scholar]
- Wan J., Zhang X.C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwar V., Oved S., Dickman M.B. (2000). Antisense expression of the calmodulin gene from Colletotrichum trifolii impairs prepenetration development(1). FEMS Microbiol. Lett. 191: 213–219 [DOI] [PubMed] [Google Scholar]
- Widmann C., Gibson S., Jarpe M.B., Johnson G.L. (1999). Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79: 143–180 [DOI] [PubMed] [Google Scholar]
- Wilson R.A., Talbot N.J. (2009). Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7: 185–195 [DOI] [PubMed] [Google Scholar]
- Wong Sak Hoi J., Dumas B. (2010). Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell 9: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Hettenhausen C., Meldau S., Baldwin I.T. (2007). Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.R. (2000). Map kinases in fungal pathogens. Fungal Genet. Biol. 31: 137–152 [DOI] [PubMed] [Google Scholar]
- Xu J.R., Hamer J.E. (1996). MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10: 2696–2706 [DOI] [PubMed] [Google Scholar]
- Xu J.R., Staiger C.J., Hamer J.E. (1998). Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA 95: 12713–12718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C., Park G., Choi W., Zheng L., Dean R.A., Xu J.R. (2002). Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 14: 2107–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Kawaguchi H., Sakata Y., Kominami K., Hirano M., Shima H., Akada R., Yamashita I. (1990). Initiation of meiosis and sporulation in Saccharomyces cerevisiae requires a novel protein kinase homologue. Mol. Gen. Genet. 221: 176–186 [DOI] [PubMed] [Google Scholar]
- Yoshimi A., Kojima K., Takano Y., Tanaka C. (2005). Group III histidine kinase is a positive regulator of Hog1-type mitogen-activated protein kinase in filamentous fungi. Eukaryot. Cell 4: 1820–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Xue C., Kong L., Li G., Xu J.R. (2011). A Pmk1-interacting gene is involved in appressorium differentiation and plant infection in Magnaporthe oryzae. Eukaryot. Cell 10: 1062–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Kim Y., Park G., Xu J.R. (2005). A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 17: 1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Mehrabi R., Xu J.R. (2007). Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 6: 1701–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Xu J.R. (2007). A highly conserved MAPK-docking site in Mst7 is essential for Pmk1 activation in Magnaporthe grisea. Mol. Microbiol. 63: 881–894 [DOI] [PubMed] [Google Scholar]
- Zhang Y., McCormick S. (2009). AGCVIII kinases: at the crossroads of cellular signaling. Trends Plant Sci. 14: 689–695 [DOI] [PubMed] [Google Scholar]
- Zheng L., Campbell M., Murphy J., Lam S., Xu J.R. (2000). The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant Microbe Interact. 13: 724–732 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.