Abstract
Background: Rice plant diseases play a major role as biological constraints on production. One of such rice disease is bacterial leaf blight, caused by Xanthomonas oryzae pv. Oryzae (Xoo). The diffusible signal factor (DSF) synthesized by Xoo has a major role in virulence to rice plant. The DSF synthase RpfF protein, which is related to crotonase superfamily is responsible for the maintaining concentration of DSF. DSF-dependent quorum sensing (QS) system adopts protein- protein interaction mechanism to auto regulates the production of DSF. The antibacterial activity of pesticides against Xoo has not yet been completely understood. Three dimensional structure of RpfF protein was predicted using homology modeling method by MODELLER 9V9 software, SWISS MODEL and GENO3D online tools and structures were validated by Ramachandran plot, TM-Score and RMSD. 3D structure of RpfF (accession number AAL06345) was predicted using DSF synthase of Xanthomonas campestris pv. campestris (Xcc) (PDB ID: 3M6M) as a template. The stereo chemical check reveals the structure developed from the modeller was the best one and the potential ligand binding sites were identified by CASTp Server. The predicted RpfF model provides insight into its structure, active sites and aid in the development of novel inhibitors to control bacterial leaf blight in rice plant. DSF synthase RpfF protein could be used as a novel target to control infection
Keywords: Xanthomonas oryzae, RpfF, DSF, Homology modeling
Background
Bacterial blight (BB) of rice, caused by Xanthomonas oryzae pv. oryzae (Xoo) is one of the oldest known diseases and was first noticed by the farmers of Japan in 1884 [1]. BB is a vascular disease resulting in a systemic infection [2]. The principle host for Xoo is rice plant and enters its host through hydathodes or wounds around leaf edges and then multiplies and spreads in xylem vessels, causing disease [3]. Symptoms are observed at the tillering stage, disease incidence increases with plant growth, peaking at the flowering stage. Kresek is the more destructive manifestation of the disease, wherein the leaves of the entire plant turn pale yellow and wilt during the seedling to the early tillering stage, resulting in a partial or total crop failure [4]. The disease is known to occur in epidemic proportions in many parts of the world, incurring severe crop loss of up to 50%. Crop loss assessment studies have revealed that this disease reduces grain yield to varying levels, depending on the stage of the crop, degree of cultivar susceptibility and the conduciveness of the environment in which it occurs. The severity and significance of damages caused by infection have necessitated the development of strategies to control and manage the disease, so as to reduce crop loss and to avert an epidemic. Many gram-negative pathogenic bacteria are highly dependent on secretary systems for virulence [5]. The secretary pathways mediate the secretion of proteins (substrate virulence effectors) into the extracellular milieu and into the host cells themselves, wherein the effectors facilitate virulence [6]. Ablation of the secretary pathway in plant pathogen systems often results in the loss of pathogenesis or severe reduction of virulence [7]. Diffusible signal factor (DSF) is a long chain fatty acid and chemical name is ‘cis-11-methyl-2-dodecenoic acid’. The DSF -dependent quorum sensing (QS) mechanism in Xoo allows sensing their population density and synchronizing individual activity into cooperative community behavior. This appears to provide bacterial pathogens an obvious competitive advantage over their hosts in pathogen-host interaction [8]. DSF family signals have been reported in a range of plant and human bacterial pathogens, including Xanthomonas oryzae pv. Oryzae (Xoo), Xanthomonas campestris pv. Campestris (Xcc), Xyllela fastidiosa, Stenotrophomonas maltophilia and Burkholderia cenocepacia. The DSF signalling system comprises several key regulatory proteins and a second messenger cyclic-di-GMP (c-di-GMP). Among them, the RpfC/RpfG two component system is involved in sensing and transduction of DSF signal through a conserved phosphorelay mechanism in Xoo.
The putative enoyl-CoA hydratase RpfF is a key enzyme for DSF biosynthesis. Functional analysis of RpfF or RpfC mutants in different bacterial species suggests that the general role of the DSF-signaling system in the modulation of virulence seems to be conserved, but the regulatory mechanisms and DSFdependent traits may differ among taxa [9]. Since the DSF QS system is involved in the virulence to rice plant, the blocking DSF synthase RpfF could be a possible target protein to control the infection. The previous study elucidate that the threedimensional structure (3D) of DSF synthase RpfF protein from Xanthomonas oryzae pv. oryzae is remains unknown. A number of techniques have been developed to predict 3D structure of macromolecules which may play a pivotal role to find out their activity. Homology modelling is one of the techniques used to predict the 3D structure of proteins, relying heavily on resources such as pattern/function and sequence. In view of the above, the present study was aimed to predict the three dimensional structure of RpfF protein through homology modeling method.
Methodology
GY Sequence retrieval:
The sequence of DSF synthase RpfF protein (accession no: AAL06345; length: 289 residues), from Xanthomonas oryzae pv. oryzae was retrieved from NCBI Entrez http://www.ncbi.nlm.nih.gov.
Structure prediction and evaluation:
Template search was performed using BLASTP program against PDB database http://www.rcsb.org/pdb. Sequence analysis was performed using CLC Sequence viewer. RpfF protein model was generated by MODELLER 9V9. SWISSMODEL and GENO3D were also used to check the structure reliability [10, 12]. Stereo chemical quality of predicted models was improved by subjecting energy minimization protocol to correct bond angles and bond lengths. Energy minimization for 3D structures was performed by using YASARA, which runs molecular dynamics simulations of models in explicit solvent, using a new partly knowledge-based all atom force field derived from Amber. Model quality was assessed using variety of validation tools, such as RAMPAGE, TM- Score and RMSD. RAMPAGE was used in validation of protein structure and models by verifying the parameters like Ramachandran plot quality. TM-score is an algorithm to calculate the similarity of topologies of two protein structures. A single score between 0-1 is assigned to each comparison based on statistics. RMSD value indicates the degree to which two three dimensional structures are similar [13].
Ligand binding site prediction:
An analysis of the ligand binding pockets of the predicted structure was performed using CASTp [14]. As determined by CASTp using a 1.4 Å radius probe, the internal cavity surface volume of the ligand binding sites were calculated.
Discussion
Protein Blast search against PDB data base revealed 96 percent sequence identity with experimental structure of PDB ID: 3M6M with 2.5 Å resolutions, from Xanthomonas Campestris Pv. Campestris. The pair wise sequence alignment shown in the (Figure 1). Homolgy model was constructed through three tools viz MODELLER9v9, Geno3D and SwissModel. The predicted model obtained by using three software validated by RAMPAGE, TM-Score and RMSD, were reliable structures. These results Table 1 & Table 2 (see supplementary material) revealed that the model generated through modeller was most acceptable and it was used for further investigation. RpfF model from Modeller showed psi and phi torsion angles 97.8% and 1.6% residues in favorable and allowed regions respectively and it showed TM-score and RMSD 0.7749 and 1.520 respectively. Topology analyses elucidated twelve α-helices and eight anti- parallel β sheets (Figure 2). Active site prediction performed using CASTp server showed RpfF has 37 pockets. Based on the high surface area (640.1) and volume (772.1), pocket ID 37 was chosen to be a potential cavity in the structure. The amino acid residues involved in potential ligand binding site were LEU84, GLY85, GLY86, LEU88, PHE91, ILE95, TYR106, ALA107, CYS110, VAL111, LEU136, GLY137, GLY138, GLU141, PRO160, GLU161, LEU163, LEU164, LEU166, PRO168, GLY169, MET170, THR255, TRP258, ALA262, LEU265, THR272, MET273, LEU276 (Figure 3).
Figure 1.

Sequence alignment between target and template.
Figure 2.
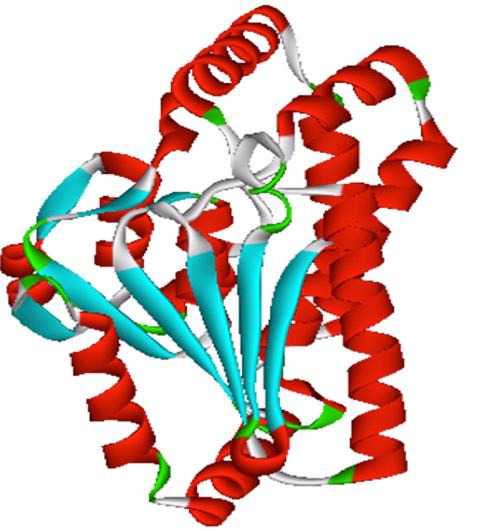
3D structure of DSF synthase RpfF protein from Xanthomonas oryzae pv. Oryzae
Figure 3.
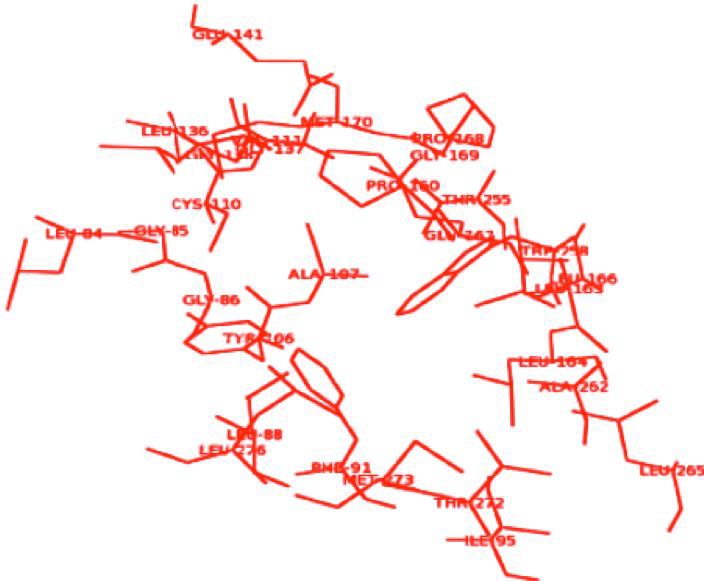
Potential ligand binding pocket residues in RpfF model from Xoo
Conclusion
The study of quorum sensing pathways to identify suitable targets against infectious agents is an effective strategy. The plethora of target enzymes is to be discovered for rice diseases. Molecular model of DSF synthase RpfF from Xanthomonas oryzae pv. Oryzae is predicted using three homology modeling methods which are reliable and it may be proceed for further drug designing studies. Blocking RpfF protein can inhibit the DSF-dependent quorum sensing mechanism and production of required DSF and bacterial population will be altered. This mechanism could control bacterial infection in rice plants. The predicted ligand binding sites in the model will help to develop RpfF specific novel inhibitors that can control the disease conditions.
Supplementary material
Footnotes
Citation:Reddy et al, Bioinformation 8(11): 504-507 (2012)
References
- 1.PJE Velusamy, et al. Can J Microbiol. 2006;52:56. [Google Scholar]
- 2.Y Tagami, T Mizukami. Minist Agric Jpn. 1962;10:1. [Google Scholar]
- 3.SH Tsuge, et al. Phytopathology. 2004;94:478. [Google Scholar]
- 4.TW Mew, et al. Annu Rev Phytopathol. 1967;7:51. [Google Scholar]
- 5.BJ Staskawicz, et al. Science. 2001;292:2285. doi: 10.1126/science.1062013. [DOI] [PubMed] [Google Scholar]
- 6.D Buttner, U Bonas. EMBO J. 2002;21:5313. doi: 10.1093/emboj/cdf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Bing, F Frank. The Ame Phytopath Society. 2004;17:1192. [Google Scholar]
- 8.SB Von Bodman, et al. Annu Rev Phytopathol. 2003;41:455. doi: 10.1146/annurev.phyto.41.052002.095652. [DOI] [PubMed] [Google Scholar]
- 9.YW He, et al. BMC Microbiol. 2010;10:187. doi: 10.1186/1471-2180-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. http://www.ncbi.nlm.nih.gov.
- 11. http://www.rcsb.org/pdb.
- 12.A Fiser, et al. Protein Sci. 2000;9:1753. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A Sali, Blundelll J Mol Biol. 1993;234:779. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 14.Combet, et al. Bioinformatics. 2002;18:213. doi: 10.1093/bioinformatics/18.1.213. [DOI] [PubMed] [Google Scholar]
- 15.K Arnold, et al. Bioinformatics. 2006;22:195. [Google Scholar]
- 16.Y Zhang, J Skolnick. Proteins. 2004;57:702. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 17.J Dundas, et al. Nucleic Acids Res. 2006;34:W116. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


