Methylketones are insecticidal compounds that accumulate in certain plants as defense against herbivorous pests. Methylketone biosynthesis is a two-step process within the plant, with the second step, decarboxylation of 3-keto acids, catalyzed by Methylketone Synthase1 (MKS1). This study focuses on understanding the enzymatic mechanism of MKS1, which differs from proteins of the same structural class.
Abstract
Specialized methylketone-containing metabolites accumulate in certain plants, in particular wild tomatoes in which they serve as toxic compounds against chewing insects. In Solanum habrochaites f. glabratum, methylketone biosynthesis occurs in the plastids of glandular trichomes and begins with intermediates of de novo fatty acid synthesis. These fatty-acyl intermediates are converted via sequential reactions catalyzed by Methylketone Synthase2 (MKS2) and MKS1 to produce the n-1 methylketone. We report crystal structures of S. habrochaites MKS1, an atypical member of the α/β-hydrolase superfamily. Sequence comparisons revealed the MKS1 catalytic triad, Ala-His-Asn, as divergent to the traditional α/β-hydrolase triad, Ser-His-Asp. Determination of the MKS1 structure points to a novel enzymatic mechanism dependent upon residues Thr-18 and His-243, confirmed by biochemical assays. Structural analysis further reveals a tunnel leading from the active site consisting mostly of hydrophobic residues, an environment well suited for fatty-acyl chain binding. We confirmed the importance of this substrate binding mode by substituting several amino acids leading to an alteration in the acyl-chain length preference of MKS1. Furthermore, we employ structure-guided mutagenesis and functional assays to demonstrate that MKS1, unlike enzymes from this hydrolase superfamily, is not an efficient hydrolase but instead catalyzes the decarboxylation of 3-keto acids.
INTRODUCTION
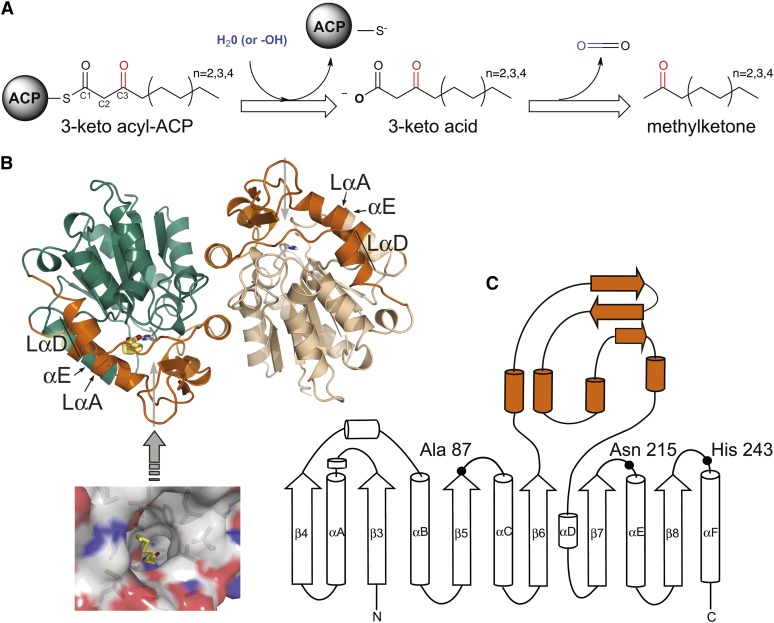
Methylketones, such as 2-undecanone (C11), 2-tridecanone (C13), and 2-pentadecanone (C15), are semivolatile, insecticidal compounds produced by a number of disparate plants, most notably wild species of tomato (Williams et al., 1980). Methylketones derive from intermediates of fatty acid synthesis (Fridman et al., 2005). Typically, fatty acid biosynthesis is terminated by cleavage of the thioester linking a fully reduced fatty acyl chain to the acyl carrier protein (ACP). For methylketone production, 3-keto acyl-ACP intermediates are diverted prior to reduction of the 3-carbonyl moiety and are instead converted via two sequential enzyme-catalyzed reactions: First, hydrolysis of the ACP-linked 3-keto acyl group, and, second, decarboxylation of the resultant 3-keto acid (Figure 1A).
Figure 1.
Methylketone Biosynthesis and MKS1 Three-Dimensional Structure.
(A) General reaction scheme proposed for the production of methylketones.
(B) Two monomers make up the crystallographic asymmetric unit: monomer A (green) and monomer B (tan). The lid domain is colored orange in each monomer, and His-243 is shown as sticks. Gray arrows point to the opening of the central tunnel. Bound fatty acid is colored yellow and is also displayed in a surface rendering of monomer A viewed from the direction of the gray arrow (inset).
(C) Topology diagram of the MKS1 secondary structural elements. To preserve the conventional numbering of the core α/β-hydrolase fold, MKS1 β-strands start with β-strand 3. The lid domain (orange) is inserted between β-strands 6 and 7.
Methylketone Synthase1 (MKS1) was first identified in Solanum habrochaites f. glabratum (formerly Lycopersicon hirsutum f. glabratum). In a comparison of trichome EST databases from methylketone producing and nonproducing tomato lines, a high abundance of MKS1 mRNA was seen only in the methylketone-producing line. Subsequently, in vitro assays confirmed that MKS1 uses 3-keto acyl-ACPs to biosynthesize methylketones, although with very low efficiency (Fridman et al., 2005). More recent genetic analyses revealed a second locus in S. habrochaites, MKS2, with expression levels also strongly correlated with methylketone production in the trichomes (Ben-Israel et al., 2009). Notably, MKS2 belongs to a family of hydrolase enzymes possessing the hot dog fold, which includes thioesterases such as 4-hydroxybenzoyl-CoA thioesterase and various putative fatty acyl hydrolases (Benning et al., 1998; Thoden et al., 2002; Dillon and Bateman, 2004). Recently, we obtained biochemical data showing that MKS2 catalyzes the first step of methylketone synthesis, cleavage of the thioester linkage between the 3-keto acyl group and the ACP, and that MKS1 catalyzes the subsequent decarboxylation of the resulting 3-keto acid (Yu et al., 2010).
These results indicate that the low-level thioesterase activity of MKS1 (Fridman et al., 2005) is not its physiological function. Instead, MKS1 is a 3-keto acid decarboxylase (Yu et al., 2010). Comparative sequence analyses place MKS1 within a diverse superfamily of enzymes possessing the α/β-hydrolase fold, which is characterized by a central eight-stranded β-sheet surrounded by α-helices. Hydrolases of this superfamily typically share only low levels of sequence identity and encompass diverging catalytic activities, including thioesterase, peptidase, general hydrolase, and lyase activities (Holmquist, 2000). Although functionally divergent, these α/β-hydrolases often retain crucial active-site residues forming a Ser-His-Asp catalytic triad for general acid-base and nucleophilic catalysis and an adjacent oxyanion hole for transition state stabilization (Nardini and Dijkstra, 1999). MKS1 clusters most closely with a subclass of α/β-hydrolases defined by its founding member, Hydroxy Nitrile Lyase (HNL) (Wagner et al., 1996; Gruber et al., 1999). To date, other characterized members of the HNL subfamily include Salicylic Acid Binding Protein 2 (SABP2; also referred to as methyl salicylate esterase) (Forouhar et al., 2005), Methyl Jasmonate Esterase (Stuhlfelder et al., 2004), and Polyneuridine Aldehyde Esterase (PNAE) (Dogru et al., 2000; Yang et al., 2009), and all possess the canonical Ser-His-Asp catalytic triad.
Unexpectedly, MKS1 lacks two key active-site residues presumed to be essential for hydrolase activity. In place of the commonly found nucleophilic Ser residue, MKS1 retains an Ala; and in place of the more typical Asp counterion for the His catalytic base, MKS1 possesses an Asn. Thus, the MKS1 active-site configuration appears to be evolutionarily unique, even after close examination of related paralogs from Solanum species; therefore, MKS1 must have emerged recently within the Solanum genus (Yu et al., 2010). Furthermore, the absence of an intact catalytic triad raises the possibility that residues from previously unanticipated areas of the protein sequence contribute to the MKS1 catalytic mechanism. Indeed, with other α/β-hydrolase family members, active-site reconfigurations are observed and include the repositioned acidic residues of the triads in the haloalkane dehalogenase from Sphingomonas paucimobilis and the thioesterase domain of human fatty acid synthase (Hynková et al., 1999; Chakravarty et al., 2004).
Given this apparent adaptation by an α/β-hydrolase fold for new catalytic machinery, we determined crystal structures of MKS1, in apo form and in complex with a number of substrates, products, and analogs. We obtained detailed three-dimensional snapshots of an active site with the capacity to (1) bind long-chain 3-keto acid substrates and (2) use an unanticipated mechanism for catalyzing decarboxylation of the 3-keto acid substrates. In tandem with these structural studies, we quantitatively demonstrated that MKS1 catalyzes the efficient decarboxylative release of methylketones from 3-keto acids with a narrow distribution of chain lengths.
RESULTS
MKS1 Three-Dimensional Structures
Three-dimensional structures of MKS1-WT (for wild-type) in its apo form and in complex with various small-molecule ligands (methyl-3-hydroxydodecanoate [M3D], 3-hydroxyoctanoate [3HO], and 2-tridecanone [2TD]) (see Supplemental Figure 1 online) were determined at resolutions ranging from 1.9 to 2.5 Å. The structures of two MKS1 variants containing single amino acid substitutions, H243A and T18A, were also obtained (see Supplemental Table 1 online).
As expected, the MKS1 structure generally conforms to the canonical α/β-hydrolase fold (Figures 1B and 1C), with the most substantial difference arising from an N-terminal truncation. As a result, the central core of MKS1 is a six-stranded parallel β-sheet surrounded by a peripheral set of α-helices. The two MKS1 molecules present in the asymmetric unit form an intimate dimer related by a noncrystallographic twofold axis (Figure 1B). The dimeric state of MKS1 is consistent with its behavior in solution as evidenced by its elution from a size-exclusion column. A dimeric state is also observed in related family members (Wagner et al., 1996; Forouhar et al., 2005; Yang et al., 2009). The two monomers within the MKS1 noncrystallographic dimer differ significantly in one region, encompassing part of helix αE and the preceding loop (Figure 1B). In monomer A, an additional helical turn in αE forces the preceding loop to flip outwards into an atypical open conformation. We are currently investigating whether these structural differences arise from crystal-packing influences or reflect the inherent and possibly functionally relevant dynamics of MKS1.
A large insertion between β-strands 6 and 7 of the MKS1 core domain (Figure 1C) constitutes a lid subdomain, a commonly occurring feature of α/β-hydrolases. This lid typically modulates substrate preference (Nardini and Dijkstra, 1999), and in MKS1, it forms part of a central tunnel lined by nonpolar residues. The hydrophobic nature of the tunnel and its 16-Å length are well suited for accommodating the aliphatic chain of MKS1 substrates (Figure 2A). Notably, in all of the MKS1 crystal structures determined to date, the central tunnel is occupied by what appears to be an apolar small-molecule ligand. For the cases in which no substrate or product analog was intentionally soaked into preexisting crystals, the bound ligand is inferred to be a fatty acid acquired during protein expression in the Escherichia coli host (see Supplemental Figure 2 and Supplemental Methods 1 online). Indeed, the serendipitous binding of endogenous E. coli fatty acids further supports the proposed functional role of the central tunnel (Figure 1B, inset).
Figure 2.
Comparison of MKS1 with Other Members of the HNL Family.
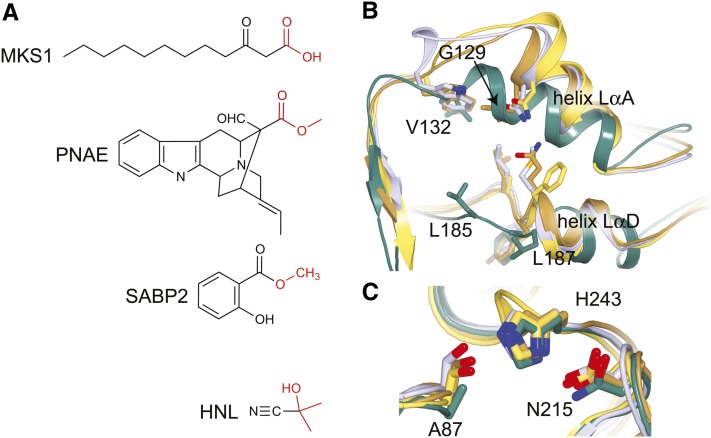
(A) Substrates of MKS1, PNAE, HNL, and SABP2.
(B) View looking down the central tunnel shows helices LαA and LαD of the lid domain. The loops leading up to the lid domain are positioned differently in MKS1 (green), HNL (purple), SABP2 (gold), and PNAE (yellow). Residues of presumed importance are labeled in MKS1 with corresponding residues in HNL, SABP2, and PNAE shown.
(C) Comparison of the HNL, SABP2, PNAE, and MKS1 active sites. While HNL, SABP2, and PNAE conserve a Ser-His-Asp active site, MKS1 deviates with Ala-His-Asn at the structurally equivalent sites. Colored as in (B). Labels correspond to the MKS1 sequence.
Three-dimensional structures have been determined for several other members of the HNL subfamily: HNL from both Hevea brasiliensis (Wagner et al., 1996; Gruber et al., 1999) and Manihot esculenta (Lauble et al., 2001), SABP2 (Forouhar et al., 2005) from tobacco (Nicotiana tabacum), and PNAE from Rauvolfia serpentina (Yang et al., 2009). Like MKS1, all possess a truncated α/β-hydrolase fold and lid domain insertion. Focusing here on the typical closed conformation of helix αE (as observed in MKS1 monomer B), MKS1 differs from HNL, SABP2, and PNAE by 1.4, 1.3, and 1.4 Å (root mean square deviation values over all equivalent Cα atoms, after structural superposition), respectively. The major differences between MKS1 and the other HNL subfamily members center around two α-helices (denoted LαA and LαD) forming part of the lid domain and contributing to the walls of the central tunnel. The specific positioning of these helices in MKS1 is responsible in part for the size of its tunnel (∼8 Å in diameter and 16 Å in length) (Figure 2B). For example, Leu-185 and Leu-187 from helix LαD border the central tunnel in MKS1, whereas with the more inward positioning of this helix in HNL, SABP2 and PNAE, the equivalent residues in these proteins protrude into the tunnel and would sterically occlude the extended aliphatic chain of MKS1 ligands. In HNL, Trp-128, which resides in the loop following LαA, has been implicated in conferring substrate specificity (Lauble et al., 2002). In MKS1, the corresponding residue, Val-132, likewise participates in forming the substrate binding tunnel and despite its smaller size has an equivalent spatial position to HNL Trp-128 due to the substantial displacement of the carboxy-end of the MKS1 LαA helix (Figure 2B).
Previously characterized members of the HNL subfamily recognize substrates that do not contain a long aliphatic tail like that of the MKS1 substrate (Figure 2A). Although HNL is reportedly active against medium-chain-length ketones, the activity decreases substantially with increasing chain length (Forster et al., 1996). Thus, the less voluminous substrate binding pockets of these enzymes can be readily rationalized. On the other hand, MKS1 must physiologically accommodate fatty acyl chains of up to 16 carbons in length, consistent with its more unobstructed central tunnel compared with the other HNL family members.
An Unexpected α/β-Hydrolase Active Site
The catalytic residues of α/β-hydrolases typically lie in highly conserved regions of the protein core (Figure 1C). The nucleophilic residue (Ser or Cys) is situated after β-strand 5 projecting from a sharp turn termed the nucleophilic elbow (Nardini and Dijkstra, 1999), which is characterized by a conserved Gly-Xaa-Nuc-Xaa-Gly sequence motif (Holmquist, 2000). Oddly, the corresponding sequence in MKS1 is Gly-His-Ala-Leu-Gly. Our structure determinations of MKS1 confirm that Ala-87 indeed resides at the nucleophilic elbow, clearly evidence for the marked divergence of the MKS1 catalytic residues from a typical α/β-hydrolase. A second, less radical departure from the canonical catalytic triad is the substitution by Asn for the acidic residue (Asp or Glu), which is situated on a turn linking αE and β-strand 7 (Nardini and Dijkstra, 1999). The acidic moiety normally provides the carboxylate counterion for the catalytic His. In MKS1 monomer B, the substituting residue Asn-215 is positioned identically to the catalytic Asp of HNL and SABP2, whereas in MKS1 monomer A, Asn-215 is remote from the active site due to substantial displacement of helix αE. The lone residue of the canonical catalytic triad that is retained in MKS1 is the His (His-243), which lies on a loop following β-strand 8 and in the α/β-hydrolases serves as the general base for activation of the nucleophilic Ser or Cys. An overlay of HNL, SABP2, and MKS1 highlights the structural differences between the active sites of these α/β-hydrolase (Figure 2C).
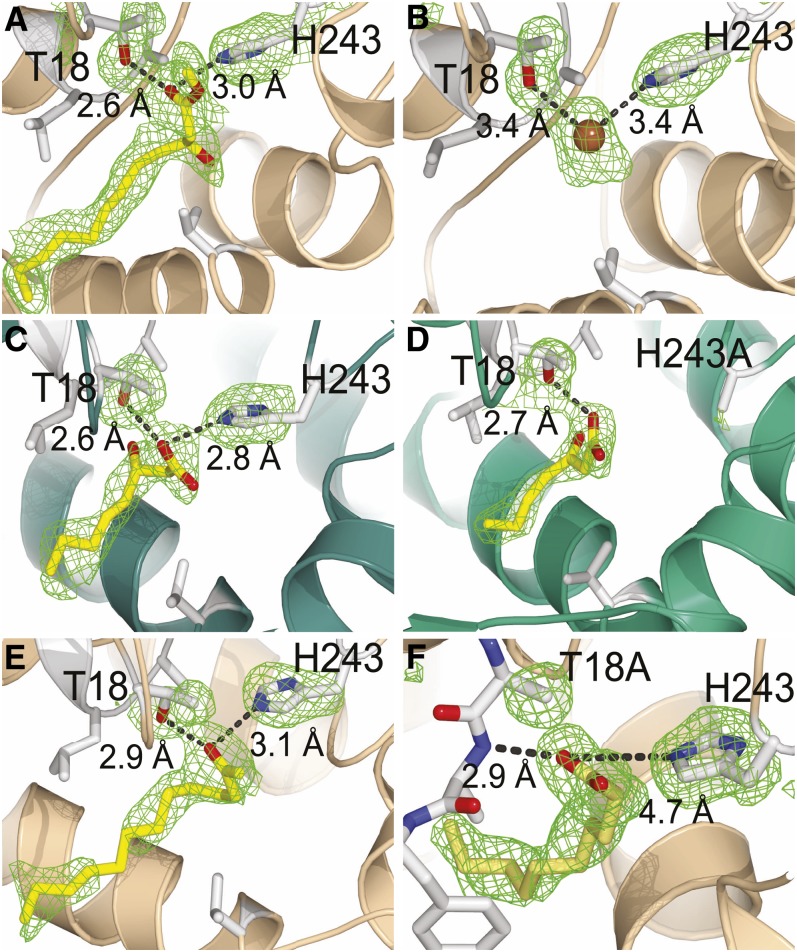
To definitively map the ligand binding and catalytic sites of MKS1, we employed small-molecule analogs with structural similarity to either substrate or product for soaking into preexisting MKS1 crystals. Structures were obtained for MKS1-WT complexed singly with two substrate mimics, M3D and 3HO, and a natural product, 2TD. The MKS1-H243A variant complexed with a potential substrate, 3-ketoheptanoate (3KH), while the MKS1-T18A variant sequestered a fatty acid acquired during protein expression in E. coli (Figure 3; see Supplemental Figure 1 online). The MKS1 crystallographic asymmetric unit contains a dimer and thus provides two corroboratory views of each bound ligand. However, in some cases, the occupancy of ligand binding differed between the two monomers.
Figure 3.
Small-Molecule Complexes of MKS1 Reaction Mimics and Active-Site Variants.
Each small molecule soaking experiment resulted in identical positioning of the analogs within the MKS1 structure, with the acyl tail located in the hydrophobic central tunnel (toward the bottom left of images) and the polar head group located in hydrogen bonding distance to Thr-18 and/or His-243. M3D (A), monomer B of 3HO complex showing a bromide within the active site (B), 3HO in monomer A (C), 3KH within H243A active site (D), 2TD (E), serendipitous binding of a fatty acid within T18A active site (F). Monomer A is colored green, and monomer B is colored tan. Electron density (simulated annealing omit map for ligand and active-site residues contoured at 2.5σ for all except [A] contoured at 1.5σ) surrounding residues 18 and 243 and bound ligand is shown in green.
Two features of the ligand interactions with MKS1 are common to all of the complexes. First, the ligand aliphatic acyl tail occupies the hydrophobic central tunnel. Second, the C1 polar head group is positioned approximately equidistant between two residues, His-243 and Thr-18, at the base of the tunnel. Most notably, His-243, which corresponds to the conserved His of the canonical α/β-hydrolase catalytic triad, forms a hydrogen bond with the α-carboxylate of the substrate-like ligands (Figures 3A and 3C) or with the 2-keto group of the 2TD product (Figure 3E). Interestingly, comparison of these hydrogen bonding patterns (and the absence of interactions of the 3-hydroxy group of the substrate analogs, as discussed further below) point to the inward shift in position of the substrate/product that accompanies the decarboxylation reaction. As indicated by the structure of the H243A MKS1-variant complexed with 3KH, native-like substrate binding is maintained despite the loss of His-243, a result that emphasizes the potential importance of hydrogen bond interactions formed between Thr-18 and the substrate carboxylate group (Figure 3D). Indeed, in the absence of Thr-18, a fatty acid ligand binds at an alternative position (Figure 3F), such that its carboxylate group forms a hydrogen bond interaction not with His-243, but instead with the backbone amide of the nearby residue Ala-19. This backbone amide is shielded by Thr-18 in wild-type MKS1, suggesting that Thr-18 plays a role in establishing the proper positioning of the MKS1 substrate prior to efficient decarboxylation
The 3-hydroxyl groups of both 3HO (Figure 3C) and M3D (Figure 3A) in complex with MKS1 lack direct hydrogen bond interactions with any MKS1 side chain or backbone atom. This hydroxyl group mimics the 3-keto group of the true MKS1 substrate, and its isolation within the active site is mechanistically intriguing, especially given its discrimination between fatty acyl and 3-keto acyl substrates, both emanating from fatty acid biosynthesis. Nevertheless, crystal soaking experiments demonstrate that MKS1 selectively sequesters only the S enantiomer from a racemic mixture of M3D (Figure 3A), a result consistent with the stereoselectivity previously described for the structurally related HNLs (Forster et al., 1996).
In monomer B of the MKS1-3HO complex, a strong electron-dense peak within the active site could not be accounted for by atoms of the 3HO analog, a water molecule, or a fatty acid. Although the origin of this excess density cannot be unequivocally identified, the best fitting model is a bromide anion, likely derived from the crystallization solution, bound at a position corresponding to that of the carbonyl group of the above-mentioned analogs (Figure 3B). The coordination of halide ions to proteins is generally nonspecific; however, a similar coordination to that found in MKS1 has been reported (Dauter and Dauter, 2001). As discussed below, the negatively charged bromide ion may mimic an activated water molecule (a hydroxide ion).
MKS1 Is an Efficient Decarboxylase
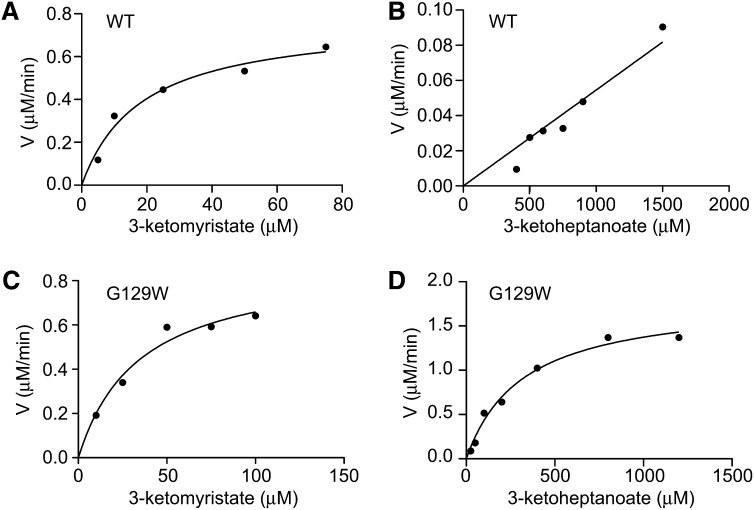
Through genetic analyses in tomato (Solanum lycopersicum) plants, Ben-Israel et al. (2009) showed that methylketone production required the expression of both MKS1 and MKS2 (Ben-Israel et al., 2009). We recently demonstrated MKS1 decarboxylase activity using both a heterologous in vivo system as well as in vitro biochemical assays (Yu et al., 2010). Here, we show that activity with the substrate 3-ketomyristate conforms to typical saturation kinetics (hyperbolic curve fitting r2 = 0.961) (Figure 4A; see Supplemental Methods 1 online). Derived kinetic parameters obtained from nonlinear regression analysis for Km and Kcat are 18.4 µM and 227.9 min−1, respectively (Table 1). MKS1 is much less active with 3-ketoheptanoate as substrate, exhibiting no observable saturation at the limit of substrate solubility (Figure 4B) affording only an estimated Km value >1500 µM (Table 1). The 14-carbon 3-ketomyristate provides a significantly longer aliphatic tail than the seven-carbon 3-ketoheptanoate, suggesting that a long hydrocarbon moiety dominates the affinity of MKS1 for its substrate.
Figure 4.
MKS1 and MKS1-G129W Steady State Kinetic Comparisons of Varying Chain Length 3-Keto Acid Substrates.
MKS1-WT with 3-ketomyristate (A) or 3-ketoheptanoate (B) as substrate. MKS1-G129W with 3-ketomyristate (C) or 3-ketoheptanoate (D) as substrate. WT, the wild type.
Table 1. Steady State Kinetic Constants for the MKS1 Wild Type and G129W Tunnel-Blocking Variant.
| Substrate | MKS1 |
G129W |
||||
|---|---|---|---|---|---|---|
| Kcat | Km | Kcat/Km | Kcat | Km | Kcat/Km | |
| 3-ketomyristate | 227.9 | 18.4 | 206,667 | 257.5 | 34.49 | 124,432 |
| 3-ketoheptanoate | – | >>1500 | 11 | 10.53 | 319.2 | 550 |
Values for Kcat, Km, and Kcat/Km are min−1, µM, and M−1⋅s−1, respectively.
MKS1 Catalytic Residues Functionally Defined
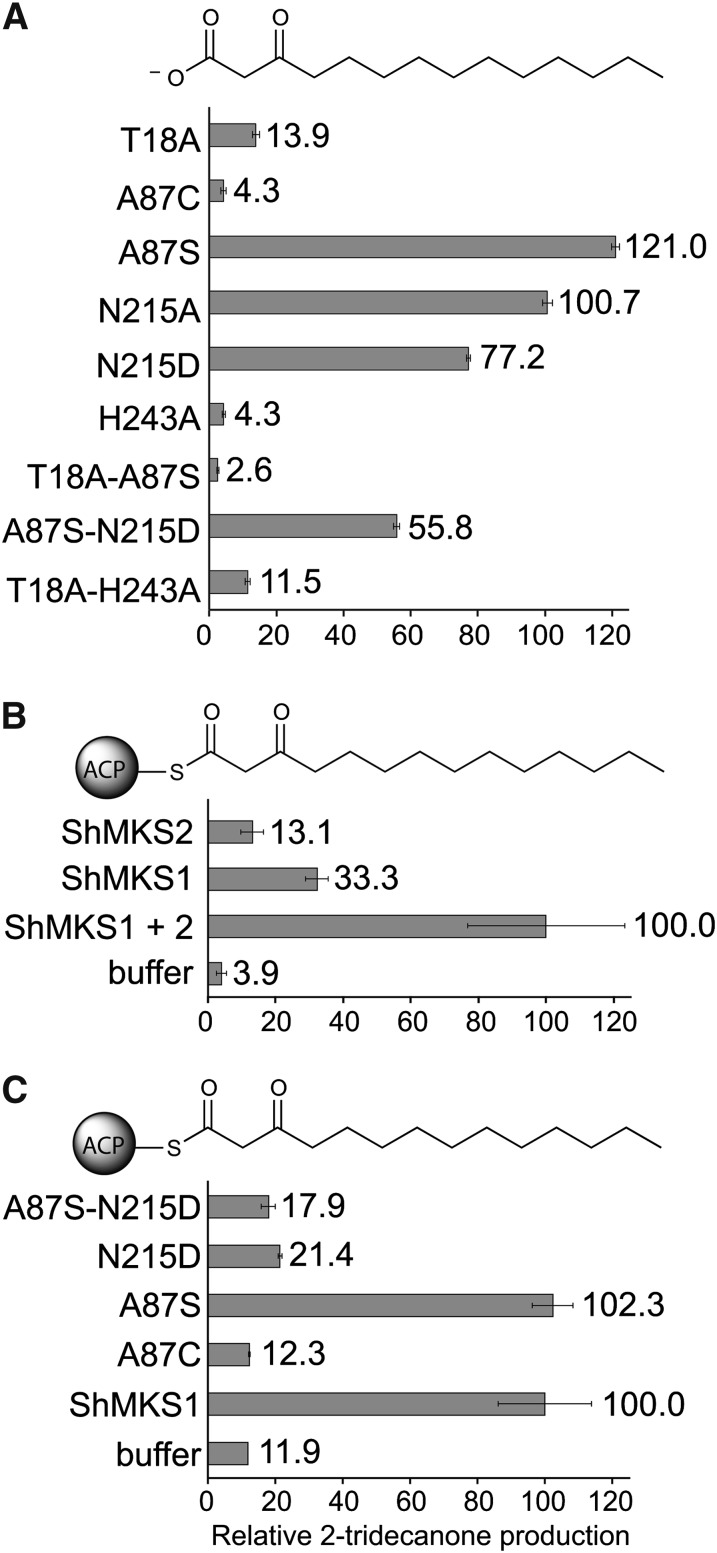
Three-dimensional structures of MKS1 reveal an active site consisting of a long central tunnel with His-243 at its foundation, thus pointing to a central role of this residue in catalysis (Figure 3). Other potential catalytic candidates are Thr-18, due to its proximity to His-243 and the carboxylate group of bound substrate analogs, and Asn-215 due to its equivalence to the acidic residue of the canonical α/β-hydrolase catalytic triad. Both of these residues as well as Ala-87 were targeted for amino acid substitution. We then measured the enzymatic activity of the variant MKS1 proteins with 3-ketomyristate as substrate (Figure 5A), in which the activities of the variants are expressed relative to the activity of wild-type MKS1). T18A and H243A variants retain only 13.9 and 4.3% activity, respectively. Unlike the T18A and H243A variants, the MKS1-N215A variant retains a wild-type level of activity. By contrast, in α/β-hydrolases such as PNAE that possess the canonical catalytic triad, substitutions for the conserved Asp abolish activity (Dogru et al., 2000).
Figure 5.
Enzymatic Activity of MKS1 and Active-Site Variants.
Substrates are shown at the top of each panel.
(A) Decarboxylase activities using 3-ketomyristate as substrate are shown as a percentage of the wild type.
(B) and (C) Combined thioesterase and decarboxylase activities of MKS2, MKS1, and MKS1 active-site variants. See Methods for details of synthesis.
(B) Activities are shown relative to the combined activities of MKS2 and MKS1 using 3-ketomyristoyl-ACP as substrate.
(C) Activities of MKS1 variants are shown relative to MKS1-WT using 3-ketomyristoyl-ACP as substrate. Error bars represent se (n = 3).
Several amino acid substitutions were introduced in attempts to restore in MKS1 the typical α/β-hydrolase catalytic triad (Figure 5A). First, Ala-87 was mutated to Ser to recapitulate the canonical nucleophilic residue. Second, Asn-215 was replaced with an Asp, which is more typical of Ser-containing α/β-hydrolases. Finally, to assemble the alternative Cys-His-Asn catalytic triad in MKS1, Ala-87 was mutated to Cys. Both N215D and A87S-N215D show a decrease in activity, while paradoxically, A87S decarboxylation activity is higher than the wild type. However, with the T18A-A87S variant, A87S is not able to compensate for the loss of activity caused by the T18A substitution. The A87C variant has little to no activity.
Simplistic View of an α/β-Hydrolase Proves Ineffectual
Although the combined activities of MKS2 (thioesterase) and MKS1 (decarboxylase) are required for efficient conversion of 3-keto acyl-ACP to methylketones in vivo and in vitro (Yu et al., 2010), MKS1 alone possesses low-level thioesterase activity (Figure 5B). However, presumably due to the Ala-87 replacement of the canonical nucleophile, MKS1 apparently lacks a suitable nucleophile for promoting thioesterase activity. As previously mentioned, an active-site bromide ion (occurring in monomer B of the 3HO complex) may mimic a hydroxide ion (Figure 3B), and it could be argued that MKS1 employs a His-243–activated water molecule as the nucleophile for any low-level thioesterase activity. Measurements of thioesterase activity show that MKS2 in the absence of MKS1 has markedly reduced activity (indeed, less than MKS1 alone) reflecting the slow spontaneous decarboxylation of the 3-keto acid (Figure 5B).
We reasoned that restoration of the canonical α/β-hydrolase catalytic triad within MKS1, as in the A87S-N215D variant, may confer full thioesterase activity (Figure 5C). However, this simplistic engineering of the MKS1 active site was clearly insufficient, as evident most directly from the lack of increased activity of the A87S-N215D variant. As was seen with MKS1 decarboxylase activity, the N215D variant alone has a negative effect on combined thioesterase/decarboxylase activity that is not compensated for by the A87S substitution. Partially restored catalytic triads Ser-His-Asn (variant A87S) and Cys-His-Asn (variant A87C) likewise do not transform MKS1 into a more active enzyme. Thus, the substitutions within the canonical α/β-hydrolase catalytic triad are not solely responsible for the ineffectual thioesterase activity of MKS1.
Structure-Guided Engineering of the MKS1 Active Site
Guided by the three-dimensional structures of MKS1 bound to ligands with aliphatic chains of various lengths (Figure 3), we introduced a series of amino acid substitutions aimed at obstructing the elongated central tunnel at specific locations, thereby restricting the chain length of the ligands that can be accommodated. Initially, amino acid residues lining the central tunnel were identified, and at each of these sites, the bulky residues Phe, Met, and Trp were modeled and visually assessed for effectiveness in obstructing the tunnel. In addition, dual-site channel blockage was targeted with the variants L88W-V132W and A19M-A128M, on the basis of their opposing positions within the tunnel of Leu-88 with Val-132 and of Ala-19 with Ala-128. For each of the potential channel-blocking amino acid substitutions in MKS1, we judged the approximate site of impingement along the ligand aliphatic chain (Table 2), with reference to the structure of the C12 M3D-ligand bound to MKS1 (Figure 3A).
Table 2. Activities of Tunnel-Blocking Mutants Given as a Percentage of the Wild Type.
| Variant | Approx. Block Site | Substrate |
|
|---|---|---|---|
| 3-Ketomyristate | 3-Ketoheptanoate | ||
| A19F | C2 | 18.3 ± 0.4 | 23.1 ± 0.9 |
| A19M | C2 | 56.8 ± 1.1 | 40.8 ± 1.7 |
| A19M-A128M | C2 + C6 | 26.5 ± 2.2 | n.d. |
| V132W | C3 | 22.3 ± 0.5 | n.d. |
| L88W-V132W | C3 | 15.3 ± 1.3 | n.d. |
| L88W | C3 | 95.4 ± 7.4 | 350.0 ± 15.4 |
| G129W | C5 | 143.1 ± 11.7 | 4408.0 ± 47.7 |
| A128M | C6 | 131.4 ± 8.3 | 100.4 ± 11.8 |
| A128W | C6 | 2.8 ± 0.2 | n.d. |
| C125W | C11 | 136.2 ± 9.7 | 3183.4 ± 55.2 |
3-ketomyristate (C14) or 3-ketoheptanoate (C7) was used as a substrate to assess the effectiveness of each substitution. Assays were performed as described in Methods. n.d., not detectable; ± represent se values (n = 3).
We measured the activities of these MKS1 variants against chemically synthesized seven- and 14-carbon 3-keto acids, 3-ketoheptanoate, and 3-ketomyristate (Table 2). The substitutions at Ala-19, Leu-88, and Val-132 should impinge on C2 and C3, and in general, result in decreased turnover of both 3-ketomyristate and 3-ketoheptanoate. The doubly blocked L88W-V132W and A19M-A128M show additional decreases in activity relative to the singly substituted variants A19M and V132W. Although Ala-128 replacement by Met (targeting C6) had little to no effect on activity against either substrate, replacement by Trp drastically reduced both activities. An unusual C3-targeting variant is Leu-88 to Trp, which exhibits normal 3-ketomyristate turnover but increased 3-ketoheptanoate turnover (Table 2) and, thus, a clear shift in affinity for shorter chain substrates without a negative consequence for longer substrates.
Moreover, two amino acid substitutions targeted to impinge on C5 and C11 of the substrate acyl chain show the most promise in reversing substrate preference toward the shorter 3-ketoheptanoate (Table 2). C125W and G129W exhibit increased activity against 3-ketoheptanoate by 32- and 44-fold, respectively, relative to wild-type MKS1. The G129W variant was characterized further through steady state kinetic analyses (Figures 4C and 4D, Table 1). In comparison to the wild type (Figures 4A and 4B, Table 1), G129W has an increased Km with the longer chain 3-ketomyristate and a decreased Km with the shorter chain 3-ketoheptanoate. Both Cys-125 and Gly-129 lie within helix LαA, a site located within the lid domain and, in particular for Gly-129, previously identified as structurally divergent from other HNL family members (Figure 2B). Despite this structural divergence, a Gly at position 129 conserves the overall tunnel surface characteristics as other HNL members maintain larger amino acids. It is easy to envision why replacement with a Trp occludes the MKS1 central binding tunnel. These results, while consistent overall with our structural mapping of the MKS1 central tunnel, establish the potential for rational engineering of the substrate preferences of MKS1.
DISCUSSION
Through structural and functional analyses, we show that although MKS1 possesses the polypeptide chain fold of the α/β-hydrolases (Figure 1), it lacks the hydrolytic activities typifying even the most closely related enzymes of this family. MKS1 is instead characterized by its catalytically efficient 3-keto acid decarboxylase activity (Figure 4, Table 1). Furthermore, the simplistic restoration to MKS1 of the canonical α/β-hydrolase catalytic triad failed to confer hydrolase (thioesterase) activity against ACP-linked substrates (Figure 5C). Indeed, several MKS1 homologs recently discovered in the cultivated tomato genome (http://solgenomics.net/) retain at position 87 (Ala in MKS1) a Ser, the nucleophilic residue characteristic of the canonical α/β-hydrolase catalytic triad (Yu et al., 2010) (see Supplemental Figure 3 online).
Detailed analysis of the catalytic mechanism of MKS1 requires first a consideration of the chemical properties of the 3-keto acid substrate. During the decarboxylative cleavage of the C1-C2 bond, a negative charge ultimately develops in the form of a C2 carbanion. Many decarboxlases employ cofactors to stabilize carbanion formation (Begley and Ealick, 2004), although cofactor-independent decarboxylases have been recently discovered (Appleby et al., 2000; Harris et al., 2000; Miller et al., 2000; Wu et al., 2000; Cendron et al., 2007; Kim et al., 2007; Chan et al., 2009). 3-Keto acids carry an inherent electron sink in the 3-carbonyl group, a property underlying their implied instability (Jencks, 1969). In fact, the supposed facility of decarboxylation of the 3-keto acid intermediates of methylketone biosynthesis led to the reasonable assumption that methylketone formation results from spontaneous decarboxylation of the intermediate released by thioester cleavage of the fatty-acyl ACP (Fridman et al., 2005). While it is true that 3-keto acids are prone to slow spontaneous decarboxylation (Hay and Bond, 1967), the kinetic requirements of ecological responses in methylketone-producing plants more likely demand an enzymatically catalyzed decarboxylation activity.
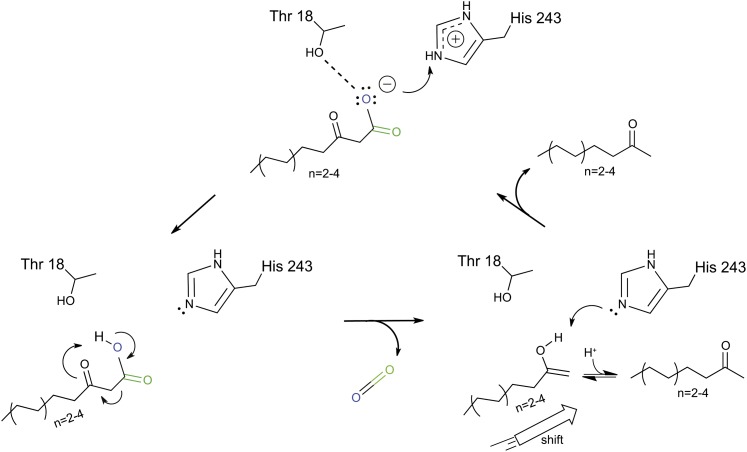
Our structural analyses of substrates and substrate analogs complexed with MKS1 highlight important catalytic roles in particular for Thr-18 and His-243 (Figure 3). We suggest that Thr-18 establishes the proper positioning of the 3-keto acid substrate within the MKS1 active site. Most importantly, His-243 acts as a proton donor to the dissociated 3-keto acid (carboxylate). Protonation accelerates the decarboxylation of the 3-keto acid through a concerted mechanism leading to formation of an enol at the keto electron sink, and subsequent tautomerization generates the methylketone product (Figure 6). In H. brasiliensis (Gruber et al., 2004) and M. esculenta (Lauble et al., 2001) HNL enzymes, which retain the canonical Ser-His-Asp catalytic triad, a Thr corresponding to MKS1-Thr 18 has also been shown to be important for enzymatic activity. It is evident that in MKS1, Asn-215 is not absolutely required for activity, as replacement by Ala has little effect (Figure 5A). However, replacement with a negatively charged Asp decreases MKS1 decarboxylase activity (Figure 5A), due possibly to a repulsive interaction with the negative charge carried by the dissociated form of the 3-keto acid substrate. Thus, the uncharged Asn perhaps has been evolutionarily selected for in MKS1.
Figure 6.
Proposed MKS1 Decarboxylase Reaction Mechanism.
The 3-keto acid substrate is positioned between Thr-18 and His-243 within the MKS1 active site. His-243 acts as a proton donor, thus facilitating decarboxylation through a concerted mechanism. Block arrow represents the shift in product orientation within the active site in relation to the substrate.
[See online article for color version of this figure.]
Decarboxylases are not prevalent within the α/β-hydrolase enzyme family. In fact, to our knowledge, only one other α/β-hydrolase has been shown to have decarboxylase activity, fermentation-respiration switch A (FrsA) protein, which catalyzes the decarboxylation of pyruvate to form acetaldehyde in E. coli during a switch to fermentative growth. In FrsA, one of its two domains is an α/β-hydrolase module, which is characterized by two unusual amino acid substitutions, an Arg at the nucleophilic elbow and the absence of the conserved His. The FrsA active site is postulated instead to reside within a cavity found between its two domains (Lee et al., 2011). The previously discussed bifunctional enzyme PNAE (which retains the typical α/β-hydrolase catalytic triad) from R. serpentina catalyzes the production of 16-epi-vellosimine via deesterification and subsequent decarboxylation. The PNAE esterase activity has been enzymatically and structurally examined (Dogru et al., 2000; Yang et al., 2009). The final decarboxylative step is thought to occur via polarization of the keto group facilitated by the active site His (Yang et al., 2009).
The emergence of decarboxylation activity in MKS1 is independent of FrsA and PNAE and must therefore represent convergent evolution. We base this conclusion on at least two lines of evidence. First, MKS1 from S. habrochaites and its apparent ortholog in S. lycoperiscum, Sl MKS1a, are closely related to proteins that are bona fide methyl esterases or other proteins with hydrolase activity (see Supplemental Figure 3 online). Second, the MKS1 active site configuration is novel to the α/β-hydrolase family. The highly conserved His residue and required Thr residue enable the efficient decarboxylation of fatty acyl 3-ketoacids at ambient temperatures.
While MKS1 appears to be able to hydrolyze fatty acyl-ACP substrates at extremely low levels, it is not clear if this activity was the ancestral activity. From the phylogenetic analysis, it is more likely that it evolved a decarboxylase activity from an esterase activity (see Supplemental Figure 3 and Supplemental Data Set 1 online). Perhaps the original substrate was a fatty acid derivative, and the emergence of the MKS1 decarboxylating activity on MKS2-liberated fatty acyl 3-keto acids was selected because the product was less toxic to the plant and more toxic to pests. Trichome-specific expression of both MKS1 and MKS2 must have evolved as well, since the derivation of methylketones from intermediates of fatty acid synthesis must be limited to special structures as to not significantly impair the production of fatty acids.
When in plant evolution the MKS1 ancestral protein acquired its decarboxylating activity is not clear. The cultivated tomato, S. lycopersicum, makes small amounts of methylketones in its trichomes, and the recent release of its genome sequence revealed that it has four functional genes related to S. habrochaites MKS1 (Sl MKS1a, MKS1b, MKS1d, and MKS1e) (Yu et al., 2010). To date, neither the activity of the proteins encoded by these four genes, including Sl MKS1a, the apparent ortholog of Sh MKS1, nor their organ-specific expression have been reported. Notably, they all have a Thr (Sl MKS1a, Sl MKS1d, and Sl MKS1e) or Ser (Sl MKS1b) at the equivalent position of Sh MKS1-Thr 18 and with the exception of MKS1a contain an Asn at the traditional acid position. It is likely that additional MKS1 sequences also exist in S. habrochaites, and perhaps other Solanum species, but their genome sequences have not yet been determined. While these sequences provide a wealth of information to study the detailed evolution of MKS1 sequence and activity, the presence of methylketone production in both S. habrochaites and S. lycopersicum must mean that MKS1 decarboxylase activity evolved prior to the split of the two lineages; therefore, informative variants of ancestral MKS1 genes prior to the acquisition of decarboxylase activity must be sought in more distant lineages, perhaps outside Solanum. However, it is worth noting that to date MKS1-related sequences outside Solanum all encode proteins that are >50% divergent from the Solanum MKS1 sequences.
Methylketones afford a chemical scaffold for the synthesis of useful biorenewable compounds (Nikolau et al., 2008), and as such the engineering of MKS1 for the recognition of substrates with specific alkyl-chain lengths is an important step toward the use of MKS1 in chemoenzymatic applications. Using a structure-guided mutagenesis strategy, we found two variants (C125W and G129W) with substantially increased activities toward shorter chain substrates (Table 2). We postulated that by sterically occluding the central acyl binding tunnel at defined sites, we could specifically disfavor the binding of long-chain substrates, while promoting the binding of shorter chain substrates. The G129W MKS1 variant, in comparison to wild-type MKS1, has increased Km values for the C12 substrate 3-ketomyristate and decreased Km values for the C7 substrate 3-ketoheptanoate. Our targeted engineering of MKS1 successfully demonstrates that MKS1 substrate preference is amenable to manipulation (Table 1) and the potential utility of MKS1 as a biocatalyst for the production of specific compounds.
METHODS
Protein Purification and Mutagenesis
Wild-type MKS1 was cloned as previously described (Fridman et al., 2005) and placed in a Gateway-compatible vector with an N-terminal poly-His tag and thrombin cleavage site for tag removal (pHis9GW) (O’Maille et al., 2004). Escherichia coli (BL21) cells harboring the MKS1 expression vector were grown in Terrific Broth at 37°C until reaching an OD600 of 1.0 at which time 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) was added. Cells were then grown overnight at 18°C and harvested by centrifugation.
For protein used in crystallization trials, cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 20 mM imidazole, 1% [v/v] Tween 20, 10% [v/v] glycerol, and 20 mM 2-mercaptoethanol) and lysed by sonication. MKS1 was purified from E. coli by passing soluble lysate over Ni2+-nitriloacetic acid agarose (Qiagen) and eluting with lysate buffer supplemented with 250 mM imidazole. The N-terminal tag was removed by treatment with thrombin while dialyzing overnight in 50 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 20 mM 2-mercaptoethanol. MKS1 was further purified by size-exclusion chromatography using a Superdex 75 HR16/60 column, which showed that MKS1-WT and variant proteins were dimeric in solution under isotonic and near neutral pH conditions. Proteins were subsequently dialyzed into crystallization buffer (12.5 mM Tris-HCl, pH 8.0, 125 mM NaCl, and 2 mM DTT) and concentrated to 10 to 17 mg/mL. For protein used in enzymatic assays, purification procedures were as above except Tris-HCl was excluded as the buffer and replaced with a phosphate buffer, pH 6.8, at the same concentration. Site-directed mutagenesis was performed on MKS1pHis9GW using the QuikChange protocol (Stratagene), and mutant proteins were expressed and purified as described for wild-type protein.
MKS1 variants exhibited the same stability and gel filtration elution profile as wild-type MKS1, with two notable exceptions the single mutant A87C and the double mutant T18A-A87S, which showed a degree of instability evidenced by low overall yields, continual aggregation during isolation, and anomalous behavior using gel filtration chromatography.
Crystallization and Data Collection
Crystallization of MKS1 was performed at 4°C by vapor diffusion in hanging drops using a 1:1 mixture of protein (10-17 mg/mL in 12.5 mM Tris-HCl, pH 8.0, 125 mM NaCl, and 2 mM DTT) and reservoir solution (15 to 24% [w/v] polyethylene glycol 8000, 0.3 M NaBr, and 2 mM DTT, in a pH range including 0.1 M PIPES-Na+, pH 6.5, 0.1 M MOPSO-Na+, pH 7.0, or 0.1 M HEPES-Na+, pH 7.5). Crystals grew within 3 d and were improved by streak seeding. Small molecule complexes were obtained by soaking crystals for 2 to 24 h in reservoir solutions supplemented with 1 mM 2-tridecanone, 3HO, 3-ketoheptanoate, or M3D.
Prior to freezing in liquid N2, crystals were transferred to a cryoprotectant solution consisting of 10% (w/v) Suc in reservoir solution. X-ray diffraction data were collected at the Advanced Light Source Lawrence Berkeley National Laboratory on beam lines 8.2.1 and 8.2.2 and at the Stanford Synchrotron Radiation Light Source on beam lines 1-5 and 9-2. Data were indexed, integrated, and scaled using XDS and XSCALE (Kabsch, 1993) or HKL2000 (Otwinowski and Minor, 1997). Initial phases were obtained by molecular replacement using a monomeric MKS1 monomeric homology model based on the reported crystal structure of HNL (7YAS) and calculated using Modeler (Sali and Blundell, 1993) together with the program Phaser (McCoy et al., 2005) as part of the CCP4 suite of programs (Winn et al., 2011). A twofold noncrystallographic symmetry restraint was used to carry out initial structure refinement using CNS (Brünger et al., 1998). Noncrystallographic symmetry restrictions were removed for all subsequent refinements, which employed a combination of CNS and Refmac refinements (Vagin et al., 2004) along with a common test set of reflections for assessment of Free-R values. The quality of the 3STX data set was degraded by the presence of strong ice rings in the diffraction images, as reflected in the less than ideal overall completeness and free R value. Coot was used for visualization of calculated electron density maps and manual rebuilding (Emsley et al., 2010). Waters were added using Coot’s Find Waters command as well as through manual inspection of the electron density maps. Pymol (DeLano, 2002) was used to render all structure related figures.
Decarboxylase Assay and Steady State Kinetics
Decarboxylase assays were performed using synthesized 3-ketomyristate and 3-ketoheptanoate for both specific activity comparative measurements and more detailed steady state kinetics analyses. A typical 500-μL reaction contained wild-type or mutant MKS1 (2.5 μg for 3-ketomyristate or 5 μg for 3-ketoheptanoate), 3-ketoheptanoate (1 mM) or 3-ketomyristate (0.1 mM), and 1,3-bis(tris(hydroxymethyl)methylamino) propane (20 mM, pH 7.0). Assays were performed as previously described (Yu et al., 2010). For 2-hexanone detection, methods as described above were used with the following variations in gas chromatography programming: An initial oven temperature of 37°C was held for 12 min; temperature was ramped at a rate of 10°C/min until 75°C, followed by a second ramp of 50°C/min to 280°C, which was held for 5 min.
The steady state kinetic assay for MKS1 and MKS1-G129W variant were performed in 500-μL reaction volumes in assay buffer (described above) with 3.44 nM (for 3-ketomyristate) or 0.344 μΜ (for 3-ketoheptanoate) enzyme and variable concentration of 3-ketomyristate (5 to 100 μΜ) or 3-ketoheptanoate (25 to 1600 μΜ). Reactions were mixed at room temperature (23°C) in 2-mL screw-top glass vials, after variable incubation time, reactions were quenched with 25 μL 3 n NaOH. The products were extracted with 500 μL of hexane or ethyl acetate with appropriate internal standards by vigorous vortexing, followed by gas chromatography–mass spectrometry analysis (as above). Km and Kcat were determined by GraphPad Prism 5 software using nonlinear regression for the Michaelis-Menten equation.
Thioesterase Assays
To produce the 3-ketomyristoyl-ACP needed for the coupled thioesterase/decarboxylase assays, three separate protein purifications were performed to assemble the requisite assay components. First, Solanum habrochaites holo-ACP.2 serves as the carrier for the 3-keto acid. Second, E. coli FabD is a malonyl transferase used to transfer the malonyl moiety from malonyl-CoA to ACP.2. Third, 3-ketoacyl-ACP synthase from Mycobacterium tuberculosis (FabH) uses lauroyl-CoA as a starter unit in a Claisen condensation with malonyl-ACP.2 to produce the ShMKS1 substrate 3-ketomyristoyl-ACP.
The mature peptide sequence of the ACP.2 protein, excluding conserved transit peptide at the N termini, was obtained based on sequence alignment of the full-length peptide (from the S. habrochaites EST database) (Fridman et al., 2005) to other plant ACPs. The following primers were used to perform the first round of amplification of mature ACP.2 coding sequence from leaf cDNA: forward, 5′-CTGGTTCCGCGTGGTTCCATGGCTAAGCCAGAGACT-3′; reverse, 5′-CAAGAAAGCTGGGTCTCAAGCTTTCTTTGAAAT-3′. The following primers were used to perform the second round of amplification with introduction of the Gateway-compatible att-B1 and att-B2 flanking sequences: forward, 5′- GGGGACAAGTTTGTACAAAAAAGCAGGCTTACTGGTTCCGCGTGGTTCC-3′; reverse, 5′- GGGGACCACTTTGTACAAGAAAGCTGGGT-3′. PCR conditions were 30 s at 94°C, followed by 25 cycles of 20 s at 94°C, 20 s at 56°C, 40 s at 68°C, and additional 10 min at 68°C. The final PCR product was used as a template in Gateway BP and LR clonase (Invitrogen) reactions, resulting in the insertion of Sh ACP.2 into pHis9GW.
ACP.2 was expressed/purified from E. coli in a Tris-HCl buffer as described above for MKS1. Recombinant ACP.2 was pantetheinylated by the E. coli host as visualized by native PAGE analysis supplemented with urea as described in Post-Beittenmiller et al. (1991). FabD (courtesy of Michael Burkart, Chemistry and Biochemistry Department, University of California, San Diego) and FabH (courtesy of Kevin Reynolds, Chemistry Department, Portland State University) were obtained as His-tagged clones in IPTG-inducible expression vectors. The FabD clone was put into E. coli (BL21 DE3) cells and grown in Terrific Broth at 37°C until reaching an OD600 of 0.5, at which time 0.5 mM IPTG was added. Cells continued to grow overnight at 18°C and were harvested and purified in a Tris-HCl buffer as described above for MKS1. FabH was purified as described by Scarsdale et al. (2001). Purified FabH and FabD were dialyzed into 12.5 mM Tris-HCl, pH 8.0, 125 mM NaCl, and 2 mM DTT and concentrated to 2.0 and 2.5 mg/mL, respectively.
To enzymatically synthesize 3-ketomyristoyl-ACP, a combined FabD/FabH reaction was run overnight, for ∼16.5 h at 37°C. Assays were assembled in a master mix that included 1,3-bis(tris(hydroxymethyl)methylamino) propane (20 mM, pH 7.0), malonyl-CoA (0.2 mM), lauroyl-CoA (0.2 mM), ACP.2 (0.1 mM), FabD (10 μg), and MtFabH (10 μg) per 200-μL reaction volume. The overnight reaction was aliquoted prior to addition of wild-type or mutant MKS1. After addition of MKS1 (30 ng in Figure 5C or 2.5 μg in Figure 5B), reactions proceeded for 30 min at room temperature, supplemented with 2-undecanone (2 μM) as an internal standard, and then extracted three times with 500 μL of hexanes.
Hexane extracts were pooled, concentrated to 200 μL at room temperature by N2 gas, and analyzed on a Hewlett-Packard 6890 gas chromatograph coupled to a 5973 mass-selective detector equipped with an HP-5MS capillary column (0.25-mm i.d., 30-m length, and 0.25-μm film). Carrier gas (He) flow rate was 1.5 mL/min, and the mass-selective detector was operated at 70 eV. Injections (5 μL) were performed in splitless mode with an inlet temperature of 280°C. The gas chromatograph was programmed as follows: initial oven temperature of 60°C was held for 2 min, temperature was ramped at a rate of 5°C/min until 200°C, followed by a second ramp of 50°C/min to 280°C, which was held for 5 min. Retention times were 14.0 and 19.1 min for 2-undecanone and 2-tridecanone, respectively.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GU987105.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Small-Molecule Ligands Used as Substrate Mimics, with the Exception of 2-Tridecanone, Which Is a MKS1 Natural Product.
Supplemental Figure 2. MKS1 Active Site Serendipitously Binds Fatty Acid during Heterologous Production in E. coli.
Supplemental Figure 3. Maximum Likelihood Phylogenetic Tree Including S. habrochaites MKS1 and Other Closely Related Proteins.
Supplemental Table 1. Data Collection and Refinement Statistics for the MKS1 Wild Type and Variants, with and without Analogs.
Supplemental Methods 1. Supplemental Methods for 3-Keto Acid Synthesis, Fatty Acid Composition Determination, and Phylogenetic Tree Construction.
Supplemental Data Set 1. Alignment Output from MEGA 5 Software Used to Construct Phylogenetic Tree in Supplemental Figure 3.
Supplementary Material
Acknowledgments
We thank Mike Burkart (University of California, San Diego) for providing the FabD and Svp clones and Kevin Reynolds (Portland State University) for the FabH clone. We thank Gordon Louie (The Salk Institute) for his valuable assistance in crystallographic related work and careful reading of the manuscript. We also thank Marianne Bowman (The Salk Institute) for advice on protein purification strategies and Tom Baiga, Zheng Xiang, and Jonathan Melnick (all of The Salk Institute) for helpful discussions related to the chemical synthesis of ligands. We acknowledge the U.S. National Institutes of Health for which M.E.A. was a postdoctoral fellow (F32 GM076997-03). This material is based in part upon work supported by the National Science Foundation under Award EEC-0813570 (J.P.N. and E.P.) and Award MCB-0645794 (J.P.N.). Portions of this research were conducted at the Advanced Light Source and Stanford Synchrotron Radiation Light Source. We thank the staff at the Stanford Synchrotron Radiation Light Source and Advanced Light Source for assistance with x-ray data collection.
AUTHOR CONTRIBUTIONS
M.E.A. purified and crystallized proteins and solved the structures, with assistance on the initial structure from M.B.A. M.E.A. performed fatty acid analyses and coupled thioesterase/decarboxylase assays. J.R. and Y.G. synthesized chemical substrates. Y.G. performed decarboxylase assays, steady state kinetic analyses, and chain-length dependence assays. E.F. isolated MKS1 and ACP.2 from S. habrochaites. All authors contributed to experimental design and interpretation. M.E.A. wrote the article with input from all authors.
Glossary
- ACP
acyl carrier protein
- M3D
methyl-3-hydroxydodecanoate
- 3HO
3-hydroxyoctanoate
- IPTG
isopropyl-β-d-thiogalactoside
- IPTG
2TD
References
- Appleby T.C., Kinsland C., Begley T.P., Ealick S.E. (2000). The crystal structure and mechanism of orotidine 5′-monophosphate decarboxylase. Proc. Natl. Acad. Sci. USA 97: 2005–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley T.P., Ealick S.E. (2004). Enzymatic reactions involving novel mechanisms of carbanion stabilization. Curr. Opin. Chem. Biol. 8: 508–515 [DOI] [PubMed] [Google Scholar]
- Ben-Israel I., Yu G., Austin M.B., Bhuiyan N., Auldridge M., Nguyen T., Schauvinhold I., Noel J.P., Pichersky E., Fridman E. (2009). Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol. 151: 1952–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning M.M., Wesenberg G., Liu R., Taylor K.L., Dunaway-Mariano D., Holden H.M. (1998). The three-dimensional structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. strain CBS-3. J. Biol. Chem. 273: 33572–33579 [DOI] [PubMed] [Google Scholar]
- Brünger A.T., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Cendron L., Berni R., Folli C., Ramazzina I., Percudani R., Zanotti G. (2007). The structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase provides insights into the mechanism of uric acid degradation. J. Biol. Chem. 282: 18182–18189 [DOI] [PubMed] [Google Scholar]
- Chakravarty B., Gu Z., Chirala S.S., Wakil S.J., Quiocho F.A. (2004). Human fatty acid synthase: Structure and substrate selectivity of the thioesterase domain. Proc. Natl. Acad. Sci. USA 101: 15567–15572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., Wood B.M., Fedorov A.A., Fedorov E.V., Imker H.J., Amyes T.L., Richard J.P., Almo S.C., Gerlt J.A. (2009). Mechanism of the orotidine 5′-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization. Biochemistry 48: 5518–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauter Z., Dauter M. (2001). Entering a new phase: Using solvent halide ions in protein structure determination. Structure 9: R21–R26 [DOI] [PubMed] [Google Scholar]
- DeLano W.L. (2002). The Pymol Molecular Graphics System. (San Carlos, CA: DeLano Scientific) [Google Scholar]
- Dillon S.C., Bateman A. (2004). The hotdog fold: Wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics 5: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogru E., Warzecha H., Seibel F., Haebel S., Lottspeich F., Stöckigt J. (2000). The gene encoding polyneuridine aldehyde esterase of monoterpenoid indole alkaloid biosynthesis in plants is an ortholog of the alpha/betahydrolase super family. Eur. J. Biochem. 267: 1397–1406 [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhar F., Yang Y., Kumar D., Chen Y., Fridman E., Park S.W., Chiang Y., Acton T.B., Montelione G.T., Pichersky E., Klessig D.F., Tong L. (2005). Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. USA 102: 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster S., Roos J., Effenberger F., Wajant H., Sprauer A. (1996). The first recombinant hydroxynitril lyase and its application in the synthesis of (S)-cyanohydrins. Angew. Chem. Int. Ed. Engl. 35: 437–439 [Google Scholar]
- Fridman E., Wang J., Iijima Y., Froehlich J.E., Gang D.R., Ohlrogge J., Pichersky E. (2005). Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17: 1252–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber K., Gartler G., Krammer B., Schwab H., Kratky C. (2004). Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: The three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236. J. Biol. Chem. 279: 20501–20510 [DOI] [PubMed] [Google Scholar]
- Gruber K., Gugganig M., Wagner U.G., Kratky C. (1999). Atomic resolution crystal structure of hydroxynitrile lyase from Hevea brasiliensis. Biol. Chem. 380: 993–1000 [DOI] [PubMed] [Google Scholar]
- Harris P., Navarro Poulsen J.C., Jensen K.F., Larsen S. (2000). Structural basis for the catalytic mechanism of a proficient enzyme: Orotidine 5′-monophosphate decarboxylase. Biochemistry 39: 4217–4224 [DOI] [PubMed] [Google Scholar]
- Hay R.W., Bond M.A. (1967). Kinetics of the decarboxylation of acetoacetic acid. Aust. J. Chem. 20: 1823–1828 [Google Scholar]
- Holmquist M. (2000). Alpha/beta-hydrolase fold enzymes: Structures, functions and mechanisms. Curr. Protein Pept. Sci. 1: 209–235 [DOI] [PubMed] [Google Scholar]
- Hynková K., Nagata Y., Takagi M., Damborský J. (1999). Identification of the catalytic triad in the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. FEBS Lett. 446: 177–181 [DOI] [PubMed] [Google Scholar]
- Jencks W.P. (1969). Catalysis in Chemistry and Enzymology. (New York: McGraw-Hill Book Company) [Google Scholar]
- Kabsch W. (1993). Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26: 795–800 [Google Scholar]
- Kim K., Park J., Rhee S. (2007). Structural and functional basis for (S)-allantoin formation in the ureide pathway. J. Biol. Chem. 282: 23457–23464 [DOI] [PubMed] [Google Scholar]
- Lauble H., Förster S., Miehlich B., Wajant H., Effenberger F. (2001). Structure of hydroxynitrile lyase from Manihot esculenta in complex with substrates acetone and chloroacetone: Implications for the mechanism of cyanogenesis. Acta Crystallogr. D Biol. Crystallogr. 57: 194–200 [DOI] [PubMed] [Google Scholar]
- Lauble H., Miehlich B., Förster S., Kobler C., Wajant H., Effenberger F. (2002). Structure determinants of substrate specificity of hydroxynitrile lyase from Manihot esculenta. Protein Sci. 11: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.J., Jeong C.S., An Y.J., Lee H.J., Park S.J., Seok Y.J., Kim P., Lee J.H., Lee K.H., Cha S.S. (2011). FrsA functions as a cofactor-independent decarboxylase to control metabolic flux. Nat. Chem. Biol. 7: 434–436 [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Storoni L.C., Read R.J. (2005). Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61: 458–464 [DOI] [PubMed] [Google Scholar]
- Miller B.G., Hassell A.M., Wolfenden R., Milburn M.V., Short S.A. (2000). Anatomy of a proficient enzyme: The structure of orotidine 5′-monophosphate decarboxylase in the presence and absence of a potential transition state analog. Proc. Natl. Acad. Sci. USA 97: 2011–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardini M., Dijkstra B.W. (1999). Alpha/beta hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 9: 732–737 [DOI] [PubMed] [Google Scholar]
- Nikolau B.J., Perera M.A., Brachova L., Shanks B. (2008). Platform biochemicals for a biorenewable chemical industry. Plant J. 54: 536–545 [DOI] [PubMed] [Google Scholar]
- O’Maille P.E., Tsai M.D., Greenhagen B.T., Chappell J., Noel J.P. (2004). Gene library synthesis by structure-based combinatorial protein engineering. Methods Enzymol. 388: 75–91 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997). Processing of X-ray diffraction data collected in oscillation mode. (New York: Academic Press) [Google Scholar]
- Post-Beittenmiller D., Jaworski J.G., Ohlrogge J.B. (1991). In vivo pools of free and acylated acyl carrier proteins in spinach. Evidence for sites of regulation of fatty acid biosynthesis. J. Biol. Chem. 266: 1858–1865 [PubMed] [Google Scholar]
- Sali A., Blundell T.L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Scarsdale J.N., Kazanina G., He X., Reynolds K.A., Wright H.T. (2001). Crystal structure of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III. J. Biol. Chem. 276: 20516–20522 [DOI] [PubMed] [Google Scholar]
- Stuhlfelder C., Mueller M.J., Warzecha H. (2004). Cloning and expression of a tomato cDNA encoding a methyl jasmonate cleaving esterase. Eur. J. Biochem. 271: 2976–2983 [DOI] [PubMed] [Google Scholar]
- Thoden J.B., Holden H.M., Zhuang Z., Dunaway-Mariano D. (2002). X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase. J. Biol. Chem. 277: 27468–27476 [DOI] [PubMed] [Google Scholar]
- Vagin A.A., Steiner R.A., Lebedev A.A., Potterton L., McNicholas S., Long F., Murshudov G.N. (2004). REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60: 2184–2195 [DOI] [PubMed] [Google Scholar]
- Wagner U.G., Hasslacher M., Griengl H., Schwab H., Kratky C. (1996). Mechanism of cyanogenesis: The crystal structure of hydroxynitrile lyase from Hevea brasiliensis. Structure 4: 811–822 [DOI] [PubMed] [Google Scholar]
- Williams W.G., Kennedy G.G., Yamamoto R.T., Thacker J.D., Bordner J. (1980). 2-Tridecanone: A naturally occurring insecticide from the wild tomato Lycopersicon hirsutum f.glabratum. Science 207: 888–889 [DOI] [PubMed] [Google Scholar]
- Winn M.D., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Mo Y., Gao J., Pai E.F. (2000). Electrostatic stress in catalysis: Structure and mechanism of the enzyme orotidine monophosphate decarboxylase. Proc. Natl. Acad. Sci. USA 97: 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Hill M., Wang M., Panjikar S., Stöckigt J. (2009). Structural basis and enzymatic mechanism of the biosynthesis of C9- from C10-monoterpenoid indole alkaloids. Angew. Chem. Int. Ed. Engl. 48: 5211–5213 [DOI] [PubMed] [Google Scholar]
- Yu G., Nguyen T.T., Guo Y., Schauvinhold I., Auldridge M.E., Bhuiyan N., Ben-Israel I., Iijima Y., Fridman E., Noel J.P., Pichersky E. (2010). Enzymatic functions of wild tomato methylketone synthases 1 and 2. Plant Physiol. 154: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.