Abstract
Background
Systolic heart failure (HF) is a complex systemic syndrome that can result from a wide variety of clinical conditions and gene mutations. Despite phenotypic similarities, characterized by ventricular dilatation and reduced contractility, the extent of common and divergent gene expression between different forms of HF remains a matter of intense debate.
Methods and Results
Using a meta-analysis of 28 experimental (mouse, rat, dog) and human HF microarray studies, we demonstrate that gene expression changes are characterized by a coordinated and reciprocal regulation of major metabolic and signaling pathways. In response to a wide variety of stressors in animal models of HF, including ischemia, pressure overload, tachypacing, chronic isoproterenol infusion, Chagas disease, and transgenic mouse models, major metabolic pathways are invariably downregulated, while cell signaling pathways are upregulated. In contrast to this uniform transcriptional pattern observed in experimental animal models which recapitulates a fetal gene expression program, human HF microarray studies displayed a greater heterogeneity, with some studies even showing upregulation of metabolic and downregulation of signaling pathways in end-stage human hearts. These discrepant results between animal and human studies are due to a number of factors, prominently cardiac disease and variable exposure to cold cardioplegic solution in non-failing human samples which can downregulate transcripts involved in oxidative phosphorylation (OXPHOS) within the first 6h, thus mimicking gene expression patterns observed in failing samples. Additionally, beta-blockers and ACE-inhibitor use in end-stage human HF was associated with higher levels of myocardial OXPHOS transcripts, thus partially reversing the fetal gene expression pattern. In human failing samples, downregulation of metabolism was associated with hemodynamic markers of disease severity.
Conclusions
Irrespective of the etiology, gene expression in failing myocardium is characterized by downregulation of metabolic transcripts and concomitant upregulation of cell signaling pathways. Gene expression changes along this metabolic-signaling axis in mammalian myocardium are a consistent feature in the heterogeneous transcriptional response observed in phenotypically similar models of HF.
Keywords: Heart Failure, Fetal Gene Program, Oxidative Phosphorylation
Systolic heart failure (HF), a leading cause of mortality in the Western world, is a complex clinical syndrome of diverse etiologies, characterized by impaired contractile performance of the left ventricle (LV) and commonly, dilatation of the cardiac chambers. Studies of individual signaling pathways at the genetic level have provided significant insight into the development of HF. However, it has become increasingly evident that, despite a common phenotype, various HF models and end-stage human HF display divergent mRNA expression profiles,1 thus limiting our understanding of the transcriptional basis of this complex cardiovascular syndrome. Depending on the underlying etiology, cardiac hypertrophy and failure involve distinct and shared signaling pathways which ultimately lead to a common HF phenotype.1–5 Additionally, important differences have been noted between experimental models of LV dysfunction and end-stage human HF. While experimental models have reliably shown a downregulation of major metabolic pathways in failing myocardium,1, 2, 6–9 opposite results have been documented for end-stage human HF.10–15 In order to explore these apparent discrepancies, we performed a comparison of animal models of LV dysfunction and human end-stage HF using a comprehensive pathway analysis.
Previously, we have identified a common theme across heterogeneous microarray studies, exemplified by a robust genome-wide inverse regulation of metabolic and cell signaling pathways: We found that upregulation of cell signaling pathways was accompanied by downregulation of cell metabolic transcriptional activity (and vice versa).16 Importantly, this coordinated transcriptional pattern occurred in a wide variety of physiological and pathophysiological conditions and was identified across 20 human and animal tissue types examined. Moreover, the differences in metabolic gene expression predicted the magnitude of differences for signaling and all other pathways, i.e. tissue samples with similar expression levels of metabolic transcripts did not show any differences in gene expression for all other pathways. In addition, this transcriptional pattern predicted a profound effect on the proteome, evident by differences in structure, stability and post-translational modifications of proteins belonging to signaling and metabolic pathways, respectively. Previously, we demonstrated the general occurrence of this transcriptional pattern across a wide range of different tissues.16 In the present manuscript, we tested the hypothesis that the inverse regulation of metabolic and signaling pathways will help to refine the distinction between diseased and non-diseased hearts, by examining 28 microarray datasets of failing and non-failing myocardial tissue. Furthermore, we sought to delineate the transcriptional networks involved in the myocardial response to a wide variety of stressors that are shared in experimental animal models of LV dysfunction and human HF.
Methods
Gene Expression Data
Public datasets were obtained from the GEO database.17 734 microarray studies containing the word “heart” in the study description were identified. Of these, we considered only studies for our meta-analysis that (a) compared two different sets of heart samples, i.e. studies comparing myocardial tissue to other organs were excluded (~300 studies remaining), (b) contained at least 5 samples per group (~100 studies), and (c) used microarrays with whole-genome coverage (≥ 20,000 transcripts). Series matrix files of all 68 studies that fulfilled these criteria were downloaded from GEO and used in the original format submitted by the authors (normalized data). 15 studies were subsequently excluded as testing for differentially expressed genes yielded only very few transcripts (<200 genes), thus precluding a comprehensive pathway analysis. An additional 5 studies were excluded as the mean signal intensity between the two experimental groups differed by more than 20%, raising concerns regarding data normalization and/or technical problems with hybridization. The remaining 48 studies of myocardial tissue included 28 HF studies which are listed in Supplemental Table 1. In order to demonstrate that the transcriptional theme identified in the current study applies to a wide range of disease conditions, we also included several additional microarray studies which examined rare conditions for which studies with whole-genome coverage (e.g. comparison of human fetal and adult myocardium) or with more than 5 samples per group were not available (e.g. Chagas disease and effect of ACE-inhibitor treatment in HF).
Statistical Analysis
All 48 studies were first analyzed separately using the original Series Matrix File data format provided in GEO. As a result, data in the current study included different methods of normalization (quantile, MAS5, Lowess, VSN). To determine differentially expressed genes, two-class Significance Analysis of Microarrays (SAM) was used.18 All samples in a given control or experimental group were used, as detailed in Supplemental Table 1. Differences in gene expression were regarded as statistically significant if a false discovery rate (FDR) of q<0.05 was achieved. Comparison of different studies was then based on functional annotation of differentially expressed genes using the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways database.19 Overrepresentation of specific KEGG pathways in a gene set was statistically analyzed by the Database for Annotation, Visualization and Integrated Discovery (DAVID).20 In order to include the maximum number of KEGG pathways per study, the default parameters for threshold were changed (count = 1, EASE-score = 10). Subsequently, we focused our analysis on 160 major KEGG pathways that were consistently represented across all 48 studies. In order to enable comparison of KEGG pathways representing a different number of genes, the net expression of a pathway was defined as the number of up- minus downregulated transcripts expressed as percentage of the total number of genes within a KEGG pathway.16 Net expression levels of the 160 KEGG pathways were then color-coded and visualized using the Genesis software package.21
Non-parametric tests (Kolmogorov-Smirnoff and Wilcoxon) incorporated into StatView (SAS Institute Inc., NC, USA) were used to determine statistical significance for clinical parameters and individual genes in the largest human HF microarray dataset (GSE5406).
Analysis and graphical representations of protein-protein interactions were performed using the “Unified Human Interactome Database” (UniHI4 Express)22 which contains over ~253980 distinct interactions between ~22300 unique proteins. UniHI Express allows the filtering of protein-protein-interactions based on gene expression in selected tissues and thus enables the construction of tissue-specific networks, by integrating gene expression data from the “Human Gene Atlas” (http://biogps.gnf.org). Interacting partners of the list of query proteins are displayed if they have an RNA expression in heart tissue of more than 1000 arbitrary units (expression summaries for 44,775 transcripts were derived utilizing the MAS5 algorithm by Affymetrix). The display was restricted to direct interactions between query proteins.
Semiquantitative real-time RT-PCR
Adult rat hearts were either immediately snap-frozen or perfused with St. Thomas Hospital cardioplegic solution23 and stored at 4°C for 4–10 hours. RNA isolation, reverse transcription and SYBR-green semiquantitative RT-PCR were carried out as previously described.24, 25 Primers were designed using the NCBI primer design tool “Primer-BLAST”. Specificity of the primers was confirmed by a single band of the PCR product on an agarose gel and a single peak of the dissociation curve (SYBR-Green RT-PCR). Gene expression was normalized to ribosomal protein 18S and expressed as ratio of gene expression at the 4h or 10h time-points relative to control, i.e. snap-frozen tissue.
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
Inverse Regulation of Metabolic and Cell Signaling Pathways
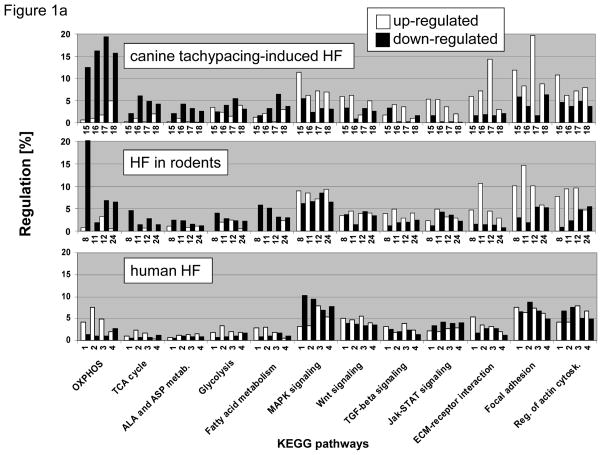
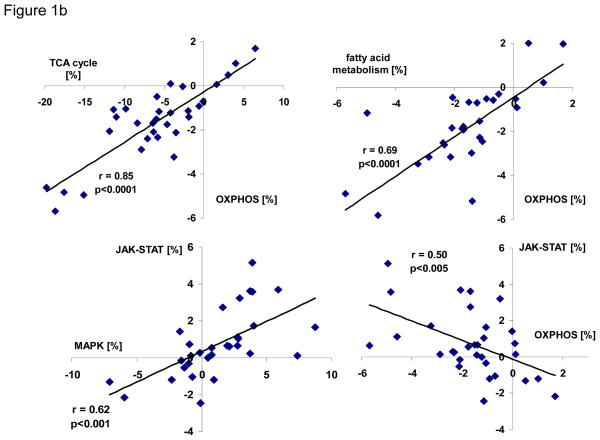
Comparison of changes in KEGG pathway expression in experimental models of LV dysfunction revealed important differences when compared to analogous studies of end-stage human HF. In animal models of HF, transcripts encoding metabolic pathways, including oxidative phosphorylation (OXPHOS), glucose-, fatty acid- and amino acid metabolism were greatly downregulated, while various cell signaling pathways and extracellular matrix components were upregulated. In contrast, in end-stage failing human hearts, expression of metabolic pathways was variable; however, metabolic and major signaling pathways (e.g. MAPK and JAK-STAT-pathways) showed a reciprocal pattern of expression (Figure 1a). When the net expression between the JAK-STAT- and oxidative phosphorylation (OXPHOS) pathways was compared across all 28 human and animal microarray HF studies, i.e. the number of up- minus downregulated genes within a study in relation to total number of genes per given KEGG pathway, a negative correlation was found (Figure 1b, right lower panel), whereas TCA-cycle, fatty acid metabolism and OXPHOS pathways showed a positive correlation (Figure 1b, upper panels). To extend this observation beyond individual pathways, the net expression of all 160 KEGG pathways present in mammalian myocardium was color-coded and depicted as a heat map where KEGG pathways were sorted based on their similarity to OXPHOS expression. A highly coordinated transcriptional response of mammalian myocardium becomes apparent with inverse regulation of metabolic and signaling processes (Figure 2). It is of interest to note that the tight regulation extended beyond KEGG pathways important for metabolic and signaling functions, as evident by the positive correlation between OXPHOS and transcripts belonging to the “proteasome”. In contrast, structural components important for cell-cell contact (e.g. “cell adhesion molecules”, “tight junctions”, “adherens junctions”, “focal adhesion”) and ribosome were negatively correlated with OXPHOS (Supplemental Figure 1).
Figure 1. A) KEGG pathway analysis of heart failure (HF).
Transcripts up- and downregulated in HF compared to non-failing (NF) ventricular myocardium are represented by white and black columns, respectively and displayed on the y-axis as percentage of the total number of transcripts per given KEGG pathway. Four different microarray studies of canine tachypacing-induced HF microarray studies are presented in the top panel (studies #15–18), four different HF models in rodents are shown in the middle graph (studies #8, #11, #12, #24), and four different microarray studies of human HF are displayed in the bottom panel (studies #1–4). A detailed description of these studies is given in Supplemental Table 1. B) When the net regulation, i.e. the number of up- minus downregulated genes within a study expressed as percentage of genes per given KEGG pathway (this corresponds to white minus black columns of Figure 1a), was compared between TCA-cycle, fatty acid metabolism and OXPHOS, a positive correlation was found (upper panels, p<0.05 for all correlations), while JAK-STAT-signaling and OXPHOS show a negative correlation (lower right panel). The correlation plots include all 28 animal and human HF microarray studies listed in Supplemental Table 1.
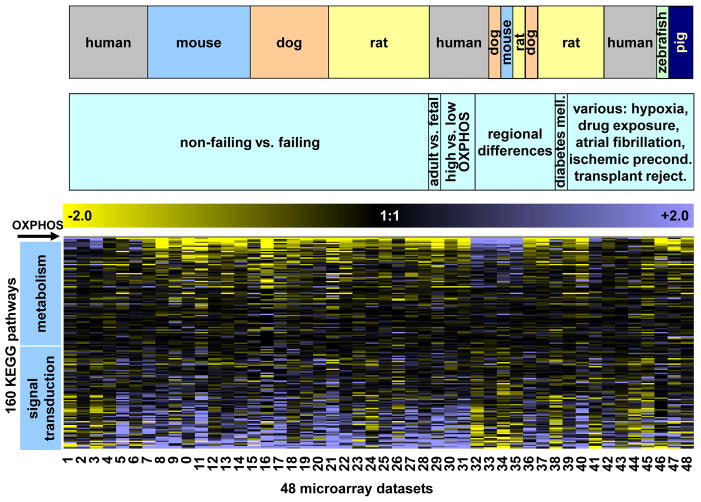
Figure 2. Comprehensive analysis of 160 KEGG pathways expressed in 48 different myocardial gene expression datasets.
The net expression of a KEGG pathway (number of up- minus downregulated genes within a study in relation to total number of genes per given KEGG pathway, see also Figure 1), is color-coded with yellow and blue representing low and high expression of the pathway, respectively. The net expression of 160 different KEGG pathways is depicted on the y-axis and sorted according to their similarity of gene expression to “oxidative phosphorylation” which is represented by the first row (labeled OXPHOS). 48 different microarray studies, described in detail in Supplemental Table 1, are shown on the x-axis: Samples #1–#28 are from datasets comparing non-failing vs. failing myocardial samples. Within these heart failure samples, samples from the four different species were grouped together. For samples #29–#48, we grouped samples according to the pathophysiology (e.g. regional differences, high vs. low OXPHOS, etc.) and then by species. Across a wide range of diverse myocardial gene expression datasets, metabolic and biosynthesis pathways were consistently positively correlated to each other and negatively correlated with the expression of cell signaling pathways.
Taken together, these data suggest that, irrespective of the etiology of HF, gene expression changes in mammalian myocardium are characterized by an inverse transcriptional regulation of major metabolic and cell signaling pathways. Importantly, transcriptional changes along this developmental axis accounted for >80% of transcriptional alterations in HF (as defined by the number of genes in KEGG pathways that show a statistically significant Pearson correlation coefficient to OXPHOS KEGG pathway expression, p<0.05). In addition to HF, an inverse regulation of metabolic and cell signaling pathways was also observed in atrial fibrillation, and regional myocardial gene expression patterns (atrial vs. ventricular gene expression), highlighting that this coordinated transcriptional pattern occurred in a wide variety of physiological and pathophysiological conditions. For instance, Figure 2 shows that metabolic pathways are more abundant in ventricular compared to atrial myocardium (datasets #32–34; blue indicates higher expression in the ventricle and yellow indicates lower expression in the atria). Likewise, metabolic pathways were upregulated in atrial myocardium from patients with chronic atrial fibrillation compared with patients in normal sinus rhythm (Figure 2, dataset #44).
Variability of the Fetal Gene Expression Pattern in Human Myocardium
Reactivation of the fetal gene program is regarded as a hallmark of HF, and is characterized by alterations in the expression of contractile α- and β-myosin heavy chain (MHC) isoforms, downregulation of metabolic genes and upregulation of natriuretic peptides.26 Additionally, we found that the highly coordinated regulation of metabolic and cell signaling pathways shows a striking similarity to the fetal gene program (Figure 2, dataset #29). In contrast to the high concordance between the fetal gene program and experimental models of HF, we noted substantial variability in expression of a fetal gene program in end-stage human myocardium. Out of six human HF datasets examined, a fetal gene expression program comparable to experimental animal HF models was only observed twice (Figure 2, datasets #4 and #5), while the remaining studies showed either the opposite pattern or very little change in gene expression for cell signaling and metabolic pathways. No significant etiology-specific gene expression patterns were observed when ischemic and non-ischemic HF samples were analyzed separately in the two largest human microarray datasets (Supplemental Figure 2).
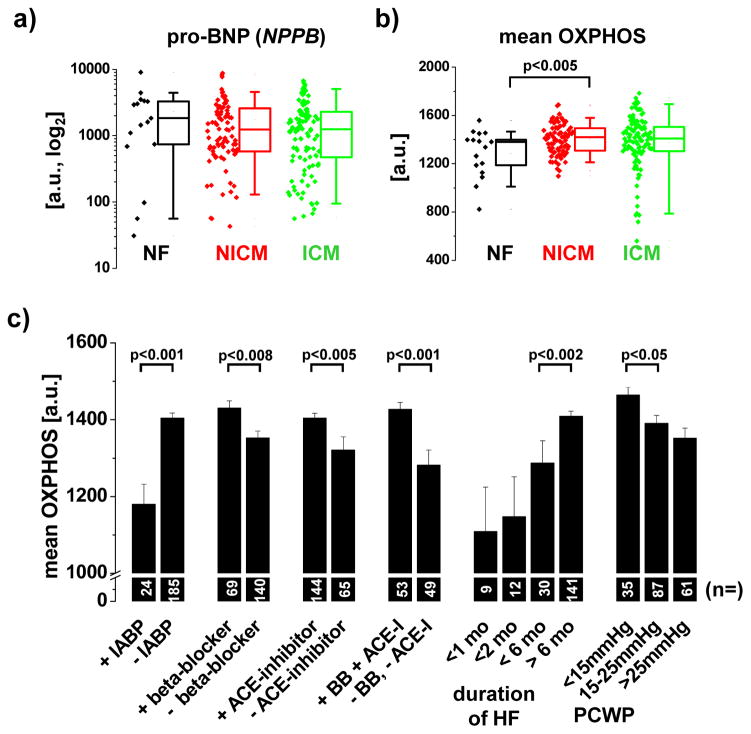
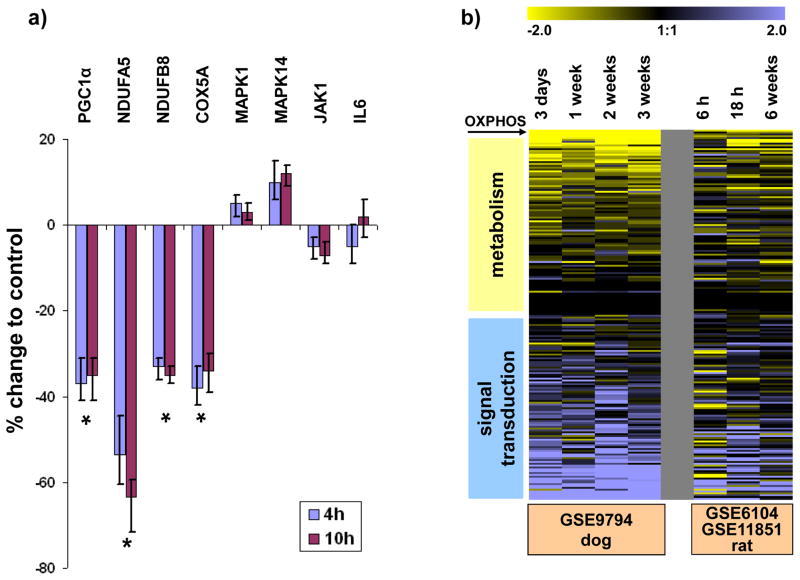
The striking difference between myocardial gene expression in human and experimental HF is likely to be multifactorial. We tested the hypotheses that the nature of human controls, differences in tissue preparation and medication use contribute to heterogeneity of results in human tissue relative to animal models. Control human hearts, while non-failing, usually display some pathology, e.g. cardiac hypertrophy,27 valvular heart disease or coronary atherosclerosis, so that these organs are deemed unsuitable for transplantation; indeed some of the highest pro-brain natriuretic peptide (pro-BNP) values in the largest human dataset (GSE5406, dataset #3) were found in the group of “non-failing” myocardial samples (Figure 3a and Supplemental Figure 3). Thus, some of these “non-failing” samples are not appropriate controls, but rather skew the observed transcriptional pattern. In addition, different tissue handling of non-failing tissue samples in human vs. experimental models could account for the observed differences. While in experimental HF models, tissue samples are typically directly snap-frozen, human myocardial samples are usually perfused with and stored in cold cardioplegic solution in order to limit ischemic tissue damage. When we perfused non-failing rat hearts with cold (4°C) St. Thomas Hospital cardioplegic solution,23 we found significant downregulation of OXPHOS gene expression after 4 hours (Figure 4a). Thus, downregulation of OXPHOS gene expression, caused by tissue handling of human donor hearts, could account for the different gene expression profiles observed in human HF datasets as compared to experimental HF models. The rapid downregulation of OXPHOS gene expression was also noted in two animal models, examining the early vs. late transcriptional changes in HF: Downregulation of metabolic and upregulation of signaling pathways was observed as early as 6 hours after the onset of volume-overload in right ventricular myocardium in rats and 3 days after initiation of tachypacing in dogs (i.e. earliest time point examined), consistent with the notion that gene expression changes in myocardium are dynamic and can occur rapidly after a perturbation of the physiological environment (Figure 4b).
Figure 3. Correlation of OXPHOS gene expression and clinical parameters in human HF.
The analysis utilizes the largest publicly available microarray dataset of human HF (GSE5406, dataset #3). A and B) In 210 human HF samples, pro-BNP (NPPB) and OXPHOS expression (average expression of all genes comprised in the KEGG pathway of oxidative phosphorylation) displayed a high variability, independent of the etiology of HF (NF = non-failing, NICM = non-ischemic cardiomyopathy, ICM = ischemic cardiomyopathy). Boxplots delineate the median value as well as the 25th and 75th percentiles. Raw data for NPPB and OXPHOS are plotted as diamonds next to the boxplots. C) Patients on mechanical circulatory support (IABP, intra-aortic balloon pump) had lower myocardial expression of OXPHOS genes compared to more hemodynamically stable patients. Likewise, patients with a rapidly progressive course of HF (less than one month between clinical onset of HF and cardiac transplantation) and a higher pulmonary capillary wedge pressure (PCWP) had significantly lower OXPHOS expression compared to patients with a longer clinical course and lower PCWP, respectively. In contrast, use of beta-blockers and ACE-inhibitors was associated with higher myocardial OXPHOS levels. The number of patients is indicated on the respective columns.
Figure 4. A) Effect of hypothermia and cardioplegic solution on myocardial gene expression in rats.
Expression of eight transcripts was measured with SYBR-green semiquantitative RT-PCR in control hearts that were immediately snap-frozen after harvest or perfused with St. Thomas Hospital cardioplegic solution and stored at 4°C for 4–10 hours. Significant downregulation of four OXPHOS transcripts (PGC1α, NDUFA5, NDUFB5, COX5A) was noted in left ventricular myocardium perfused with cold cardioplegic solution (p<0.05, unpaired t-test, n=4 hearts each group), while there was no statistically significant change in mRNA expression for transcripts involved in signal transduction pathways (MAPK1, MAPK14, JAK1, IL6). B) Concordant regulation of pathways in early and late HF. Characteristic HF-induced gene expression changes develop as early as 3 days after initiation of tachypacing in dogs (left panel) or 6 hours after RV pressure overload induced by experimental pulmonary embolism (PE) in rats.
Finally, the heterogeneity of the fetal gene program across different human HF datasets prompted us to test the hypothesis that the fetal gene expression profile, defined by upregulation of signaling and downregulation of metabolism, is variably expressed among HF patients. Using the mean expression of >100 OXPHOS transcripts as a surrogate of OXPHOS gene expression, we found that OXPHOS expression displays substantial variability across 210 human samples in the largest publicly available HF dataset (GSE5406, dataset #3, Figure 3b). Consistent with an upregulation of metabolic pathways in human HF in this study, we noted that non-failing samples had a significantly lower expression of OXPHOS transcripts compared to non-ischemic failing samples. Importantly, by comparing KEGG pathways in patients showing the lowest and the highest OXPHOS expression, we obtained a pattern similar to the fetal gene expression program and to experimental models of HF, highlighting that metabolic processes (including OXPHOS) and cell signaling pathways are inversely regulated (Figure 2, datasets #30 and #31).
Ultimately, the finding of greater heterogeneity of OXPHOS expression in human HF compared to experimental animal models (Figure 2) raises the question why the fetal gene program is present in myocardium of only few HF patients, while it is pronounced in virtually all animal HF models examined. While we didn’t find any significant effect of age, gender, ethnicity, or diabetes mellitus on OXPHOS expression in 210 patients (GSE5406, dataset #3, data not shown), we found a significant correlation between OXPHOS and various clinical parameters of disease severity. For instance, expression of OXPHOS and pro-BNP (NPPB) were significantly negatively correlated (Supplemental Figure 4). In addition, patients requiring mechanical circulatory support (IABP, intra-aortic balloon pump) had lower myocardial expression of OXPHOS genes compared to hemodynamically stable patients (Figure 3c and Supplemental Table 2). Likewise, patients with a rapidly progressive course of HF who required cardiac transplantation within one month of the onset of clinical symptoms had significantly lower myocardial OXPHOS expression compared to patients with a longer clinical course. Similarly, a higher pulmonary capillary wedge pressure (PCWP) was associated with reduced OXPHOS expression, while the use of beta-blockers and ACE-inhibitors lead to higher myocardial OXPHOS levels (Figure 3c). Taken together, these results suggest that expression of a fetal gene expression program in HF is linked to disease severity.
Network Analysis of Individual Transcripts Common to Human and Experimental HF Models
In order to identify genes which might regulate the highly coordinated transcriptional program between metabolic and signaling pathways, we first sought to identify a core set of transcripts which are consistently regulated across different HF models. By limiting our analysis to 10 HF datasets showing the largest differences between metabolic and signaling pathways in HF (Figure 2, datasets 7, 8, 11, 15–17, 25, 28, 30, 31), we found 135 and 545 transcripts to be consistently up- and downregulated, respectively, in at least 5 of the 10 HF studies examined (Table 1 and Supplemental Table 3). Downregulated transcripts included known marker genes of HF, including β1-adrenergic receptor (ADRB1), endothelin receptor (EDNRA), ryanodine receptor (RYR2), sarcoplasmatic Ca2+-ATPase SERCA2 (ATP2A2) and its regulator phospholamban (PLN), PGC1α (PPARGC1A) and more than two dozen OXPHOS transcripts (NDUFA1, -A2, -A3, -A5, -A7, -A12, -AB1, -AF1, -B8, -B9, -B10, -B11, -S1, -S2, -S3, -S4, -S6, -S8, -V1, -V2, ATP5A1, -5C1, -5D, -5F1, -5G1, -5J). Upregulated transcripts reflected prominent extracellular remodeling in HF and included various collagen transcripts (COL1A2, 4A1, -4A2, -5A1, -5A2, -6A3, -16A1), connective-tissue growth factor (CTGF), fibronectin 1 (FN1) and asporin (ASPN).
Table 1.
Transcripts consistently regulated across 10 HF studies.
| number of studies | common UP-regulated genes | common DOWN-regulated genes |
|---|---|---|
| 10 | - | - |
| 9 | - | 7* |
| 8 | 1* | 31* |
| 7 | 7* | 67* |
| 6 | 25* | 139 |
| 5 | 102 | 301 |
| regulated in at least 5/10 studies | Σ135 | Σ545 |
query genes for protein-protein interaction network (Figure 6)
Next, we hypothesized that the robust reciprocal transcriptional regulation between metabolic and signaling pathways would also be evident in network models that incorporate prior knowledge of protein-protein interactions. In particular, we were interested to define a set of proteins that mediate the cross-talk between signaling and metabolic pathways. Therefore, we searched for protein-protein interaction partners for the 90 most consistently up- and downregulated transcripts using the Unified Human Interactome database (UniHI4)22.
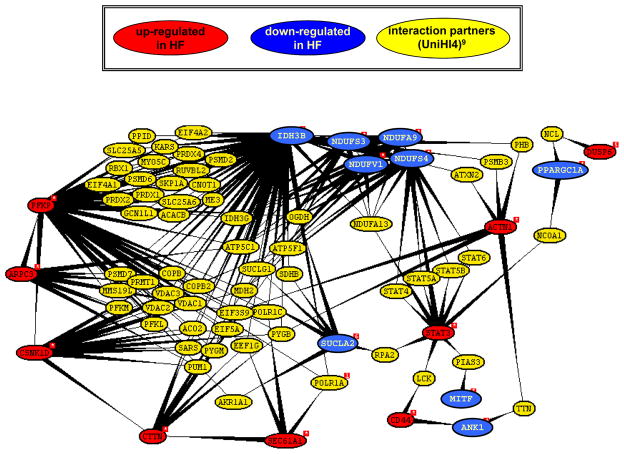
Consistent with the scale free dynamics of biological networks, we identified a small fraction of 18 highly interconnected nodes (“hubs”) linking the 90 most consistently up- and downregulated transcripts (Figure 5). The resulting protein-protein interaction network included the upregulated transcripts phosphofructokinase (PFKP), casein kinase 1δ (CSNK1D), cortactin (CTTN), actin related protein 2/3 complex, subunit 3, 21kDa (ARPC3) and signal transducer and activator of transcription 3 (STAT3) on the one hand and the downregulated OXPHOS transcripts (NDUFA9, -S3, -S4, V1) and IDH3B, encoding for the TCA-cycle enzyme isocitrate dehydrogenase 3 beta, on the other hand. Importantly, many of the predicted interaction partners, were also part of the common HF genes listed in Supplemental Table 3 and included peroxiredoxin 2 (PRDX2), the voltage-dependent anion-selective channel proteins 1, 2, and 3 (VDAC1, -2, -3) as well as the metabolic genes aconitase 2 (ACO2), isocitrate dehydrogenase 3 gamma (IDH3G), malate dehydrogenase 2 (MDH2), succinate dehydrogenase complex, subunit B (SDHB), alpha subunit of succinate-CoA ligase (SUCLG1), subunits of the F0-F1 ATPase (ATP5A1, -F1) and proteasomal subunits (PSMD7).
Figure 5. Graphical representations of protein interactions.
A query was conducted with 90 transcripts consistently regulated in HF (listed in Supplemental Table 3) using the Unified Human Interactome database (UniHI4)22. Interacting partners of the list of query proteins are displayed if they have an expression in heart tissue of more than 1000 units (expression summaries for 44,775 transcripts were derived utilizing the MAS5 algorithm by Affymetrix).22 The display was restricted to direct interactions (yellow proteins) between query proteins (red and blue for up- and downregulated transcripts, respectively). Network analysis revealed that 18 out of 90 transcripts formed a network with multiple interconnected nodes (hubs) between up- and downregulated genes. Importantly, many of the predicted interaction partners, were also part of the common HF genes listed in Supplemental Table 3.
Discussion
Inverse Regulation of Metabolic and Cell Signaling Pathways
Cells must coordinate adjustments in genome expression to accommodate changes in their environment. Using a meta-analysis of 48 myocardial microarray studies, we show an inverse correlation between signaling and metabolic pathways in mammalian myocardium (Figures 1 and 2). This transcriptional program is present in response to a wide range of divergent environmental stimuli, such as tachycardia, pressure overload, and ischemia in the ventricular myocardium of small and large mammals and constitutes a reversal of the well-defined developmental transition from fetal to adult phenotype. In addition to the myocardium, we previously found the inverse regulation of signaling and metabolism to be present in 20 extramyocardial tissues,16 suggesting a canonical theme in mammalian transcriptional regulation. This is supported by the fact that the changes in steady-state mRNA levels also have a profound effect on the expressed proteome, as KEGG cell signaling pathways are characterized by an increased magnitude of intrinsically unstructured proteins (IUPs) compared to metabolic and biosynthetic pathways.16 IUPs are defined by a lack of a rigid 3D structure and conformational flexibility, which facilitates interaction with multiple targets. These properties are generally ascribed to proteins that mediate signaling, transcription and coordinate regulatory events, where binding to multiple partners in the context of high-specificity/low-affinity interactions play a crucial role.28 Of note, an increase of IUPs has been associated with perturbed cellular signaling in a wide range of pathological conditions such as cancer, diabetes, and neurodegenerative diseases. The upregulation of signaling pathways and IUPs in HF is therefore in good agreement with these findings.
Network analysis identified STAT3 as an important “hub”, linking signaling pathways directly to OXPHOS activity (Figure 5). STAT3 is required for optimal function of the electron transport chain (ETC) in the inner mitochondrial membrane. In mice that do not express STAT3 in the heart, there were selective defects in the activities of complexes I and II of the ETC.29 Thus, upregulation of STAT3 could represent an attempt to compensate for downregulation of OXPHOS activity. While these data indicate that JAK-STAT signaling might be important for maintaining OXPHOS activity, there is also evidence to support the thesis that upregulation of signaling pathways might contribute to downregulation of OXPHOS and metabolic pathways. In fact, in transgenic mouse models overexpressing the p38 MAPK activator MAPK kinase 6 (MKK6), a profound downregulation of OXPHOS and metabolic gene expression was noted.30 Likewise, Crunkhorn and colleagues have shown that PGC1α, a major regulator of mitochondrial biogenesis and respiration in cardiac muscle,31–33 can be downregulated by overexpressing p38-MAPK.34 Thus, upregulation of MAPK-signaling seems to be sufficient to downregulate OXPHOS activity. The different effects of specific signaling pathways (JAK-STAT vs. MAPK) on OXPHOS gene expression underscore that metabolic genes are regulated in the context of complex signaling networks. Further studies are clearly necessary to elucidate the relative importance of various signaling pathways in controlling OXPHOS and metabolic gene expression.
In summary, we provide evidence for inverse regulation of metabolic and cell signaling pathways in a wide variety of physiological and pathophysiological conditions in the myocardium, including HF, atrial fibrillation, cardiac development, and regional gene expression patterns, suggesting a common theme for myocardial transcriptional plasticity.
Variability of the Fetal Gene Expression Pattern in Human Myocardium
This meta-analysis revealed two findings related to human HF microarray datasets. First, we noted that experimental animal models and human HF showed divergent myocardial expression patterns. The reasons for this divergence are almost certainly multi-factorial and include differences in the duration of HF and a multitude of confounding clinical variables that contribute to heterogeneity in human studies (age, sex, ethnicity, genetic variation, medication, etiology of HF, diabetes, etc.) compared to well-controlled animal models. For instance, patients with a longer clinical course of HF showed higher myocardial OXPHOS mRNA levels than patients with a more acute disease progression. Therefore, compensatory mechanisms might be operative, partly reversing the fetal gene expression pattern in these patients. Likewise, the use of beta-blockers and ACE-inhibitors was associated with higher myocardial OXPHOS levels (Figure 3c). In contrast, most animal studies employ relatively acute HF models without any treatment. Consistent with the human data, a partial reversal of the fetal gene expression pattern was observed in rats treated with ACE-inhibitors and thyroid hormone analogs after a myocardial infarction (Figure 2, dataset #23: infarct only, and #24: infarct + ACE-inhibitor/T3 analogs).
Alternatively, one has to keep in mind that the human non-failing samples used as controls often display a variable degree of cardiac pathology27 and that handling of human and animal non-failing myocardial tissue samples differs substantially. In well-controlled laboratory studies myocardial specimens are snap-frozen immediately after explantation, while human non-failing samples are perfused with hypothermic cardioplegic solutions in order to limit ischemic damage and generally stored for longer times before freezing. This time delay and the tissue preservation techniques are critical factors contributing to alterations of the myocardial gene expression signature, as documented by pronounced downregulation of OXPHOS gene expression within 4 hours of storage in cold cardioplegic solution. Given that the number of failing tissue samples usually greatly outnumbers the number of NF samples, even a few non-failing samples with a “hypertrophic” or “fetal” gene expression pattern will substantially influence the observed gene expression pattern. Consistent with the idea that non-failing human samples often display a greater heterogeneity in gene expression than failing samples, we previously reported that only 1 of 196 failing, but 3 of 14 non-failing samples were misclassified in dataset GSE5406 using a genomic classifier of human HF that achieved an overall prediction accuracy of >96% in several independent human HF microarray studies.4, 10 Our results highlight the difficulties of interpreting results from “real-world” human clinical samples and stress the importance of well-controlled animal models to elucidate disease mechanisms.
Second, we found great variability in the levels of OXPHOS across failing HF specimens. In myocardium, reversal to a fetal gene expression pattern is largely seen as an adaptive process that facilitates short-term survival.26 For instance, the switch in expression of contractile α- and β-MHC isoforms, considered to be a hallmark of the fetal gene expression program, reflects a relative increase of the slow β-over the fast α-MHC isoform in order to economize on energy expenditure.35 Likewise, upregulation of the natriuretic peptides is thought to increase sodium diuresis and counterbalance the effect of increased levels of catecholamines. While return to a fetal gene program seems to be essential for short-term adaptation to cardiac stress, it could prove to be maladaptive in the long run, leading to activation of multiple signaling pathways and an increase in the number of IUPs which are strongly associated with various disease processes. In agreement with this notion, we found that decreased expression of OXPHOS mRNA was associated with the severity and prognosis of HF (Figures 3c–3e) in independent studies. Therefore, it provides the rationale for exploring the potential of OXPHOS and metabolic genes to serve as biomarkers of heart disease and response to therapy in prospective studies.
Supplementary Material
Acknowledgments
Funding Sources
The work was supported by NIH P01 HL077180, HL072488, R33 HL087345 and RC1HL099892 to G.F.T., R01 AG17022 to K.B.M., R01 HL088577 and R21 HL092379 to T.P.C., R01 DA023879 to C.F. and NIH T32 HL007227 to A.S.B. G.F.T. is the Michel Mirowski M.D. Professor of Cardiology.
Footnotes
Disclosures
None
References
- 1.Sharma UC, Pokharel S, Evelo CT, Maessen JG. A systematic review of large scale and heterogeneous gene array data in heart failure. J Mol Cell Cardiol. 2005;38:425–432. doi: 10.1016/j.yjmcc.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 2.Akavia UD, Benayahu D. Meta-analysis and profiling of cardiac expression modules. Physiol Genomics. 2008;35:305–315. doi: 10.1152/physiolgenomics.90248.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronow BJ, Toyokawa T, Canning A, Haghighi K, Delling U, Kranias E, Molkentin JD, Dorn GW., 2nd Divergent transcriptional responses to independent genetic causes of cardiac hypertrophy. Physiol Genomics. 2001;6:19–28. doi: 10.1152/physiolgenomics.2001.6.1.19. [DOI] [PubMed] [Google Scholar]
- 4.Kuner R, Barth AS, Ruschhaupt M, Buness A, Zwermann L, Kreuzer E, Steinbeck G, Poustka A, Sultmann H, Nabauer M. Genomic analysis reveals poor separation of human cardiomyopathies of ischemic and nonischemic etiologies. Physiol Genomics. 2008;34:88–94. doi: 10.1152/physiolgenomics.00299.2007. [DOI] [PubMed] [Google Scholar]
- 5.Strom CC, Kruhoffer M, Knudsen S, Stensgaard-Hansen F, Jonassen TE, Orntoft TF, Haunso S, Sheikh SP. Identification of a core set of genes that signifies pathways underlying cardiac hypertrophy. Comp Funct Genomics. 2004;5:459–470. doi: 10.1002/cfg.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barth AS, Aiba T, Halperin V, DiSilvestre D, Chakir K, Colantuoni C, Tunin RS, Dimaano VL, Yu W, Abraham TP, Kass DA, Tomaselli GF. Cardiac resynchronization therapy corrects dyssynchrony-induced regional gene expression changes on a genomic level. Circ Cardiovasc Genet. 2009;2:371–378. doi: 10.1161/CIRCGENETICS.108.832345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanton LW, Garrard LJ, Damm D, Garrick BL, Lam A, Kapoun AM, Zheng Q, Protter AA, Schreiner GF, White RT. Altered patterns of gene expression in response to myocardial infarction. Circ Res. 2000;86:939–945. doi: 10.1161/01.res.86.9.939. [DOI] [PubMed] [Google Scholar]
- 8.Ueno S, Ohki R, Hashimoto T, Takizawa T, Takeuchi K, Yamashita Y, Ota J, Choi YL, Wada T, Koinuma K, Yamamoto K, Ikeda U, Shimada K, Mano H. DNA microarray analysis of in vivo progression mechanism of heart failure. Biochem Biophys Res Commun. 2003;307:771–777. doi: 10.1016/s0006-291x(03)01252-x. [DOI] [PubMed] [Google Scholar]
- 9.Wagner RA, Tabibiazar R, Powers J, Bernstein D, Quertermous T. Genome-wide expression profiling of a cardiac pressure overload model identifies major metabolic and signaling pathway responses. J Mol Cell Cardiol. 2004;37:1159–1170. doi: 10.1016/j.yjmcc.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Barth AS, Kuner R, Buness A, Ruschhaupt M, Merk S, Zwermann L, Kaab S, Kreuzer E, Steinbeck G, Mansmann U, Poustka A, Nabauer M, Sultmann H. Identification of a common gene expression signature in dilated cardiomyopathy across independent microarray studies. J Am Coll Cardiol. 2006;48:1610–1617. doi: 10.1016/j.jacc.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 11.Grzeskowiak R, Witt H, Drungowski M, Thermann R, Hennig S, Perrot A, Osterziel KJ, Klingbiel D, Scheid S, Spang R, Lehrach H, Ruiz P. Expression profiling of human idiopathic dilated cardiomyopathy. Cardiovasc Res. 2003;59:400–411. doi: 10.1016/s0008-6363(03)00426-7. [DOI] [PubMed] [Google Scholar]
- 12.Hwang JJ, Allen PD, Tseng GC, Lam CW, Fananapazir L, Dzau VJ, Liew CC. Microarray gene expression profiles in dilated and hypertrophic cardiomyopathic end-stage heart failure. Physiol Genomics. 2002;10:31–44. [Google Scholar]
- 13.Sanoudou D, Vafiadaki E, Arvanitis DA, Kranias E, Kontrogianni-Konstantopoulos A. Array lessons from the heart: focus on the genome and transcriptome of cardiomyopathies. Physiol Genomics. 2005;21:131–143. doi: 10.1152/physiolgenomics.00259.2004. [DOI] [PubMed] [Google Scholar]
- 14.Steenman M, Chen YW, Le Cunff M, Lamirault G, Varro A, Hoffman E, Leger JJ. Transcriptomal analysis of failing and nonfailing human hearts. Physiol Genomics. 2003;12:97–112. doi: 10.1152/physiolgenomics.00148.2002. [DOI] [PubMed] [Google Scholar]
- 15.Yung CK, Halperin VL, Tomaselli GF, Winslow RL. Gene expression profiles in end-stage human idiopathic dilated cardiomyopathy: altered expression of apoptotic and cytoskeletal genes. Genomics. 2004;83:281–297. doi: 10.1016/j.ygeno.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Barth AS, Kumordzie A, Colantuoni C, Margulies KB, Cappola TP, Tomaselli GF. Reciprocal regulation of metabolic and signaling pathways. BMC Genomics. 2010;11:197. doi: 10.1186/1471-2164-11-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 21.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 22.Chaurasia G, Malhotra S, Russ J, Schnoegl S, Hanig C, Wanker EE, Futschik ME. UniHI 4: new tools for query, analysis and visualization of the human protein-protein interactome. Nucleic Acids Res. 2009;37:D657–660. doi: 10.1093/nar/gkn841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kober IM, Obermayr RP, Brull T, Ehsani N, Schneider B, Spieckermann PG. Comparison of the solutions of Bretschneider, St. Thomas’ Hospital and the National Institutes of Health for cardioplegic protection during moderate hypothermic arrest. Eur Surg Res. 1998;30:243–251. doi: 10.1159/000008583. [DOI] [PubMed] [Google Scholar]
- 24.Barth AS, Kizana E, Smith RR, Terrovitis J, Dong P, Leppo MK, Zhang Y, Miake J, Olson EN, Schneider JW, Abraham MR, Marban E. Lentiviral vectors bearing the cardiac promoter of the Na+-Ca2+ exchanger report cardiogenic differentiation in stem cells. Mol Ther. 2008;16:957–964. doi: 10.1038/mt.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kaab S, Hinterseer M, Kartmann H, Kreuzer E, Dugas M, Steinbeck G, Nabauer M. Reprogramming of the human atrial transcriptome in permanent atrial fibrillation: expression of a ventricular-like genomic signature. Circ Res. 2005;96:1022–1029. doi: 10.1161/01.RES.0000165480.82737.33. [DOI] [PubMed] [Google Scholar]
- 26.Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 2007;12:331–343. doi: 10.1007/s10741-007-9034-1. [DOI] [PubMed] [Google Scholar]
- 27.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., 2nd Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 29.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu XY, Avadhani N, Drake JI, Fawcett P, Lesnefsky EJ, Larner AC. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wall JA, Wei J, Ly M, Belmont P, Martindale JJ, Tran D, Sun J, Chen WJ, Yu W, Oeller P, Briggs S, Gustafsson AB, Sayen MR, Gottlieb RA, Glembotski CC. Alterations in oxidative phosphorylation complex proteins in the hearts of transgenic mice that overexpress the p38 MAP kinase activator, MAP kinase kinase 6. Am J Physiol Heart Circ Physiol. 2006;291:H2462–2472. doi: 10.1152/ajpheart.01311.2005. [DOI] [PubMed] [Google Scholar]
- 31.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: a question of balance. J Clin Invest. 2005;115:547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crunkhorn S, Dearie F, Mantzoros C, Gami H, da Silva WS, Espinoza D, Faucette R, Barry K, Bianco AC, Patti ME. Peroxisome proliferator activator receptor gamma coactivator-1 expression is reduced in obesity: potential pathogenic role of saturated fatty acids and p38 mitogen-activated protein kinase activation. J Biol Chem. 2007;282:15439–15450. doi: 10.1074/jbc.M611214200. [DOI] [PubMed] [Google Scholar]
- 35.Narolska NA, Eiras S, van Loon RB, Boontje NM, Zaremba R, Spiegelen Berg SR, Stooker W, Huybregts MA, Visser FC, van der Velden J, Stienen GJ. Myosin heavy chain composition and the economy of contraction in healthy and diseased human myocardium. J Muscle Res Cell Motil. 2005;26:39–48. doi: 10.1007/s10974-005-9005-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.