This study shows that the C-terminal tail of the preprotein receptor Toc159 possesses the physicochemical and structural properties of chloroplast transit peptides. A number of fluorescent protein fusion constructs consistently demonstrated that this sorting signal is essential for the targeting of the Toc159 receptor to the chloroplast outer membrane.
Abstract
Although Toc159 is known to be one of the key GTPase receptors for selective recognition of chloroplast preproteins, the mechanism for its targeting to the chloroplast surface remains unclear. To compare the targeting of these GTPase receptors, we identified two Toc159 isoforms and a Toc34 from Bienertia sinuspersici, a single-cell C4 species with dimorphic chloroplasts in individual chlorenchyma cells. Fluorescent protein tagging and immunogold studies revealed that the localization patterns of Toc159 were distinctive from those of Toc34, suggesting different targeting pathways. Bioinformatics analyses indicated that the C-terminal tails (CTs) of Toc159 possess physicochemical and structural properties of chloroplast transit peptides (cTPs). These results were further confirmed by fluorescent protein tagging, which showed the targeting of CT fusion proteins to the chloroplast surface. The CT of Bs Toc159 in reverse orientation functioned as a cleavable cTP that guided the fluorescent protein to the stroma. Moreover, a Bs Toc34 mutant protein was retargeted to the chloroplast envelope using the CTs of Toc159 or reverse sequences of other cTPs, suggesting their conserved functions. Together, our data show that the C terminus and the central GTPase domain represent a novel dual domain–mediated sorting mechanism that might account for the partitioning of Toc159 between the cytosol and the chloroplast envelope for preprotein recognition.
INTRODUCTION
The majority of plastid proteins are nuclear encoded, synthesized on cytosolic ribosomes as precursor proteins (preproteins), and posttranslationally imported into the organelles. Most of these preproteins possess an N-terminal cleavable chloroplastic transit peptide (cTP) that contains essential and sufficient information for chloroplast targeting in the general import pathway. This general pathway involves the translocons at the outer chloroplast membrane (Toc) and the translocons at the inner chloroplast membrane (Tic), which mediate the coordinated events of preprotein recognition and translocation at the chloroplast envelope (reviewed in Jarvis, 2008; Agne and Kessler, 2009; Andrès et al., 2010). Toc75, Toc159, and Toc34 are core components that play essential roles in the chloroplast protein import process (Kessler and Schnell, 2006). The early stages of import are mediated by two homologous GTPase proteins, Toc159 and Toc34, which are exposed on the cytosolic side of the chloroplast surface, pertinent to their roles in early preprotein recognition. The two receptors act in concert to regulate the initial interaction with cTPs and insertion of the preproteins into the β-barrel protein channel, Toc75, for translocation across the outer envelope membrane in an ATP- and GTP-dependent manner. Despite their sequence homology, Toc159 and Toc34 receptors do not exhibit functional redundancy according to the studies of Arabidopsis thaliana knockout mutants of Toc159 orthologs (Bauer et al., 2000; Ivanova et al., 2004; Kubis et al., 2004) and Toc34 orthologs (Jarvis et al., 1998; Kubis et al., 2003; Constan et al., 2004). Currently, there is no consensus on the specific role of the two receptors, leading to the emergence of two conflicting models of protein translocation (Kessler and Schnell, 2004; Jarvis, 2008). Of particular interest in this study is the targeting model, which proposes that Toc159 is the primary preprotein receptor, as evidenced by the direct cross-linking of Toc159 with cTPs (Perry and Keegstra, 1994; Ma et al., 1996) and inhibition of preprotein binding in vitro after Toc159 antibody neutralization (Hirsch et al., 1994).
Resident proteins of the suborganellar compartments that traffic across the membrane layer(s) of chloroplast envelopes generally gain access through the Toc/Tic machinery. The targeting and integration of chloroplast outer membrane proteins, on the other hand, have been characterized to a much lesser extent and apparently involve multiple pathways (reviewed in Hofmann and Theg, 2005; Bölter and Soll, 2011). To date, Toc75 is the only outer membrane protein that contains an N-terminal cleavable cTP, and this signal is uniquely accompanied by a stretch of polyglycine that potentially functions as a stop signal to prevent its translocation into the stroma (Tranel et al., 1995; Tranel and Keegstra, 1996). Although both Toc75 and its paralog, outer envelope protein 80 kD (OEP80), are targeted with the assistance of some proteinaceous components on the chloroplast surface, the targeting of OEP80 is independent of the general import pathway, in agreement with the absence of an N-terminal cleavable cTP (Inoue and Potter, 2004; Huang et al., 2011). In fact, all other identified β-barrel proteins on the chloroplast outer membrane, including OEP21 (Bölter et al., 1999), OEP24 (Pohlmeyer et al., 1998), and OEP37 (Goetze et al., 2006), do not have a predicted cTP and are spontaneously integrated into the membrane. In addition to β-barrel proteins, chloroplasts have evolved α-helical integral proteins in the outer membrane, in spite of the endosymbiotic origin from a bacterial progenitor. Depending on the location of the transmembrane helix at the N or C terminus, these proteins are commonly referred to as signal-anchored proteins (e.g., OEP7/14 and OEP64) and tail-anchored proteins (e.g., OEP9 and Toc34), respectively. The targeting of these proteins and their efficient insertion with the correct topology depend on the information embedded in the transmembrane domains and the flanking charged regions, whereas the cytosolic GTPase domain of Toc34 also plays an essential role (Chen and Schnell, 1997; Schleiff et al., 2001; Qbadou et al., 2003; Hofmann and Theg, 2005; Dhanoa et al., 2010). Although two previous studies documented that the GTPase domain mediates the targeting and insertion of Toc159 (Bauer et al., 2002; Smith et al., 2002a), this domain per se presumably does not contain any sorting signal, and the targeting specificity of this receptor to the chloroplast envelope has not been elucidated. The absence of any predicted β-barrel or α-helical transmembrane domains implies that Toc159 is targeted in a manner independent of the aforementioned pathways, critical to its function as a soluble preprotein receptor that partitions between the cytosol and the chloroplast envelope (Hiltbrunner et al., 2001). Previous protein truncation studies suggested that the specific sorting signal of Toc159 resides within its C-terminal region (Muckel and Soll, 1996; Lee et al., 2003).
Focusing on plant organelles in a single-cell C4 system such as Bienertia sinuspersici is of special interest to obtain information on the evolution of protein sorting mechanisms. Since plastid biogenesis and functions depend on the coordinated expression and import of thousands of nuclear-encoded proteins (Jarvis, 2008), the development and function of dimorphic chloroplasts in the single-cell C4 system might involve distinct import pathways to regulate the selective import of proteins and mediate plastid differentiation. In this study, we identified two Toc159 homologs from the single-cell C4 species B. sinuspersici (Akhani et al., 2005; Chuong et al., 2006). Analyses of the B. sinuspersici Toc159 sequences using various bioinformatics tools indicated that the C-terminal tails (CTs) possess general physicochemical and structural properties similar to the transit sequences of chloroplast preproteins. We used various enhanced green fluorescent protein (EGFP) fusion constructs to demonstrate that the CTs directed the passenger protein to the chloroplast envelope, as evidenced by fluorescence microscopy and immunoblot analysis following subcellular fractionation. Reciprocal fusion constructs of the CTs of Toc159 and various cTPs confirmed that these sequences behave as chloroplast-targeting signals. Considering the importance of the GTPase domain (Bauer et al., 2002; Smith et al., 2002a), we propose that a dual domain–mediated pathway may account for the partitioning of Toc159 receptors between the cytosol and the chloroplast envelope.
RESULTS
The Tripartite Structure of Toc159 Is Conserved in B. sinuspersici Homologs
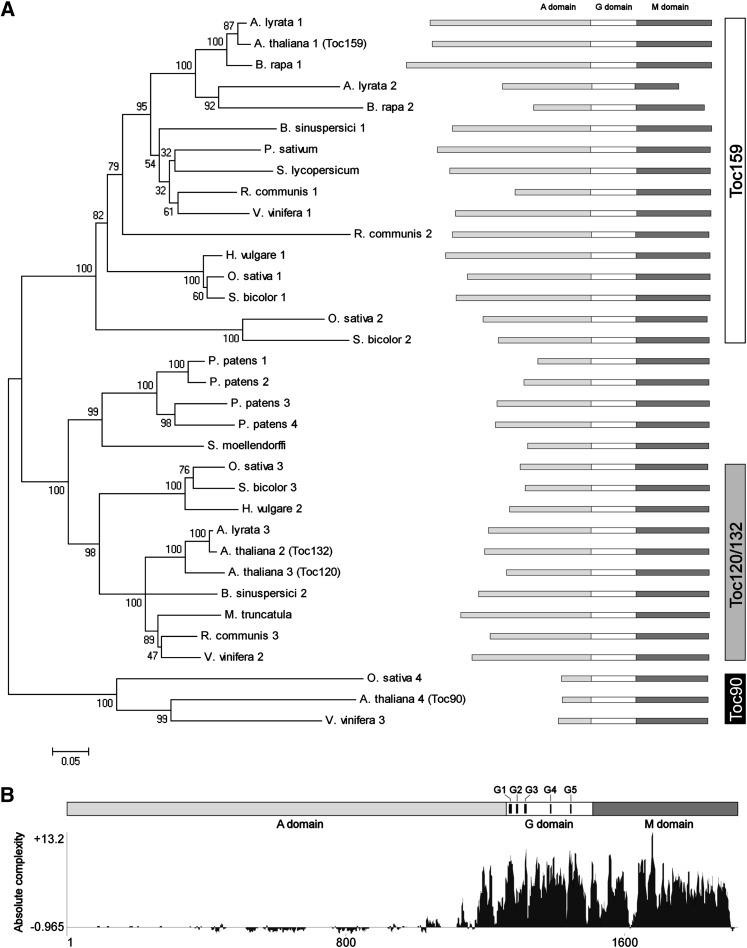
As a first step toward understanding chloroplast protein import in the single-cell C4 species, we identified the Toc159 and Toc34 homologs in B. sinuspersici. Earlier studies of preprotein recognition at the chloroplast envelope focused on the Toc components in pea (Pisum sativum) (Hirsch et al., 1994; Schnell et al., 1994). More recent research was also based on the orthologs from Arabidopsis (Li and Chen, 1997; Bölter et al., 1998; Jarvis et al., 1998; Bauer et al., 2000) and on two spinach (Spinacia oleracea) Toc34 isoforms (Voigt et al., 2005). To date, the identity of Toc receptor components in other plant species has not been studied. By conducting a BLASTp search using the primary sequences of pea Toc159 and Toc34 as query, we retrieved homologous sequences of the two protein families from various plant species, designed gene-specific primers based on the conserved regions from nucleotide sequence alignments, and successfully cloned full-length cDNAs that encode two Toc159 homologs and one Toc34 homolog. Phylogenetic analyses indicated their sequence similarities with other putative and demonstrated Toc159 (Figure 1A; see Supplemental Data Set 1 online) and Toc34 (see Supplemental Figure 1A and Supplemental Data Set 2 online) proteins.
Figure 1.
Comparison of Deduced Amino Acid Sequences from Different Putative or Reported Toc159.
(A) A neighbor-joining tree of Toc159 homologs. The alignment used for this analysis is available as Supplemental Data Set 1 online. The bootstrap values with 2000 repetitions (%) are given at the respective nodes. The distance scale (substitutions per site) is shown in the bottom left corner.
(B) The absolute complexity plot of aligned Toc159 sequences. The absolute complexity indicates the average of pairwise alignment scores of each residue as computed using the AlignX module of Vector NTI Advance 10.3.0 (Invitrogen) with the substitution matrix blosum62mt2. A more positive value indicates a higher degree of conservation. The positions of the conserved motifs G1-G5 of the GTPase superfamily are indicated.
Similar to other reported Toc159 sequences, the tripartite structure, which comprises an N-terminal acidic domain (A-domain), a central GTPase domain (G-domain), and a C-terminal membrane-anchor domain (M-domain), is conserved in the two B. sinuspersici Toc159 homologs (Figure 1A). The G-domains are highly conserved in both Toc159 (Figure 1B) and Toc34 (see Supplemental Figure 1B online) homologs, with the maximum similarities within the G1, G3, G4, and G5 motifs typically found in the GTPase superfamily (Sun et al., 2002). The M-domains of Toc159 homologs are also conserved in the plant kingdom, although the physiological relevance of these domains is unknown (Figure 1B). Despite the lack of predictable membrane-spanning structures, the C-terminal regions of Toc159 are commonly termed membrane anchors solely due to their protease resistance, which implies their embedment in the outer membrane. In fact, the amino acid sequence alignment of Toc159 and Toc34 homologs shows that the transmembrane regions of Toc34 are partly substituted by less hydrophobic residues in the sequences of Toc159 (see Supplemental Figure 2 online). The A-domains of Toc159 homologs are highly variable in length (Figure 1A) and amino acid composition (Figure 1B), correlating with the fact that this domain is fully dispensable (Lee et al., 2003) and plays an accessory role in regulating the substrate selectivity of the receptor (Inoue et al., 2010). The concept of preprotein specificity of the Toc159 receptors has arisen from the molecular genetics analysis of Arabidopsis knockout mutants (Bauer et al., 2000; Ivanova et al., 2004; Kubis et al., 2004) and in vitro binding studies (Smith et al., 2004), suggesting that Arabidopsis Toc159 ortholog is specific for more abundant photosynthetic preproteins, whereas Arabidopsis Toc132 and Toc120 orthologs are functionally redundant for targeting nonphotosynthetic, housekeeping preproteins. Pairwise homology comparison of the Toc159 sequences indicates that the two B. sinuspersici homologs share substantial identity with Arabidopsis Toc159 and Toc132, respectively (see Supplemental Table 1 online). Thus, we named the two identified proteins in this study Bs Toc159 and Bs Toc132, respectively. While this study confirmed the expression of multiple Toc159 isoforms in a plant species other than Arabidopsis, the selectivity of Toc159 for specific subsets of preproteins is predicted to be universal in higher plants, as the phylogenetic tree illustrates that multiple sequences retrieved from the same species separate into the Toc159 clade and the Toc120/132 clade, respectively (Figure 1A). The two Arabidopsis Toc34 isoforms (Toc33 and Toc34), on the other hand, fall into the same clade (see Supplemental Figure 1A online).
Expression Profiles of Bs Toc159 and Toc132 Indicate Their Substrate Specificities
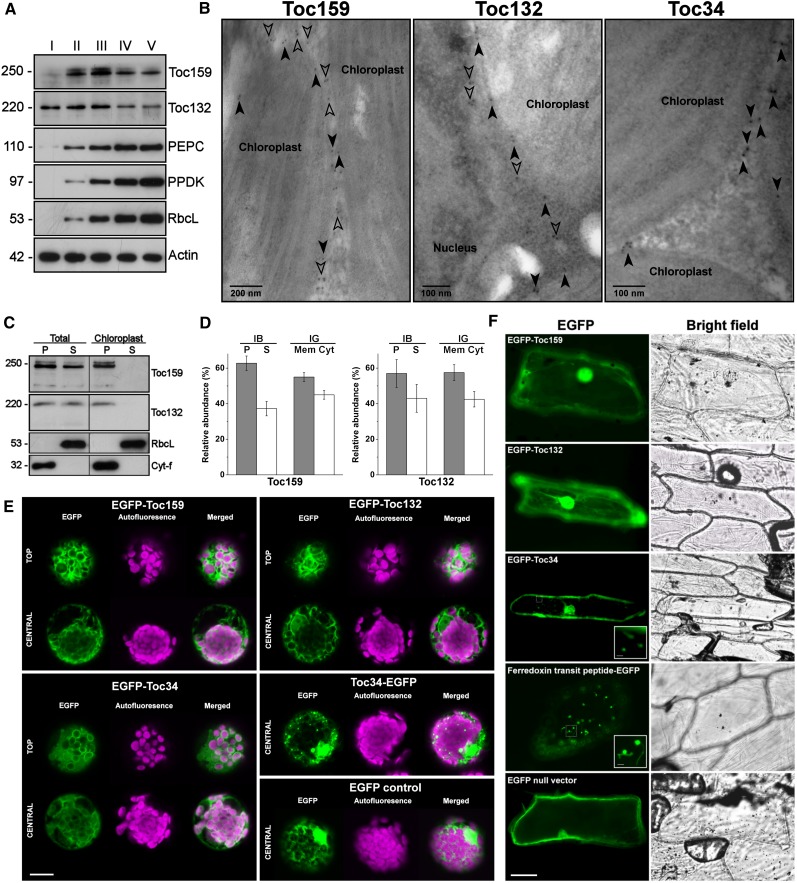
To confirm that the two identified Toc159 homolog sequences are transcriptionally active for protein expression in B. sinuspersici, we raised isoform-specific antibodies against recombinant proteins of the highly variable N-terminal A-domains, which did not have any significant similarity found in PSI-BLAST. Immunoblot analysis detected immunoreactive bands of both Toc159 isoforms in crude leaf extracts of B. sinuspersici (Figure 2A). The migration of Toc159 and Toc132 during polyacrylamide gel electrophoresis deviated from their theoretical molecular masses of 148 and 134 kD, respectively (Figure 2A). Similarly, recombinant proteins that contained A-domains, which were used for antibody production, also exhibited unexpectedly low electrophoretic mobility (see Supplemental Figure 3A online). However, the identities of these pure Escherichia coli–expressed proteins were confirmed by tandem mass spectrometry–based sequencing (see Supplemental Figure 3 online). Similar aberrant electrophoretic migration of A-domains (Richardson et al., 2009) and full-length proteins of Toc159 (Chen et al., 2000; Lee et al., 2003) has been attributed to their high acidity. Comparison of Bs Toc159 and Toc132 protein amounts at different developmental stages of leaves revealed their different expression profiles in relation to their respective functions. The leaf primordia at the earliest developmental stage (stage I) contained proplastids that had not yet expressed the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit or imported photosynthetic enzymes, such as pyruvate orthophosphate dikinase (Figure 2A). Toc132 was abundant at this stage, whereas Toc159 was undetectable (Figure 2A). A drastic increase in Toc159 was observed at stages II and III, when chloroplasts are actively accumulating photosynthetic enzymes, while the amount of Toc132 remained steady (Figure 2A). Basal levels of both Toc159 and Toc132 were maintained at stages IV and V, when leaves are primarily composed of mature chloroplasts (Figure 2A). Thus, the expression profiles confirmed previous observations in Arabidopsis that Toc159 and Toc132 exhibit substrate specificities for photosynthetic and housekeeping proteins, respectively (Bauer et al., 2000).
Figure 2.
Biochemical Characterization and Subcellular Localization of Toc159 Homologs from B. sinuspersici.
(A) Immunoblot analysis of various proteins at different stages of leaf development. For each individual blot, an equal amount of total protein per lane was used. Crude protein homogenates were extracted from leaves of various lengths at stage I (<1 mm), stage II (2 to 3 mm), stage III (5 to 6 mm), stage IV (10 to 12 mm), and stage V (>20 mm). Protein molecular masses (in kD) are shown. PEPC, phosphoenolpyruvate carboxylase; PPDK, pyruvate orthophosphate dikinase; RbcL, Rubisco large subunit.
(B) Immunogold localization of Toc proteins. Immunogold labeling of mature B. sinuspersici leaves was performed with isoform-specific antibodies raised against Toc159 and Toc132 and a commercial antibody against Toc34. Closed arrowheads indicate gold particles on the chloroplast envelopes. Open arrowheads indicate other gold particles in the interchloroplastic space.
(C) Immunoblot analysis of endogenous Toc159 and Toc132 in subcellular fractions. The total protoplasts or purified chloroplasts were subfractionated into pellet (P) and soluble (S) fractions. Detection with antibodies against Rubisco large subunit (RbcL) and cytochrome-f (Cyt-f) served as loading controls for the soluble and pellet fractions, respectively.
(D) Relative abundance of endogenous Toc159 and Toc132. From the immunoblot analysis (IB) shown in (C), immunoreactive bands of the pellet (P) and soluble (S) fractions of total protoplast extracts were analyzed by densitometric quantification. From the immunogold localization (IG) as shown (B), the relative abundance was determined by counting the number of membrane-associated (Mem) and cytosolic (Cyt) gold particles. All values are the mean of three replicates (±se).
(E) Transient expression of EGFP fusion proteins in B. sinuspersici chlorenchyma protoplasts. Toc proteins were fused to the C terminus of EGFP (EGFP-Toc159, EGFP-Toc132, and EGFP-Toc34) or the N terminus of EGFP (Toc34-EGFP) for constitutive 35S-driven expression following polyethylene glycol–mediated transfection of isolated protoplasts. Representative images showing central and/or top focal planes of EGFP fluorescence (excitation at 488 nm and emission detected at 509 nm), chlorophyll autofluorescence (excitation at 649 nm and emission detected at 666 nm), and a merge of the two channels are shown. Bar = 10 μm
(F) Transient expression of EGFP fusion proteins in onion epidermal cells. EGFP was either unmodified (null vector), fused to the N terminus of Toc159, Toc132, or Toc34, or fused to the C terminus of ferredoxin transit peptide. Bars = 40 μm and 10 μm (insets).
[See online article for color version of this figure.]
Toc159 and Toc34 Demonstrated Distinctive Subcellular Localization Patterns
Since Toc159 and Toc34 have distinctive functions and localizations in pea and Arabidopsis, we asked whether the homologs have different subcellular localization in B. sinuspersici. Immunogold micrographs revealed that the endogenous Toc34 proteins strictly reside at the chloroplast envelope (Figure 2B), in agreement with the presence of a putative transmembrane α-helix in the Toc34 sequence (see Supplemental Figure 2 online). In addition to the localization of Bs Toc159 and Toc132 to the chloroplast envelope, gold particles were approximately equally abundant in the interchloroplastic space (Figure 2B). This observation is consistent with previous subcellular localization studies showing that endogenous Toc159 or N-terminally truncated Toc159 fused to EGFP were localized to the intervening space between chloroplasts in Arabidopsis (Hiltbrunner et al., 2001; Bauer et al., 2002). The presence of both soluble and membrane-associated forms of endogenous Bs Toc159 and Toc132 was confirmed by immunoblot analysis (Figure 2C). Densitometric analysis of the immunoreactive bands indicated that ∼40% of both receptors were found in the soluble fractions (Figure 2D). Similar values of relative abundance were obtained from quantitative analysis of gold particles (Figure 2D).
To further support the notion that Toc159 is a soluble receptor that partitions between the cytosol and chloroplast envelopes, we confirmed our immunogold localization and immunoblotting data by transient expression of EGFP-tagged full-length Toc receptors in B. sinuspersici leaf-derived protoplasts (Figure 2E). Confocal laser scanning microscopy revealed fluorescent signals of EGFP-BsToc159 and EGFP-BsToc132 in the intervening space between chloroplasts (Figure 2E). The ring-like fluorescent signals of EGFP-BsToc34 (Figure 2E), on the other hand, appeared similar to the immunofluorescent signals of Toc34 (Dhanoa et al., 2010) and Toc75 (Hiltbrunner et al., 2001), both of which are integral Toc components of the outer chloroplast membrane. Fusion of Toc34 at the N terminus of EGFP (i.e., BsToc34-EGFP), however, produced punctate structures of variable size (Figure 2E). Since the C-terminal transmembrane domain of Toc34 and its flanking sequences contain important sorting information and the protein is integrated into the outer chloroplast membrane with an Nout-Cin topology (Qbadou et al., 2003; Dhanoa et al., 2010), the addition of a bulky tag (i.e., 27 kD) to the C terminus might result in mistargeting of the fusion protein and formation of insoluble aggregates. Fusion constructs of Toc159 and Toc132 with a C-terminal EGFP (i.e., BsToc159-EGFP and BsToc132-EGFP) did not produce any fluorescent signal in B. sinuspersici protoplasts in repeated experiments using different sequencing-verified clones. Concomitantly, immunoblot analysis of the transfected protoplasts indicated the absence of EGFP fusion proteins and ruled out the possibility of signal abolishment simply due to protein misfolding. We speculated that the C-terminal tags masked the important sorting information at the CTs of Toc159 and Toc132, resulting in the proteasome-mediated degradation of the mistargeted fusion proteins.
Biolistic bombardment of onion epidermal cells with the EGFP-BsToc159 and EGFP-BsToc132 constructs resulted in diffuse fluorescent signals similar to that of the EGFP null vector, whereas EGFP-Toc34 labeled punctate and occasionally elongated structures (Figure 2F). A construct of ferredoxin TP fusion with EGFP produced similar punctate structures with elongated extensions that resembled stromules (Figure 2F). Taken together, the distinctive subcellular localization patterns of Toc159 and Toc34 suggest that Toc159 might use a chloroplast envelope–targeting pathway different from that of the tail-anchored proteins, although some sorting information might similarly reside at the C terminus of Toc159.
Bioinformatics Analyses Revealed cTP Properties of Toc159 CTs
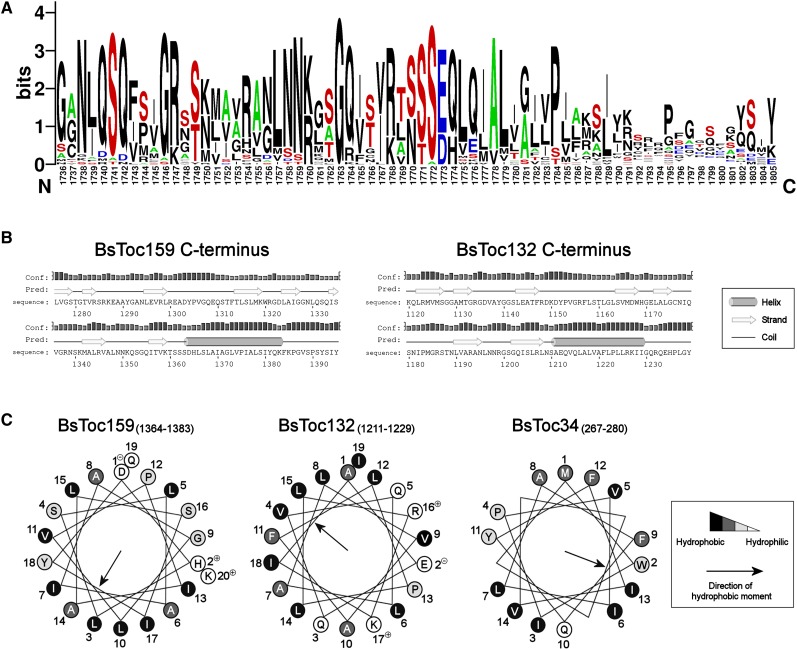
In an attempt to unravel the chloroplast envelope-sorting signals of Toc159, we analyzed the amino acid sequences of the Toc159 homologs from B. sinuspersici and other plant species using various bioinformatics tools. A recurring observation is that the CT of Toc159 exhibits features similar to those characteristic of cTPs, while other Toc159 homologs in the same region were also predicted to have moderate similarities. These similarities are generally found in the physicochemical and structural properties of cTPs, which are otherwise highly divergent in length, amino acid composition, and organization (Bruce, 2001). First, we found that hydroxylated residues (i.e., Ser and Thr) are overrepresented (20.3%) and acidic residues (i.e., Asp and Glu) are underrepresented (1.4%), leading to a basic (pI = 10) CT of Toc159 (Table 1). A sequence logo representation of the aligned CTs of Toc159 homologs also illustrates a similar trend of amino acid occurrence (Figure 3A), which is a common feature of cTPs (von Heijne et al., 1989; Patron and Waller, 2007; Zybailov et al., 2008; Li and Chiu, 2010).
Table 1. Comparison of the Amino Acid Occurrence between the Toc159 C Termini and the Transit Peptides of Chloroplast Preproteins.
| Amino Acid Occurrence (%) |
|||||||
|---|---|---|---|---|---|---|---|
| Peptide(s) | Species | Region | Residues | D-E | S-T | Calculated pI | Reference |
| Bs Toc159-1 | B. sinuspersici | C terminus | 69 | 1.4 | 20.3 | 10.21 | This study |
| Other Toc159 (n = 32) | Multiple | C terminus | 60–77 | 4.0 | 15.4 | 10.63 | This study |
| Transit peptides (n = 1028) | Arabidopsis, Oryza | N terminus | 12 | 2.4 | 26.6 | N.D. | Patron and Waller (2007) |
N.D., not determined.
Figure 3.
Bioinformatics Prediction of a cTP-Like Targeting Signal at the C terminus of Toc159.
(A) WebLogo representation of the aligned C-terminal sequences from Toc159 homologs. Amino acid sequence alignment was performed using the ClustalW algorithm and is available as Supplemental Data Set 1 online. Sequence logos were generated from the alignment using WebLogo 2.8.2 (Crooks et al., 2004). Letter height is proportional to its frequency of occurrence. The graph illustrates high frequencies of hydroxylated residues (in red) and Ala residues (in green) and low frequencies of acidic residues (in blue).
(B) Prediction of secondary structures at the C termini of Toc159 homologs. Predictions were performed using the Web-based program PSIPRED v3.0 (Jones, 1999). The height of the bar for each residue is proportional to the confidence level of prediction.
(C) Helical wheel projections of the predicted C-terminal helices of Toc159 homologs and the transmembrane helix of Toc34. Numbering designates the order of amino acid sequence. Hydrophobicities of residues are highlighted on the Kyte-Doolittle hydropathy scale (hydrophilic in white, neutral in light gray, hydrophobic in dark gray, and very hydrophobic in black).
[See online article for color version of this figure.]
Next, we also observed structural similarities. cTPs primarily form random coils in an aqueous environment and yet might adopt amphipathic α-helical structure(s) at the aqueous/lipid bilayer interface at the chloroplast envelope, although the positions and degrees of amphipathicity may vary (Bruce, 2001). Secondary structure prediction analysis suggested that the CTs of both Bs Toc159 and Toc132 have a tendency of forming an α-helix adjacent to a random coil of 10 to ∼12 residues at the C-terminal end (Figure 3B). Helical wheel projections of the predicted α-helices depict a substantial hydrophobic moment to one side of the helices, while hydrophilic residues largely comprise the other side (Figure 3C). The predicted amphipathic α-helices are somewhat conserved in the CTs of Toc159 homologs from other plant species (see Supplemental Table 2 online). We further evaluated their degrees of amphipathicity from the mean hydrophobicities and mean hydrophobic moments per residue (see Supplemental Table 2 online), using the structurally characterized α-helices of cTPs for comparison (Bruce, 2001). The resulting hydrophobic moment plot analysis depicts that all analyzed sequences have a similar range of mean hydrophobicities, while the mean hydrophobic moments of the predicted α-helices of most Toc159 homologs are slightly above the values of the characterized cTPs (see Supplemental Figure 4 online). Thus, the structural prediction data suggest that Toc159 CTs tend to form amphipathic α-helical structures analogous to those of cTPs.
In addition to the evaluation based on the physicochemical and structural criteria, we wanted to confirm whether the Toc159 CTs contain a putative TP using a neural network–based approach. The ChloroP predictor is a popular algorithm for predicting putative cTPs and is also able to predict stromal processing peptidase cleavage independently of the cTP prediction results (Emanuelsson et al., 1999). Since cTPs all occur at the N-terminal ends of preproteins (hence, the training data sets of chloroplast proteins used), we submitted the reverse sequences of full-length Toc159 homologs for ChloroP analysis. In agreement with the other bioinformatics results, ChloroP predicted a putative TP at the CT region of Toc159 (Table 2). However, the average cTP score of 32 tested Toc159 homologs is slightly below the threshold cutoff (Table 2). Interestingly, the program also predicted a cleavage site of stromal processing peptidase within the Toc159 CT sequence with a score comparable to that of Rubisco small subunit (Table 2). Taken together, the bioinformatics analyses predict a putative TP at the C terminus of Bs Toc159.
Table 2. ChloroP Prediction of Putative Chloroplast Transit Peptides.
| Protein | Orientation | cTP Scorea | cTP Length (Residues)b | Cleavage Site Scorec |
|---|---|---|---|---|
| B. sinuspersici Toc159-1 | Reverse | 0.519 | 51 | 10.81 |
| Other Toc159 (n = 32) | Reverse | 0.486 ± 0.024 | 48.3 ± 11.4 | 4.00 ± 3.30 |
| Arabidopsis Rubisco small subunit | Forward | 0.572 | 54 | 10.09 |
Prediction was performed using the ChloroP 1.1 server (Emanuelsson et al., 1999; http://www.cbs.dtu.dk/services/ChloroP).
A value of >0.5 predicts the presence of a transit peptide.
The length is defined based on the cleavage site prediction.
A more positive value indicates a higher probability of stromal processing peptidase cleavage.
Toc159 CTs Directed EGFP to the Chloroplast Envelope
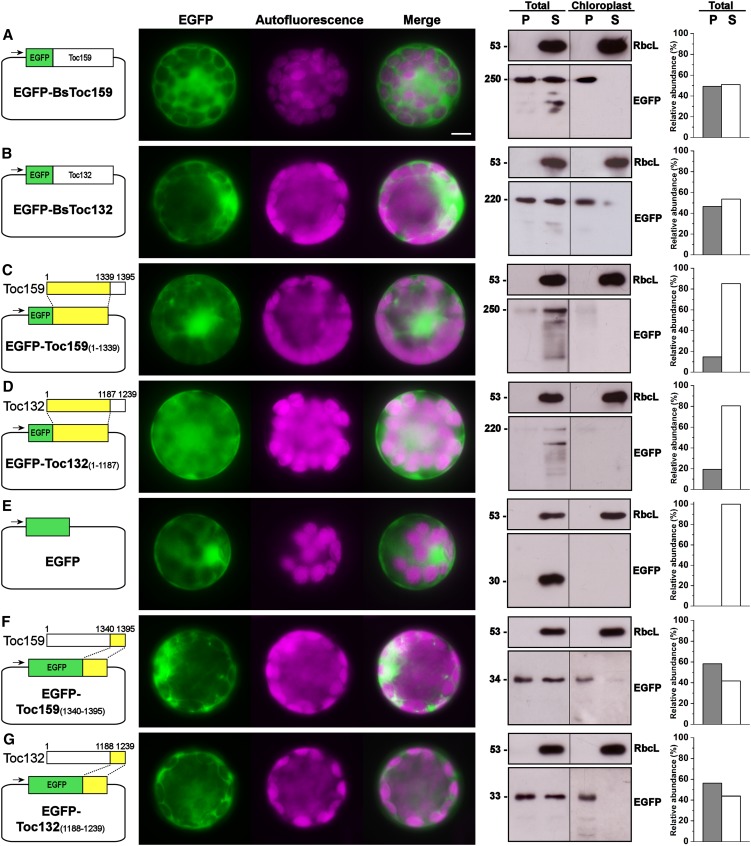
To complement our bioinformatics analyses, we evaluated the chloroplast-targeting function of Toc159 CTs by C-terminal truncation and EGFP fusion with the CTs of Bs Toc159 or Bs Toc132, of which the lengths were derived from the ChloroP prediction (Table 2) plus four additional residues. Since transient expression of EGFP fusions of full-length Bs Toc159 or Bs Toc132 in B. sinuspersici (Figure 2E) and Arabidopsis (Figures 4A and 4B) protoplasts produced identical subcellular localization patterns, we chose to express the EGFP fusion proteins in Arabidopsis protoplasts, due to the relative ease of subsequent chloroplast purification steps using standard and established protocols (Smith et al., 2002b) and given the intricate intracellular complexity of dimorphic chloroplast arrangement in B. sinuspersici. Fluorescence microscopy and immunoblot analysis of the subfractions from total transfected protoplasts and isolated chloroplasts showed that the full-length Toc159 fusion proteins were evenly distributed between the soluble and chloroplast membrane–associated fractions (Figures 4A and 4B). The C-terminally truncated mutants, on the other hand, produced diffuse fluorescent signals and were detected primarily in the soluble fractions of total protoplasts (Figures 4C and 4D), similar to the null vector control (Figure 4E). We also showed that the CTs of Bs Toc159 and Toc132 directed a significant amount of EGFP protein close to the chloroplast exterior (Figures 4F and 4G).
Figure 4.
Transient Expression of EGFP Fusion Proteins with Full-Length or Partial Sequences of Toc159.
Isolated Arabidopsis protoplasts were transfected with the EGFP fusion constructs for transient protein expression driven by the constitutive 35S promoter (left panels). For each construct, representative images of EGFP and chlorophyll fluorescence and a merge of the two channels are shown in the middle panel. The subcellular localization was confirmed by immunoblot analysis with an anti-EGFP antibody after subfractionation of the total protoplasts or purified chloroplasts into pellet (P) and soluble (S) fractions and analyzed by densitometric quantification (right panels). Detection with an antibody against Rubisco large subunit (RbcL) served as loading controls for the soluble fractions. Bar = 10 μm.
(A) Full-length Bs Toc159 fused to the C terminus of EGFP.
(B) Full-length Bs Toc132 fused to the C terminus of EGFP.
(C) C-terminally truncated Bs Toc159 fused to the C terminus of EGFP
(D) C-terminally truncated Bs Toc132 fused to the C terminus of EGFP.
(E) EGFP control in null vector.
(F) The CT of Bs Toc159 fused to the C terminus of EGFP.
(G) The CT of Bs Toc132 fused to the C terminus of EGFP.
[See online article for color version of this figure.]
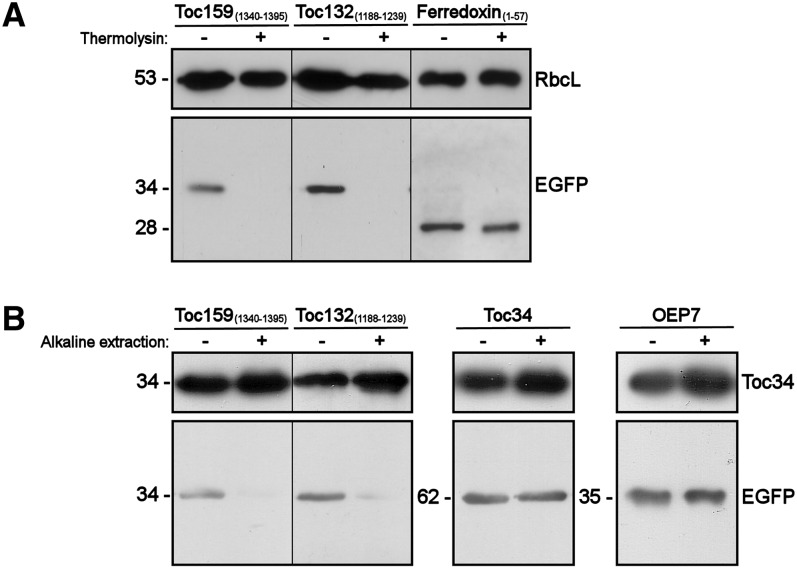
Since further subfractionation of the insoluble chloroplast fractions into envelope and thylakoid membranes was impractical due to the number of transfected protoplasts required, we sought to rule out the possibility of thylakoid membrane localization of the fusion proteins by thermolysin treatments (Figure 5A). As expected, the immunoreactive signals of both EGFP-BsToc159CT and EGFP-BsToc132CT were completely abolished after thermolysin treatments, whereas the EGFP proteins were protected from protease degradation after ferredoxin TP-directed translocation into the stroma (Figure 5A). The observation that the CTs of Toc159 and Toc132 did not guide the translocation of the passenger protein across the outer chloroplast envelope is in line with the topology of Toc159 receptors, which project the A- and G-domains to the cytosolic side. To confirm that the CTs of Toc159 and Toc132 have high affinities for the chloroplast envelope but that these segments did not traverse the outer chloroplast membrane, we treated the isolated chloroplasts with alkaline extraction prior to immunoblot analysis (Figure 5B). As expected, the immunoreactive signals of both EGFP-BsToc159CT and EGFP-BsToc132CT were abolished after the treatment, in contrast with those of the tail-anchored protein Toc34 and signal-anchored protein OEP7 (Figure 5B). In fact, the putative membrane anchor of Toc159 is likely to be found far upstream of the C-terminal end as predicted from the bulk size (e.g., ∼50 kD in pea Toc159) of the fragment being protected from protease degradation (Kessler et al., 1994). From these experiments, we conclude that Toc159 CTs function as cTP-like sorting signals capable of directing passenger proteins to the chloroplast envelope but that the CT regions do not function as a membrane anchor.
Figure 5.
Thermolysin Treatment and Alkaline Extraction of Chloroplasts Purified from Transfected Protoplasts.
(A) Thermolysin treatment. Chloroplasts were isolated from transfected protoplasts that expressed the CTs of Toc159 or Toc132 fused to the C terminus of EGFP or the transit peptide of ferredoxin fused to the N terminus of EGFP. Purified chloroplasts were incubated with or without 10 µg of thermolysin on ice for 30 min prior to reisolation of the chloroplasts on a Percoll gradient. The reisolated chloroplasts were subjected to immunoblot analysis using an anti-EGFP antibody and an antibody against Rubisco large subunit (RbcL) as loading controls.
(B) Alkaline extraction. Chloroplasts were isolated from transfected protoplasts that expressed the CTs of Toc159 or Toc132, the entire Toc34 protein fused to the C terminus of EGFP, or OEP7 fused to the N terminus of EGFP. Purified chloroplasts were incubated in carbonate buffer, pH 11.5, or HEPES buffer, pH 7.3, on ice for 10 min. The precipitated membrane pellets were subjected to immunoblot analysis using an anti-EGFP antibody and an antibody against Toc34 as loading controls.
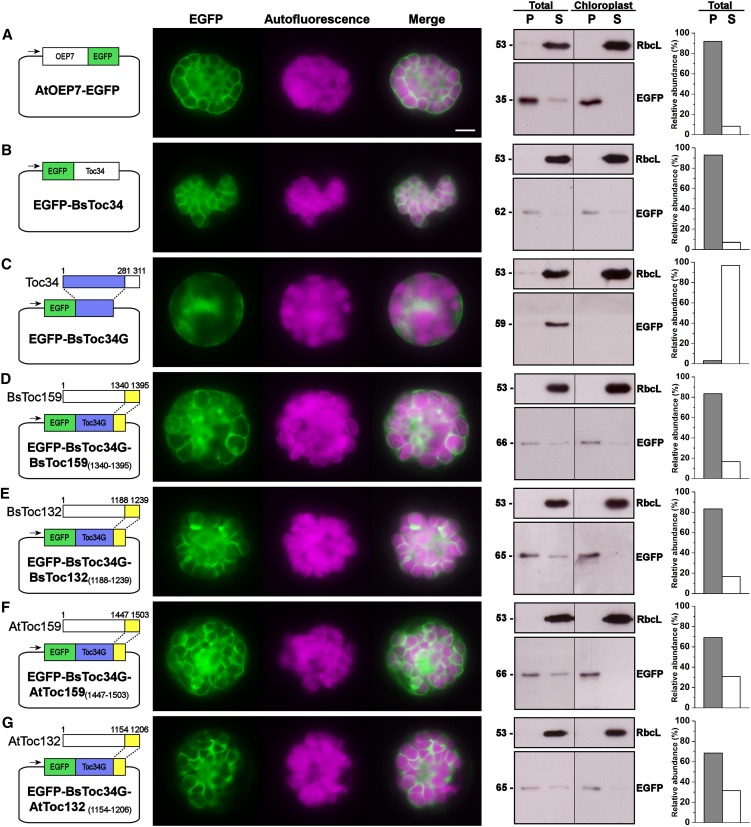
Toc159 CTs Retargeted the Toc34 Mutant to Chloroplasts
As discussed earlier, thermolysin treatment data did not implicate the protein import machinery in the translocation of Toc159 CTs across the outer chloroplast membrane (Figure 5A), and alkaline extraction data further suggested the absence of membrane anchor signals in Toc159 CTs (Figure 5B). It is therefore plausible that the binding of Toc159 CTs to the chloroplast envelope is reversible, as evidenced by the considerable amount of fusion proteins in the soluble fractions (Figures 4F and 4G). In fact, in the case of cTP targeting, the initial docking of preproteins on the chloroplast envelope is also reversible (Perry and Keegstra, 1994; Kouranov and Schnell, 1997). Thus, we further investigated whether the Toc159 CTs can target passenger proteins known to reside at the chloroplast envelope. Previous truncation studies revealed that the C-terminal hydrophilic tail of Toc34 is essential for chloroplast envelope association (Chen and Schnell, 1997; Li and Chen, 1997; Dhanoa et al., 2010). We first confirmed that, while the EGFP-tagged full-length proteins of At OEP7 and Bs Toc34 were exclusively localized to the chloroplast envelope (Figures 6A and 6B), a C-terminal truncation of the hydrophilic tail from Toc34 resulted in a diffuse signal throughout the cytosol (Figure 6C). Next, using this Toc34 mutant, we showed that the CTs of both Bs Toc159 and Toc132 retargeted the mutant Toc34 protein to the chloroplast envelope, forming ring-like fluorescent patterns, with the majority of the fusion proteins found in the membrane fractions of transfected protoplasts and isolated chloroplasts, as indicated by immunoblot analysis (Figures 6D and 6E). Similar retargeting of the Toc34 mutant to the chloroplast envelope was also achieved by fusion with the CTs of At Toc159 or At Toc132 (Figures 6F and 6G), suggesting that the chloroplast envelope-targeting signals at the Toc159 CTs are universal, at least in higher plant species. Based on these observations, we conclude that Toc159 CTs can complement the sorting signal of Toc34 CT for chloroplast envelope targeting.
Figure 6.
Retargeting of C-Terminally Truncated Toc34 to the Chloroplast Envelope by the Toc159 CTs.
Details are the same as in Figure 4.
(A) Full-length Arabidopsis OEP7 fused to the N terminus of EGFP.
(B) Full-length B. sinuspersici Toc34 fused to the C terminus of EGFP.
(C) C-terminally truncated B. sinuspersici Toc34 fused to the C terminus of EGFP (EGFP-Toc34G).
(D) The CT of B. sinuspersici Toc159 fused to the C terminus of EGFP-Toc34G.
(E) The CT of B. sinuspersici Toc132 fused to the C terminus of EGFP-Toc34G.
(F) The CT of Arabidopsis Toc159 fused to the C terminus of EGFP-Toc34G.
(G) The CT of Arabidopsis Toc132 fused to the C terminus of EGFP-Toc34G.
[See online article for color version of this figure.]
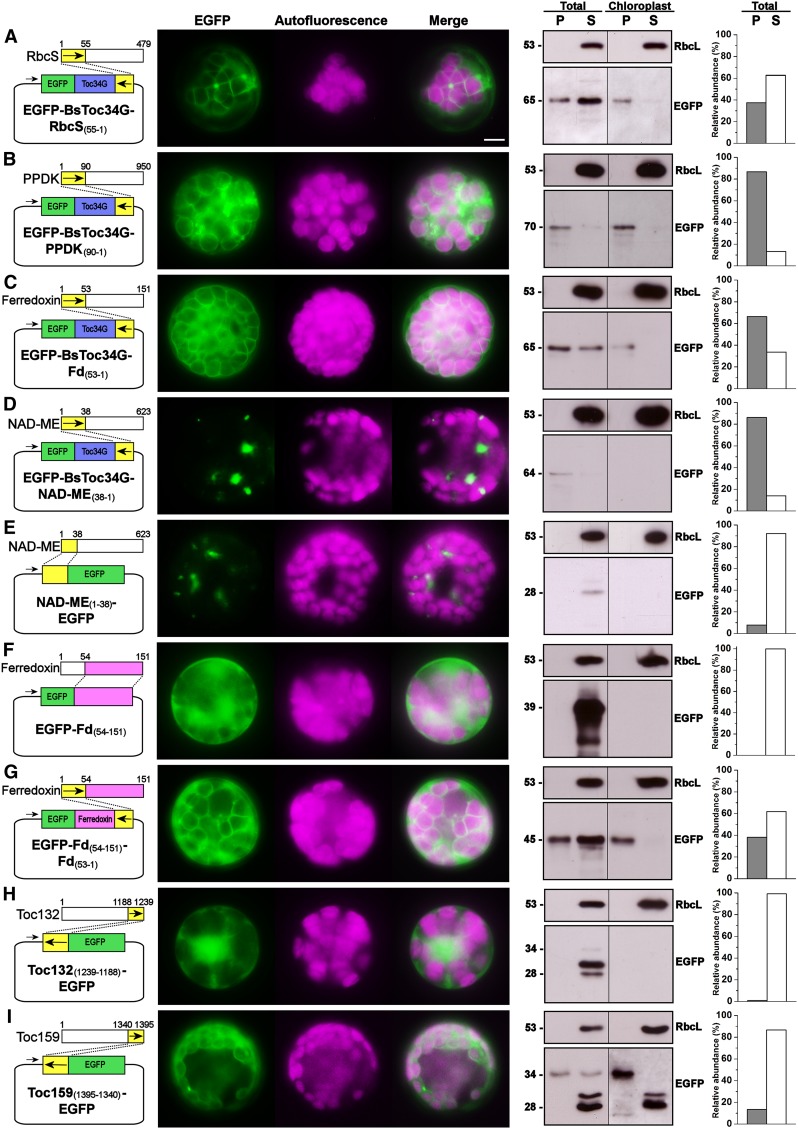
Toc159 CTs Share Similar Functionality with Other cTPs
Given the results of the bioinformatics analyses and EGFP fusion experiments, we hypothesized that the CTs of Toc159 receptors and cTPs of preproteins are in part functionally interchangeable. To test this hypothesis, we first produced reciprocal fusion constructs encoding the reverse TP sequence of Rubisco small subunit fused to the C terminus of the EGFP-tagged Toc34 truncation mutant. In spite of the lower targeting efficiency as judged by the intensities of immunoreactive bands, the association of this fusion protein with chloroplasts was evident in fluorescence microscopy and immunoblot analysis (Figure 7A). More convincingly, the Toc34 mutant was efficiently retargeted to the chloroplast envelope using the reverse TP sequences of pyruvate orthophosphate dikinase (Figure 7B) or ferredoxin (Figure 7C). Interestingly, the fusion of the Toc34 mutant with the reverse mitochondrial presequence of NAD-malic enzyme at the C terminus resulted in some punctate signals (Figure 7D), which appeared to resemble mitochondria (Figure 7E). In addition to retargeting the Toc34 mutant, the EGFP fusion with the mature protein of ferredoxin, which otherwise appeared as a diffuse cytosolic signal (Figure 7F), was partially directed to the chloroplast envelope using its own TP sequence reversely fused at the C terminus (Figure 7G).
Figure 7.
The Targeting of EGFP Fusion Proteins to the Chloroplast Envelope by the Reverse Sequences of Chloroplast Transit Peptides.
Details are the same as in Figure 4.
(A) The reverse sequence of Arabidopsis Rubisco small-subunit transit peptide fused to the C terminus of EGFP-Toc34G.
(B) The reverse sequence of B. sinuspersici pyruvate orthophosphate dikinase transit peptide fused to the C terminus of EGFP-Toc34G.
(C) The reverse sequence of B. sinuspersici ferredoxin transit peptide fused to the C terminus of EGFP-Toc34G.
(D) The reverse sequence of B. sinuspersici NAD-malic enzyme mitochondrial presequence fused to the C terminus of EGFP-Toc34G.
(E) The mitochondrial presequence of NAD-malic enzyme fused to the N terminus of EGFP.
(F) The mature protein of B. sinuspersici ferredoxin fused to the C terminus of EGFP.
(G) The reverse sequence of B. sinuspersici ferredoxin transit peptide fused to the C terminus of the fusion protein as shown in (F).
(H) The reverse sequence of the CT of B. sinuspersici Toc132 fused to the C terminus of EGFP.
(I) The reverse sequence of the CT of B. sinuspersici Toc159 fused to the C terminus of EGFP.
[See online article for color version of this figure.]
Lastly, we also produced reciprocal fusion constructs encoding the reverse sequences of Bs Toc159 CT and Toc132 CT fused at the N terminus of EGFP. Although the reversal of Toc132 CT did not produce conclusive results due to the degradation of the CT tag for some unknown reason (Figure 7H), the N-terminal fusion of EGFP with the reverse sequence of Toc159 resulted in translocation of the passenger protein into the stroma and subsequent maturation of the protein by stromal processing peptidase cleavage (Figure 7I). This observation is consistent with the ChloroP prediction suggesting that the Bs Toc159 CT is a putative cTP and possesses a cleavage site of stromal process peptidase (Table 2). Taken together, we conclude that the function of Toc159 CTs is similar to that of characteristic cTPs.
DISCUSSION
The Conserved Chloroplast Preprotein Receptors in Single-Cell C4 Species
The evolution of eukaryotic cells hinges on a sophisticated sorting mechanism to distribute newly synthesized proteins to their respective subcellular destinations. In plant cells, the acquisition of an additional endosymbiotic organelle and the massive transfer of the genetic material to the host genomes added an extra level of complexity. While uncertainties and disagreements hold for the current chloroplast preprotein import models (Jarvis, 2008), the evolution of two distinct types of chloroplasts in terrestrial single-cell C4 species poses further challenges for a thorough understanding of chloroplast biogenesis and differentiation at the biochemical level. The emerging evidence of signaling between the plastid and nuclear genomes (Nott et al., 2006) implies that the differential protein accumulation in dimorphic chloroplasts is not merely a matter of concern during organelle differentiation, but rather a highly dynamic process throughout the lifetime of the plant. This notion implicates the existence of a differential protein import mechanism that accounts for the different subsets of proteins in the dimorphic chloroplasts. As a first step, we sought to identify and characterize the protein import machinery at the chloroplast envelope in the single-cell C4 model B. sinuspersici, focusing on how the primary preprotein receptor Toc159 is targeted to the chloroplast.
We first cloned two full-length Toc159-like sequences from B. sinuspersici. Although their orthologies cannot be proven by computational methods (e.g., reciprocal BLAST search) with the lack of genome sequence information from B. sinuspersici or its close relatives, their homologies with Toc159 orthologs are strongly supported by the amino acid alignment, phylogenetic analysis, and conservation of the signature tripartite structures of Toc159 (Figure 1; see Supplemental Figure 2 online). In addition to the highly conserved central G-domain and C-terminal M-domain, Bs Toc159 and Toc132 contain an N-terminal A-domain that is highly variable in length and composition and characteristically enriched in acidic residues (one per 4.5 and 4.8 residues, respectively; see sequences in Supplemental Figure 2 online). The precise function of this intrinsically unstructured A-domain is poorly understood, but it is presumably pertinent to conferring preprotein substrate specificity of the receptor (Richardson et al., 2009; Inoue et al., 2010). In fact, the protein expression profiles of Bs Toc159 and Toc132 implicate their different substrate specificities for photosynthetic and housekeeping proteins (Figure 2A), respectively, as deduced from the previous studies on Arabidopsis Toc159 orthologs (Bauer et al., 2000; Ivanova et al., 2004; Kubis et al., 2004; Smith et al., 2004). Comparatively, the unicellular green (Chlamydomonas reinhardtii) and red (Cyanidioschizon merolae) algae apparently do not have multiple substrate-specific import pathways, since their genomes only encode a single pair of Toc159 and Toc34, the former of which has an N terminus not enriched in acidic residues (one per 8.4 and 7.1 residues, respectively; Kalanon and McFadden, 2008). Thus, the evolution of B. sinuspersici in the C4 lineage apparently has not altered the substrate-specific mechanism of preprotein recognition as elucidated in C3 species.
The Toc159 Targeting Pathway Awaited Discovery of a Sorting Signal
The mechanisms for targeting and membrane integration of the two homologous GTPase receptors (i.e., Toc159 and Toc34) remain an important facet of chloroplast preprotein import studies. In addition to immunogold localization of the endogenous receptors in B. sinuspersici leaves, transient expression of EGFP fusion proteins in B. sinuspersici and Arabidopsis leaf-derived protoplasts as well as particle-bombarded onion epidermal cells demonstrated the strict association of Toc34 with the plastids, while Toc159 appeared to be a soluble receptor, which partitioned between the interchloroplastic space and the envelope in photosynthetic cells (Figures 2B to 2F). These unique patterns of subcellular localization point to discrete sorting pathways for Toc159 and Toc34, respectively.
Similarly to other tail-anchored proteins, the targeting of Toc34 to the chloroplast envelope is dependent on its transmembrane domain plus the flanking C terminus (Chen and Schnell, 1997; Li and Chen, 1997; Qbadou et al., 2003; Dhanoa et al., 2010). Li and Chen (1997) demonstrated that this C-terminal region is sufficient for targeting and efficient insertion of a passenger protein, regardless of the location of fusion at the N or C terminus. A recent study, on the other hand, showed that GFP fusion with the transmembrane domain plus the N- and C-terminal flanking residues was localized to nonplastid punctate structures and that the correct envelope targeting required the G-domain (Dhanoa et al., 2010). The importance of the G-domain in targeting is consistent with independent studies showing that binding of GTP or GDP is required for membrane integration of Toc34 (Chen and Schnell, 1997; Qbadou et al., 2003). As expected, the assembly of its GTPase homolog, Toc159, into the Toc complex at the chloroplast envelope also depends on the G-domain (Bauer et al., 2002; Smith et al., 2002a). The efficient integration of Toc159 into the chloroplast envelope also depends on guanine nucleotide binding and is mediated by its homotypic interaction with the Toc34 G-domain (Smith et al., 2002a). While both Toc GTPases are targeted to the chloroplast envelope with the assistance of their G-domains, the essential sorting information embedded in the transmembrane domain plus the hydrophilic tail of Toc34 is missing in the sequence of Toc159 (see Supplemental Figure 2 online). However, one should expect to find equivalent signals accompanying the Toc159 G-domain, which otherwise would make it indistinguishable from diverse members of the GTPase superfamily for protein targeting to other subcellular compartments (e.g., nucleus, endoplasmic reticulum, and endomembrane system).
Since the A-domain is fully dispensable for the targeting of Toc159 (Bauer et al., 2002; Smith et al., 2002a), the putative sorting signal of Toc159 that accompanies the G-domain is expected to reside within the M-domain. In fact, earlier studies have documented that the C-terminal region or the entire M-domain of Toc159, to a certain extent, bound to the chloroplast surface in vitro and in vivo, whereas C-terminal truncations of Toc159 severely reduced their binding (Muckel and Soll, 1996; Bauer et al., 2002; Smith et al., 2002a). Wallas et al. (2003) demonstrated that the binding of At Toc159 to the chloroplast envelope was more efficient in the presence of both G- and M-domains. Interestingly, the albino phenotype of the Arabidopsis Toc159 knockout mutant (ppi2) was rescued with almost fully developed chloroplasts by overexpression of the Toc159 M-domain, which was found exclusively at the chloroplast envelope (Lee et al., 2003). Thus, numerous lines of evidence support our finding that the C-terminal end contributes to the subcellular localization of Toc159.
The C Terminus of Toc159 Is a cTP-Like Signal
Accordingly, we sought to unravel the chloroplast-sorting signal of Toc159. In view of the well-studied targeting pathways for chloroplast outer membrane proteins (Hofmann and Theg, 2005; Bölter and Soll, 2011), the sorting signals can be broadly classified into two types. Proteins with hydrophobic membrane-spanning regions (i.e., signal- and tail-anchored proteins) rely on their transmembrane α-helices and the flanking charged residues as the envelope-targeting signals (Bölter and Soll, 2011). By contrast, the preprotein translocation channel Toc75 is a β-barrel protein that is sorted to the chloroplast envelope in the presence of a cTP, which is similar to the signals generally found in other chloroplast preproteins (Tranel et al., 1995; Tranel and Keegstra, 1996). A similar cTP signal was also predicted at the N terminus of a Toc75 paralog, At OEP80 (Eckart et al., 2002), although this putative signal was later shown to be noncleavable and its physiological relevance is not known (Inoue and Potter, 2004). Given the absence of a transmembrane helix in the Toc159 sequences (see Supplemental Figure 2 online), we asked whether Toc159 has a cTP sorting signal. No N-terminal cTP was predicted, which is in agreement with the irrelevance of the N terminus in the targeting of Toc159 (Bauer et al., 2002; Smith et al., 2002a). On the other hand, bioinformatics analyses suggested that the C-terminal region of Toc159 exhibits properties of cTPs (Figure 3, Tables 1 and 2).
cTPs are highly divergent in primary structures with no consensus sequence identified but exhibit similar physicochemical and structural properties (Bruce, 2001), which were also predicted for the Toc159 CTs, in agreement with their specific association with the chloroplast envelope. First, we found that Toc159 CTs are rich in hydroxylated residues and, in contrast with the N termini, lack acidic residues, which leads to basic C-terminal ends (Figure 3A, Table 1). The basicity of cTPs has been proposed to mediate their initial ionic interaction with the anionic phospholipids at the outer chloroplast membrane, whereas the hydroxylated residues form hydrogen bonds with the Gal headgroups of the nonbilayer galactolipids, particularly the chloroplast-specific monogalactosyldiacylglycerol (MGDG) (Pinnaduwage and Bruce, 1996; Bruce, 2000). However, some studies did not support this hypothesis and suggested that MGDG may not be as important as previously indicated (Inoue et al., 2001; Schleiff et al., 2003). Furthermore, a recent molecular genetics study of Arabidopsis mutants (mgd1-1) with reduced levels of MGDG showed no significant difference in protein targeting and import efficiency between wild-type and mutant chloroplasts (Aronsson et al., 2008). Thus, the lipid-interacting activity of cTPs that forms the basis of the initial association of preproteins specifically with the chloroplast surface may involve other lipid constituents or some unknown factors. The cTP–lipid interaction can also be predicted from a structural perspective. cTPs predominantly form random coils in the cytosol but, in the membrane-mimicking environments, adopted α-helical structures that exhibited various degrees of amphipathicity (Bruce, 2001). The formation of amphipathic α-helices is an important feature of proteins at the membrane interface for seeking a surface between hydrophobic and hydrophilic phases. The Eisenberg hydrophobic moment plot exemplified one of the earliest applications of bioinformatics to predict such surface-seeking proteins (Eisenberg et al., 1984). We constructed an equivalent plot to illustrate the similar amphipathicity of the predicted α-helices from Toc159 CTs and the demonstrated cTPs (see Supplemental Figure 4 and Supplemental References 1 online). The lower values of their hydrophobic moments compared with that of surface proteins suggest weak lipid affinities of these sorting signals to the chloroplast surface (see Supplemental Figure 4 and Supplemental Table 2 online), which is plausible due to the temporary and reversible nature of the association. The bioinformatics analyses therefore predicted the tendency of the Toc159 CTs to reversibly associate with the chloroplast surface in a manner resembling the cTP–lipid interaction. The subsequent insertion of Toc159 CTs into the outer membrane, analogous to the irreversible insertion of cTPs into the protein channel for translocation, might require both Toc34 and Toc75. This notion is supported by previous findings that the Toc159 M-domain associated with Toc34 and that both Toc34 and Toc75 were essential for the in vitro insertion of Arabidopsis Toc159 into the reconstituted proteoliposomes (Wallas et al., 2003).
The cTP-like function of Toc159 CTs raises an interesting question regarding the evolutionary origin of this novel chloroplast envelope-routing signal, which was found at the C terminus. While Toc159 does not have proven homologs of prokaryotic or eukaryotic ancestry, hydrophobic cluster analysis suggested that both N- and C-terminal domains originated from a successive duplication of the central G-domain and subsequent evolution toward their distinctive functions (Hernández Torres et al., 2007). On the other hand, it has been proposed that cTPs were originally derived from the ancestral cyanobacterial genes encoding the putative secretion substrates of the voltage-gated channel SynToc75 by exon shuffling (McFadden, 1999). Thus, we speculate that the presence of a cTP-like signal in Toc159 CTs represents an example of convergent evolution, when a photosynthetic cell was under selective pressure to target the gene products to the chloroplast envelope. Indeed, despite the independent endosymbiotic evolution of mitochondria and chloroplasts, their targeting of preproteins also shares considerable similarities, from the evolutionary mechanisms and properties of the sorting signals to the nature of translocon machineries (Bruce, 2000; Schleiff and Becker, 2011). Since the neural network–based ChloroP predicted a putative cTP in Toc159 (Table 2) but not in other polypeptides retrieved from the Plant Proteomics Database (http://ppdb.tc.cornell.edu), we believe that this C-terminal signal represents a unique product from the evolution of the receptor under special circumstances that (1) the targeting could be reversible in nature enabling the receptor to partition between the cytosol and the chloroplast envelope; (2) the targeting signal must confer an Nout-Cin topology of the receptor; and (3) the targeting signal could not reside at the N terminus due to its role in conferring the receptor selectivity to different classes of preproteins (Agne and Kessler, 2010).
The cTP-Like Sorting Signal Meets the Needs of Toc159
In this study, we used a number of EGFP fusion constructs to reveal the properties of the Toc159 CTs, which appear to fulfill the sorting signal criteria for reversible targeting of the receptor to the chloroplast envelope in such an orientation that permits preprotein recognition on the cytosolic side. First, the constructs confirmed the bioinformatics analyses predicting that the Toc159 CTs exhibit properties of cTPs. We showed that C-terminal truncations abolished the chloroplast localization of Toc159 receptors, while Toc159 CTs alone sorted EGFP to the chloroplast envelope (Figure 4). In addition, the Toc159 CTs redirected the C-terminally truncated Toc34 mutant from the cytosol to the chloroplast (Figure 6). We also demonstrated that the Toc159 CTs and other cTPs were in part functionally interchangeable (Figure 7). Interestingly, when fused at the N terminus in the reverse orientation, the Toc159 CTs functioned as a cleavable cTP that mediated translocation of EGFP into the chloroplast stroma (Figure 7I). The copurified protein in the stroma extract of the isolated chloroplasts was processed to the molecular mass of EGFP (28 kD; Figure 7I), confirming the ChloroP prediction of a cleavage site for the stromal processing peptidase (Table 2). The fusion of Toc159 CTs at the C terminus of EGFP did not produce any difference between the theoretical and apparent molecular masses (Figures 4F and 4G), which is plausible since the fusion proteins in this orientation did not gain access to the stroma (Figure 5A). The assessment of whether the Toc159 CTs are processed at the chloroplast envelope requires insertion of the receptor to the outer membrane, which happens solely if both the G- and M-domains are present (Smith et al., 2002a). As a result, the subtle change in molecular masses of such fusion proteins after signal cleavage is not easily distinguishable by gel electrophoresis. The possibility of Toc159 C-terminal cleavage, however, could be addressed using other approaches, such as mass spectrometry.
In addition, we showed that a fusion of EGFP to the C terminus of Toc159 CTs was susceptible to thermolysin treatment of chloroplasts isolated from the transfected protoplasts, suggesting that the fusion proteins were exposed to the chloroplast exterior (Figure 5A). This scenario is analogous to the A- and G-domains of endogenous Toc159 being projected to the cytosolic side of the chloroplast envelope. Thus, the sorting signal at the C terminus contributes to conferring the favorable Nout-Cin topology of the receptor for preprotein recognition in the cytosol. Alkaline extraction of the isolated chloroplasts after expression of the same EGFP constructs further confirmed that the fusion proteins were not integrated into the chloroplast envelope (Figure 5B). This observation is consistent with the bioinformatics analyses predicting that the Toc159 CTs are not membrane anchors, but rather function as cTP-like sorting signals that interact reversibly with the lipid components at the chloroplast surface. This reversible interaction might account for the abundance of fusion proteins to remain in the cytosol and the occasional contamination of the stromal extract due to the detachment of the fusion proteins from the envelope during the chloroplast lysis treatment (Figure 4). In fact, Smith et al. (2002a) reported that initial docking of Arabidopsis Toc159 at the chloroplast envelope does not require bound nucleotide and the association is not stable until successful insertion occurs in a guanine nucleotide-dependent manner. Thus, the initial contacts of cTPs with the chloroplast envelope are reversible and energy independent, while subsequent insertion of the preproteins into the Toc complex is irreversible and dependent on ATP and GTP (Perry and Keegstra, 1994; Kouranov and Schnell, 1997).
Based on the affinity to the chloroplast envelope, the reversibility of the association, and the orientation of targeting, we conclude that Toc159 CT functions as a sorting signal of the receptor.
A Dual Domain–Mediated Targeting Pathway for Toc159
In our effort to identify a novel C-terminal cTP-like sorting signal, we unraveled the fundamental components that account for the subcellular localization of Toc159. Incorporating the C-terminal signaling component into the current targeting model for preprotein import into chloroplasts (Smith, 2006), we now propose a dual domain–mediated targeting pathway for Toc159. First, the C-terminal sorting signal targets the soluble preprotein-bound Toc159 receptor to the chloroplast envelope. Second, the C-terminal sorting signal mediates reversible docking on the chloroplast surface through interaction with the lipids and/or other components of the Toc complex (i.e., Toc34 and Toc75). Third, the homotypic interaction between the GTP-bound G-domains of Toc159 and Toc34 stimulates GTP hydrolysis, leading to the insertion of Toc159 into the outer membrane and initiation of preprotein translocation. Lastly, the G-domains of both Toc159 and Toc34 exchange GDP for GTP, leading to the subsequent dissociation of Toc159 receptors from the Toc complex. In the future, structural studies of the Toc159 M-domain will provide insight into its functions as a membrane anchor and a chloroplast-targeting signal. In vitro targeting assays also will be performed to further characterize the targeting pathway of Toc159, particularly in relation to the requirement for proteinaceous components, such as 14-3-3 proteins and molecular chaperones.
METHODS
Identification of Toc159 and Toc34 Homologs from Bienertia sinuspersici
Primers were designed based on the conserved regions in the alignment of open reading frames of Toc homologs from different plant species, which were initially retrieved by BLASTp search using the Toc159 and Toc34 amino acid sequences from pea (Pisum sativum). All primer sequences used for cDNA cloning can be found in the Supplemental Table 3 online. Partial cDNA fragments were amplified by RT-PCR from mature leaf cDNA of B. sinuspersici using the Protoscript II RT-PCR kit (New England Biolabs) according to the manufacturer’s instructions. The full-length cDNA clones were obtained by 5′ and 3′ rapid amplification of cDNA ends (RACE) using the First Choice RLM-RACE kit (Ambion) following the manufacturer’s protocol. A 280-bp fragment and a 1.4-kb fragment of Bs Toc159 were first identified after PCR amplification with Toc159F1/R1 and Toc159F3/R3, respectively. Toc159F5 and Toc159F6 were designed from the 280-bp sequence for 3′RACE by nested PCR. Toc159R4 was designed from the 1.4-kb sequence for PCR with Toc159-F7. Toc159R7 was designed from the resulting 1.2-kb sequence for 5′RACE by nested PCR. Full-length Bs Toc159 was amplified with Toc159F13/R11. An 825-bp fragment of Bs Toc132 was first identified after PCR amplification with Toc132F2D/R2. Toc132F5 was designed from the 825-bp sequence for 3′RACE. Toc132R4 was designed from the 825-bp sequence for PCR with Toc132F6. Toc132R5 was designed from the resulting sequence for 5′RACE. Full-length Bs Toc132 was amplified with Toc132F8/R10. A fragment of Bs Toc34 was first identified after PCR amplification with Toc34F3/R1. From the sequence, Toc34F4 and Toc34F5 were designed for 3′RACE by nested PCR, and Toc34R2 and Toc34R3 were designed for 5′RACE by nested PCR. Full-length Bs Toc34 was amplified with Toc34F8/R6.
EGFP Fusion Constructs
The EGFP fusion constructs were made by subcloning specific DNA fragments of interest into the pSAT6-35S:EGFP-N1 or -C1 vectors, which were described previously (Chung et al., 2005). Details of the primers and subcloning vector used for the generation of each EGFP fusion construct can be found in Supplemental Table 4 online. All reverse DNA sequences were designed by rearranging the codon triplets and assembled from synthetic oligonucleotides by assembly PCR reactions as described previously (Rydzanicz et al., 2005).
Antibody Production
To raise isoform-specific antibodies against the Toc159 homologs from B. sinuspersici, the highly variable N-terminal regions of Bs Toc159(1-629) and Toc132(1-482) were expressed in the BL21-Codon-Plus (DE3)-RIPL strain of Escherichia coli (Novagen). The soluble hexahistidine-tagged proteins were purified using Profinity IMAC resins (Bio-Rad) for subsequent rabbit immunization. Pure immunoglobulins were affinity purified from the antisera by reversible binding to the same antigenic recombinant proteins coupled to the activated immunoaffinity support Affi-Gel 15 (Bio-Rad). The bound immunoglobulins were eluted with 100 mM Gly-HCl, pH 2.4, and 150 mM NaCl and concentrated with a buffer exchange to PBS using Nanosep 10K ultrafiltration columns (Pall Life Sciences). A polyclonal antibody was raised against the entire EGFP protein and affinity purified by the same procedure.
Immunoblot Analysis
B. sinuspersici leaves were homogenized in extraction buffer (100 mM Tris-HCl, pH 8, 150 mM NaCl, 1% [w/v] SDS, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1% [v/v] protease inhibitor cocktail [Sigma-Aldrich]). The protein concentrations of the soluble fractions after centrifugation (20,000g, 4°C, 15 min) were determined using the Bicinchoninic Acid Protein Assay kit (Pierce). The B. sinuspersici leaf homogenates and the subfractionated samples of Arabidopsis and B. sinuspersici were separated by SDS-PAGE. The resolved proteins were transferred to polyvinylidene difluoride membranes and probed with the following primary antibodies: rabbit anti-BsToc159 (1:32,000), rabbit anti-BsToc132 (1:4,000), rabbit anti-EGFP (1:4,000), rabbit antipyruvate orthophosphate dikinase (1:4000; courtesy of Chris Chastain from Minnesota State University, Moorhead), rabbit antiphosphoenolpyruvate carboxylase (1:10,000; Chemicon), rabbit anticytochrome-f (1:1000; Agrisera), rabbit anti-Rubisco large subunit (1:10,000; Agrisera), or mouse anti-actin (1:3000; MP Biomedicals), followed by the horseradish peroxidase–conjugated anti-rabbit (Sigma-Aldrich) or anti-mouse (Sigma-Aldrich) secondary antibodies. Signals were visualized by incubating the probed membrane with Amersham ECL-Plus solution (for pyruvate orthophosphate dikinase, phosphoenolpyruvate carboxylase, cytochrome-f, and Rubisco large subunit; GE Healthcare) or Amersham ECL-Advance Solution (for Toc159, Toc132, EGFP, and actin; GE Healthcare). Luminescence was detected by exposing the membrane to Amersham Hyperfilm ECL films (GE Healthcare) and developed using a CP1000 Agfa photodeveloper (AGFA). The film was scanned and processed using Adobe Photoshop CS (Adobe Systems). The intensities of immunoreactive bands were densitometrically quantified with the gel-analyzer function of Image J software v.1.46 (National Institutes of Health).
Protoplast Isolation and Transfection
Chlorenchyma protoplasts were isolated from mature leaves of B. sinuspersici and transfected with EGFP fusion constructs using the polyethylene glycol–mediated method as described previously (Lung et al., 2011). The transfected protoplasts were examined using an Olympus FV 1000 confocal laser scanning microscope. Protoplast isolation from Arabidopsis leaves and polyethylene glycol–mediated transfection of the isolated protoplasts were performed according to the protocol provided by Yoo et al. (2007). To allow sufficient materials for subsequent subfractionation and immunoblot analysis, the standard procedures were scaled up for transfection of 160,000 protoplasts with 40 µg of plasmid DNA. The transfected protoplasts were cultured overnight in a 50-mm Petri plate with 4 mL of WI solution (4 mM MES-KOH, pH 5.7, 500 mM mannitol, and 20 mM KCl) at 23°C and a light intensity of 30 µmol m−2 s−1 and examined using a Zeiss Axio Imager D1 microscope equipped with a Zeiss AxioCam MRm camera. All images were processed and composed using Adobe Photoshop CS (Adobe Systems). Representative images were presented after similar results were obtained from at least three independent experiments.
Chloroplast Isolation from Isolated Protoplasts
The chloroplast isolation procedures were modified from Smith et al. (2002b). The isolated protoplasts of Arabidopsis and B. sinuspersici were pelleted by centrifugation (300g, 2 min) in an equal volume of W5 buffer (2 mM MES-KOH, pH 5.7, 154 mM NaCl, 125 mM CaCl2, and 5 mM KCl) and resuspended in 300 μL of HEPES-sorbitol buffer (50 mM HEPES-KOH, pH 7.3, and 330 mM sorbitol). The resuspended protoplasts were disrupted by passage through a layer of 10-µm nylon mesh in a 1-mL syringe. Intact chloroplasts were isolated from the ruptured protoplasts on a 40 to 85% Percoll gradient and further subfractionated into the membrane and stromal fractions as described previously (Smith et al., 2002b). Similarly, the total ruptured protoplasts were fractionated into insoluble and soluble fractions using the same procedures. The membrane pellets and acetone-precipitated soluble fractions were solubilized in solubilization buffer (50 mM Tris-HCl, pH 8, 5 mM EDTA, and 1% [w/v] SDS), and the protein concentrations were determined using the Bicinchoninic Acid Protein Assay kit.
Immunogold Electron Microscopy
Mature leaves of B. sinuspersici were fixed at 4°C overnight in 2% (v/v) paraformaldehyde and 1.25% (v/v) glutaraldehyde in 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) buffer, pH 7.2, and 300 mM sorbitol. The specimens were dehydrated with a graded ethanol series and embedded in London Resin White acrylic resin (Electron Microscopy Sciences). Ultrathin sections (60-nm thick) were mounted on 150-mesh nickel grids and blocked with 1% (w/v) BSA in Tris-buffered saline containing 0.1% (v/v) Tween 20 (TBST-BSA) at room temperature for 1 h. The sections were incubated with diluted anti-BsToc159 (1:150), anti-BsToc132 (1:20), or anti-Toc34 (1:150) antibodies in TBST-BSA at 4°C for 16 h. Negative controls were set up with no primary antibody. The probed sections were washed in TBST-BSA and incubated with secondary anti-rabbit antibodies (1:100) conjugated to 10-nm gold particles (Sigma-Aldrich) in TBST-BSA at room temperature for 1 h. After washing, the sections were stained with uranyl acetate and lead citrate and observed under a Philips CM-10 transmission electron microscope operated at 60 kV. Representative images were presented after similar results were obtained from at least three independent experiments. The relative abundance of gold particles representing envelope-associated and cytosolic signals in chlorenchyma cells was manually determined in 1.5 × 2-µm2 areas of multiple electron micrographs from three individual blocks of specimens at a magnification of ×64,000.
Biolistic Transformation of Onion Epidermal Cells
Particle bombardment–mediated transient expression in onion epidermal cells was performed with plasmid DNA-coated tungsten particles (1 µm; Bio-Rad) using a Biolistic PDS-1000/He particle-delivery system (Bio-Rad) at a helium pressure of 1100 p.s.i., according to the manufacturer’s instructions. The bombarded samples were incubated in Petri dishes on moist filter paper at room temperature in the dark for 16 h and observed under epifluorescence microscopy.
Phylogenetic Analysis
The amino acid sequences of Toc159 homologs were retrieved using BLASTp search. The deduced amino acid sequences were aligned using the ClustalW algorithm. The alignment used for phylogenetic analysis is available as Supplemental Data Set 2 online. The phylogenetic tree was constructed by the neighbor-joining method using the MEGA v5.05 program (Tamura et al., 2011). The reliability for internal branch was assessed using the bootstrapping method (2000 bootstrap replicates).
Accession Numbers
Sequence data of the Toc159 homologs from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Arabidopsis lyrata 1 (XP_002874910), A. lyrata 2 (XP_002874913), A. lyrata 3 (EFH62320), A. thaliana 1 (AC002330), A. thaliana 2 (AC005825), A. thaliana 3 (AB022217), A. thaliana 4 (AF296825), Brassica rapa 1 (AC232397), B. rapa 2 (AC232399), B. sinuspersici 1 (JQ739199), B. sinuspersici 2 (JQ739200), Hordeum vulgare 1 (AK371279), H. vulgare 2 (AK367377), Medicago truncatula (AC147010), Oryza sativa 1 (AK102924), O. sativa 2 (AC092557), O. sativa 3 (AC020666), O. sativa 4 (AK241335), Physcomitrella patens 1 (XP_001770227), P. patens 2 (XP_001771331), P. patens 3 (AY496562), P. patens 4 (XP_001770228), P. sativum (AF262939), Ricinus communis 1 (XP_002531885), R. communis 2 (XP_002516922), R. communis 3 (XP_002528280), Sorghum bicolor 1 (XP_002440611), S. bicolor 2 (XP_002466147), S. bicolor 3 (XP_002464976), Solanum lycopersicum (EF647601), Selaginella moellendorffi (XP_002969262), Vitis vinifera 1 (AM485197), V. vinifera 2 (AM433085), and V. vinifera 3 (AM431847).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of Deduced Amino Acid Sequences from Different Putative or Reported Toc34.
Supplemental Figure 2. Amino Acid Sequence Alignment of Toc159 and Toc34 Homologs from B. sinuspersici and Arabidopsis.
Supplemental Figure 3. Recombinant Protein Expression of Bs Toc159 and Toc132 A-Domains.
Supplemental Figure 4. Hydrophobic Moment Plot Analysis of Toc159 C-Terminal α-Helices.
Supplemental Table 1. Pairwise Homology Comparison of B. sinuspersici and Arabidopsis Toc159 Isoforms.
Supplemental Table 2. Sequence Alignment and Hydrophobic Moment Analysis of the Predicted α-Helices at the C Termini of Toc159 Homologs.
Supplemental Table 3. List of Oligonucleotides Used for cDNA Cloning.
Supplemental Table 4. List of Oligonucleotides Used for the Construction of EGFP Fusion Constructs.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 1A.
Supplemental Data Set 2. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 2A online.
Supplemental References 1. References for the Supplemental Data.
Supplementary Material
Acknowledgments
This research was supported by Natural Sciences and Engineering Research Council of Canada grants (343747-07 and 348742-07) and the University of Waterloo Start-Up Fund to S.D.X.C. S.-C.L. received additional financial support from the Ontario Graduate Scholarship Program (Government of Ontario, Canada) and President’s Graduate Scholarship (University of Waterloo, Canada). We thank Tzvi Tzfira (University of Michigan) for providing the EGFP fusion vectors pSAT6-35S:EGFP-N1 and pSAT6-35S:EGFP-C1 and Chris Chastain (Minnesota State University Moorhead, MN) for providing the anti-maize PPDK antibody. We also thank Zhenyu Cheng for technical assistance with tandem mass spectrometry.
AUTHOR CONTRIBUTIONS
S.-C.L. and S.D.X.C. designed the experiments. S.-C.L. performed the experiments and analyzed the data. S.-C.L. and S.D.X.C. wrote the article.
Glossary
- cTP
chloroplastic transit peptide
- CT
C-terminal tail
- EGFP
enhanced green fluorescent protein
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
- MGDG
monogalactosyldiacylglycerol
- RACE
rapid amplification of cDNA ends
- TBST-BSA
1% (w/v) BSA in Tris-buffered saline containing 0.1% (v/v) Tween 20
References
- Agne B., Kessler F. (2009). Protein transport in organelles: The Toc complex way of preprotein import. FEBS J. 276: 1156–1165 [DOI] [PubMed] [Google Scholar]
- Agne B., Kessler F. (2010). Modifications at the A-domain of the chloroplast import receptor Toc159. Plant Signal. Behav. 5: 1513–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhani H., Barroca J., Koteeva N., Voznesenskaya E., Franceschi V., Edwards G., Ghaffari S.M., Ziegler H. (2005). Bienertia sinuspersici (Chenopodiaceae): A new species from Southwest Asia and discovery of a third terrestrial C4 plant without Kranz anatomy. Syst. Bot. 30: 290–301 [Google Scholar]
- Andrès C., Agne B., Kessler F. (2010). The TOC complex: Preprotein gateway to the chloroplast. Biochim. Biophys. Acta 1803: 715–723 [DOI] [PubMed] [Google Scholar]
- Aronsson H., Schöttler M.A., Kelly A.A., Sundqvist C., Dörmann P., Karim S., Jarvis P. (2008). Monogalactosyldiacylglycerol deficiency in Arabidopsis affects pigment composition in the prolamellar body and impairs thylakoid membrane energization and photoprotection in leaves. Plant Physiol. 148: 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J., Chen K., Hiltbunner A., Wehrli E., Eugster M., Schnell D., Kessler F. (2000). The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403: 203–207 [DOI] [PubMed] [Google Scholar]
- Bauer J., Hiltbrunner A., Weibel P., Vidi P.-A., Alvarez-Huerta M., Smith M.D., Schnell D.J., Kessler F. (2002). Essential role of the G-domain in targeting of the protein import receptor atToc159 to the chloroplast outer membrane. J. Cell Biol. 159: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölter B., May T., Soll J. (1998). A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide. FEBS Lett. 441: 59–62 [DOI] [PubMed] [Google Scholar]
- Bölter B., Soll J. (2011). Protein import into chloroplasts: Dealing with the (membrane) integration problem. ChemBioChem 12: 1655–1661 [DOI] [PubMed] [Google Scholar]
- Bölter B., Soll J., Hill K., Hemmler R., Wagner R. (1999). A rectifying ATP-regulated solute channel in the chloroplastic outer envelope from pea. EMBO J. 18: 5505–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce B.D. (2000). Chloroplast transit peptides: Structure, function and evolution. Trends Cell Biol. 10: 440–447 [DOI] [PubMed] [Google Scholar]
- Bruce B.D. (2001). The paradox of plastid transit peptides: Conservation of function despite divergence in primary structure. Biochim. Biophys. Acta 1541: 2–21 [DOI] [PubMed] [Google Scholar]
- Chen D., Schnell D.J. (1997). Insertion of the 34-kDa chloroplast protein import component, IAP34, into the chloroplast outer membrane is dependent on its intrinsic GTP-binding capacity. J. Biol. Chem. 272: 6614–6620 [DOI] [PubMed] [Google Scholar]
- Chen K., Chen X., Schnell D.J. (2000). Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts. Plant Physiol. 122: 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.M., Frankman E.L., Tzfira T. (2005). A versatile vector system for multiple gene expression in plants. Trends Plant Sci. 10: 357–361 [DOI] [PubMed] [Google Scholar]
- Chuong S.D.X., Franceschi V.R., Edwards G.E. (2006). The cytoskeleton maintains organelle partitioning required for single-cell C4 photosynthesis in Chenopodiaceae species. Plant Cell 18: 2207–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constan D., Patel R., Keegstra K., Jarvis P. (2004). An outer envelope membrane component of the plastid protein import apparatus plays an essential role in Arabidopsis. Plant J. 38: 93–106 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanoa P.K., Richardson L.G.L., Smith M.D., Gidda S.K., Henderson M.P.A., Andrews D.W., Mullen R.T. (2010). Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS ONE 5: e10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckart K., Eichacker L., Sohrt K., Schleiff E., Heins L., Soll J. (2002). A Toc75-like protein import channel is abundant in chloroplasts. EMBO Rep. 3: 557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. (1984). Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179: 125–142 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze T.A., Philippar K., Ilkavets I., Soll J., Wagner R. (2006). OEP37 is a new member of the chloroplast outer membrane ion channels. J. Biol. Chem. 281: 17989–17998 [DOI] [PubMed] [Google Scholar]
- Hernández Torres J., Maldonado M.A.A., Chomilier J. (2007). Tandem duplications of a degenerated GTP-binding domain at the origin of GTPase receptors Toc159 and thylakoidal SRP. Biochem. Biophys. Res. Commun. 364: 325–331 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Bauer J., Vidi P.-A., Infanger S., Weibel P., Hohwy M., Kessler F. (2001). Targeting of an abundant cytosolic form of the protein import receptor at Toc159 to the outer chloroplast membrane. J. Cell Biol. 154: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Muckel E., Heemeyer F., von Heijne G., Soll J. (1994). A receptor component of the chloroplast protein translocation machinery. Science 266: 1989–1992 [DOI] [PubMed] [Google Scholar]
- Hofmann N.R., Theg S.M. (2005). Protein- and energy-mediated targeting of chloroplast outer envelope membrane proteins. Plant J. 44: 917–927 [DOI] [PubMed] [Google Scholar]
- Huang W., Ling Q., Bédard J., Lilley K., Jarvis P. (2011). In vivo analyses of the roles of essential Omp85-related proteins in the chloroplast outer envelope membrane. Plant Physiol. 157: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Rounds C., Schnell D.J. (2010). The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. Plant Cell 22: 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Demel R., de Kruijff B., Keegstra K. (2001). The N-terminal portion of the preToc75 transit peptide interacts with membrane lipids and inhibits binding and import of precursor proteins into isolated chloroplasts. Eur. J. Biochem. 268: 4036–4043 [DOI] [PubMed] [Google Scholar]
- Inoue K., Potter D. (2004). The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. Plant J. 39: 354–365 [DOI] [PubMed] [Google Scholar]
- Ivanova Y., Smith M.D., Chen K., Schnell D.J. (2004). Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol. Biol. Cell 15: 3379–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008). Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Jarvis P., Chen L.J., Li H., Peto C.A., Fankhauser C., Chory J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282: 100–103 [DOI] [PubMed] [Google Scholar]
- Jones D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Kalanon M., McFadden G.I. (2008). The chloroplast protein translocation complexes of Chlamydomonas reinhardtii: A bioinformatic comparison of Toc and Tic components in plants, green algae and red algae. Genetics 179: 95–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F., Blobel G., Patel H.A., Schnell D.J. (1994). Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 266: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Kessler F., Schnell D.J. (2004). Chloroplast protein import: Solve the GTPase riddle for entry. Trends Cell Biol. 14: 334–338 [DOI] [PubMed] [Google Scholar]
- Kessler F., Schnell D.J. (2006). The function and diversity of plastid protein import pathways: A multilane GTPase highway into plastids. Traffic 7: 248–257 [DOI] [PubMed] [Google Scholar]
- Kouranov A., Schnell D.J. (1997). Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S., Baldwin A., Patel R., Razzaq A., Dupree P., Lilley K., Kurth J., Leister D., Jarvis P. (2003). The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 15: 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis S., Patel R., Combe J., Bédard J., Kovacheva S., Lilley K., Biehl A., Leister D., Ríos G., Koncz C., Jarvis P. (2004). Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell 16: 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Kim S.J., Lee Y.J., Jin J.B., Hwang I. (2003). The M domain of atToc159 plays an essential role in the import of proteins into chloroplasts and chloroplast biogenesis. J. Biol. Chem. 278: 36794–36805 [DOI] [PubMed] [Google Scholar]
- Li H., Chen L.J. (1997). A novel chloroplastic outer membrane-targeting signal that functions at both termini of passenger polypeptides. J. Biol. Chem. 272: 10968–10974 [PubMed] [Google Scholar]
- Li H.M., Chiu C.C. (2010). Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61: 157–180 [DOI] [PubMed] [Google Scholar]
- Lung S.C., Yanagisawa M., Chuong S.D.X. (2011). Protoplast isolation and transient gene expression in the single-cell C4 species, Bienertia sinuspersici. Plant Cell Rep. 30: 473–484 [DOI] [PubMed] [Google Scholar]
- Ma Y., Kouranov A., LaSala S., Schnell D.J. (1996). Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 134: 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G.I. (1999). Endosymbiosis and evolution of the plant cell. Curr. Opin. Plant Biol. 2: 513–519 [DOI] [PubMed] [Google Scholar]
- Muckel E., Soll J. (1996). A protein import receptor of chloroplasts is inserted into the outer envelope membrane by a novel pathway. J. Biol. Chem. 271: 23846–23852 [DOI] [PubMed] [Google Scholar]
- Nott A., Jung H.-S., Koussevitzky S., Chory J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Patron N.J., Waller R.F. (2007). Transit peptide diversity and divergence: A global analysis of plastid targeting signals. Bioessays 29: 1048–1058 [DOI] [PubMed] [Google Scholar]
- Perry S.E., Keegstra K. (1994). Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnaduwage P., Bruce B.D. (1996). In vitro interaction between a chloroplast transit peptide and chloroplast outer envelope lipids is sequence-specific and lipid class-dependent. J. Biol. Chem. 271: 32907–32915 [DOI] [PubMed] [Google Scholar]
- Pohlmeyer K., Soll J., Grimm R., Hill K., Wagner R. (1998). A high-conductance solute channel in the chloroplastic outer envelope from pea. Plant Cell 10: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qbadou S., Tien R., Soll J., Schleiff E. (2003). Membrane insertion of the chloroplast outer envelope protein, Toc34: Constrains for insertion and topology. J. Cell Sci. 116: 837–846 [DOI] [PubMed] [Google Scholar]
- Richardson L.G.L., Jelokhani-Niaraki M., Smith M.D. (2009). The acidic domains of the Toc159 chloroplast preprotein receptor family are intrinsically disordered protein domains. BMC Biochem. 10: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydzanicz R., Zhao X.S., Johnson P.E. (2005). Assembly PCR oligo maker: a tool for designing oligodeoxynucleotides for constructing long DNA molecules for RNA production. Nucleic Acids Res. 33(Web Server issue): W521–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Becker T. (2011). Common ground for protein translocation: Access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 12: 48–59 [DOI] [PubMed] [Google Scholar]
- Schleiff E., Soll J., Küchler M., Kühlbrandt W., Harrer R. (2003). Characterization of the translocon of the outer envelope of chloroplasts. J. Cell Biol. 160: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E., Tien R., Salomon M., Soll J. (2001). Lipid composition of outer leaflet of chloroplast outer envelope determines topology of OEP7. Mol. Biol. Cell 12: 4090–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell D.J., Kessler F., Blobel G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266: 1007–1012 [DOI] [PubMed] [Google Scholar]
- Smith M.D. (2006). Protein import into chloroplasts: An ever-evolving story. Can. J. Bot. 84: 531–542 [Google Scholar]
- Smith M.D., Fitzpatrick L.M., Keegstra K., Schnell D.J. (2002b). In vitro analysis of chloroplast protein import. In Current Protocols in Cell Biology, K.M. Yamada, ed (New York: John Wiley & Sons), pp. 11.16.11–11.16.21. [DOI] [PubMed]
- Smith M.D., Hiltbrunner A., Kessler F., Schnell D.J. (2002a). The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP. J. Cell Biol. 159: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.D., Rounds C.M., Wang F., Chen K., Afitlhile M., Schnell D.J. (2004). atToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J. Cell Biol. 165: 323–334. [DOI] [PMC free article] [PubMed]
- Sun Y.-J., Forouhar F., Li Hm H.M., Tu S.-L., Yeh Y.-H., Kao S., Shr H.-L., Chou C.-C., Chen C., Hsiao C.-D. (2002). Crystal structure of pea Toc34, a novel GTPase of the chloroplast protein translocon. Nat. Struct. Biol. 9: 95–100 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel P.J., Froehlich J., Goyal A., Keegstra K. (1995). A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 14: 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranel P.J., Keegstra K. (1996). A novel, bipartite transit peptide targets OEP75 to the outer membrane of the chloroplastic envelope. Plant Cell 8: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A., Jakob M., Klösgen R.B., Gutensohn M. (2005). At least two Toc34 protein import receptors with different specificities are also present in spinach chloroplasts. FEBS Lett. 579: 1343–1349 [DOI] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R.G. (1989). Domain structure of mitochondrial and chloroplast targeting peptides. Eur. J. Biochem. 180: 535–545 [DOI] [PubMed] [Google Scholar]
- Wallas T.R., Smith M.D., Sanchez-Nieto S., Schnell D.J. (2003). The roles of toc34 and toc75 in targeting the toc159 preprotein receptor to chloroplasts. J. Biol. Chem. 278: 44289–44297 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zybailov B., Rutschow H., Friso G., Rudella A., Emanuelsson O., Sun Q., van Wijk K.J. (2008). Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.