Abstract
Tumor necrosis factor receptor superfamily is composed of at least 26 members in the mouse, three of which exist as a cluster within the imprinted Kcnq1 domain on chromosome 7. Tnfrsf22, 23 and 26 contain typical cystein-rich domains and Tnfrsf22 and 23 can bind ligands but have no signaling capacity. Thus, they are assumed to be decoy receptors. The developmental expression profile of these genes is unknown and knowledge of their imprinting patterns is incomplete and controversial. We found that all three genes are expressed during mouse embryonic development, and that they have a strong maternal bias, indicating that they may be affected by the KvDMR, the Kcnq1 imprinting control region. We found expression of an antisense non-coding RNA, AK155734, in embryos and some neonatal tissues. This RNA overlaps the Tnfrsf22 and possibly the Tnfrsf23 coding regions and is also expressed with a maternal bias. We were interested in exploring the evolutionary origins of the three Tnfrsf genes, because they are absent in the orthologous human Kcnq1 domain. To determine whether the genes were deleted from humans or acquired in the rodent lineage, we performed phylogenetic analyses. Our data suggest that TNFRSF sequences were duplicated and/or degenerated or eliminated from the KCNQ1 region several times during the evolution of mammals. In humans, multiple mutations (point mutations and/or deletions) have accumulated on the ancestral TNFRSF, leaving a single short non-functional sequence.
Keywords: development, evolution, expression profile, genomic imprinting, mouse, phylogenetics, tumor necrosis factor receptors
Introduction
Tumor necrosis factor receptor superfamily (TNFRSF) is composed of 29 members in the human, most of which are expressed in the immune system. These are transmembrane proteins with one to four hallmark “cysteine-rich domains” (CRDs) in the extracellular N-terminus.1 These CRDs determine ligand specificity. Less than one third of the members contain the “death domain” in the cytoplasmic tail, a protein interaction domain encoded on a single exon and that may have been captured during evolution of the family. Signaling potential varies, with some receptors having well characterized signaling motifs,2 others that may exist as soluble forms and still others that lack signaling capacity. The latter are generally assumed to be decoys that antagonize their signaling counterparts by interfering with ligand binding.
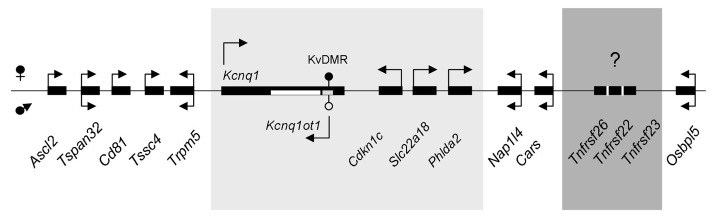
In the mouse, there are at least 26 members of the TNFR superfamily, with Tnfrsf26, 22 and 23 existing as a group within the imprinted Kcnq1 domain on chromosome 7 (Fig. 1). They contain classic CRDs, but do not have signaling capacity. Tnfrsf22 and 23 can bind TRAIL (tumor necrosis factor-related apoptosis-inducing ligand), while Tnfrsf26 remains an orphan receptor.3 These three genes are not present in the syntenic human Kcnq1 region.
Figure 1. Schematic of the Kcnq1 imprinted domain on mouse chromosome 7. Arrows indicate direction of transcription. Arrows above the genes represent maternal transcription, below the line paternal transcription and genes with two arrows have biallelic expression. Shown in the light gray box are the genes that are imprinted in both the embryo and placenta. Shown in the dark gray box are the Tnfrsf genes present in the mouse genome but absent in the human. They are transcribed in the same orientation as Cars.
In mammalian systems, the role of tumor necrosis factor superfamily (Tnfsf) and Tnfrsf genes in the adaptive immune system has been well characterized. Less well studied are their roles in embryonic development, although these may have been the ancestral functions, as they still are in invertebrates.4 Previous reports detected high levels of Tnfrsf23 expression in placenta.5 We investigated whether the Tnfrsf genes were expressed during mouse development in the embryo proper and in neonatal tissues.
Large imprinted domains, such as the Kcnq1 domain, afford a wealth of complexities in their patterns and tissue-specificity. Mechanisms and evolution of imprinting, and escape from imprinting, can be studied by comparing gene arrangements and imprinting patterns between different species. Domain-wide imprinting may have occurred by initially establishing a restricted imprinted region, which eventually spread to neighboring genes. On the other hand, duplications and rearrangements could have juxtaposed imprinted and non-imprinted genomic components and led to acquisition of imprinting as a “bystander” effect. With the advent of publicly available genomic databases and alignment tools, analysis of Tnfrsf genes can yield insights into the evolutionary history of these proteins and their relation to the remainder of the imprinted Kcnq1 domain.
Gene content and arrangement of imprinted domains is highly conserved between human and mouse, so the presence of the three Tnfrsf genes in the mouse and rat and their absence in humans suggested that they could be recent insertions and/or duplications that originated after the divergence of rodents and primates. Our hypothesis was that Tnfrsf genes could have acquired imprinting because of their insertion in the neighborhood of an imprinted region. Here, we investigate this hypothesis and study the evolution and imprinting status of this cluster during development of the mouse embryo.
Results
Tnfrsf23, 22, and 26 genes are imprinted and expressed during embryonic development
To investigate the developmental profile of the cluster throughout embryogenesis, we performed RT-PCR on C57BL/6J ES cells and embryos. Figure 2 summarizes the patterns for each of the Tnfrsf genes. The three genes are expressed in ES cells. After implantation, in E7.5, expression increases steadily as development proceeds for Tnfrsf23 and 26, whereas Tnfrsf22 peaks at E10.5 and slowly declines thereafter. Interestingly, the levels at E7.5 suggest that initiation is progressive, following the linear order in which they are located on the chromosome. All genes are expressed in placenta and in neonatal heart and liver. In agreement with previous studies,3 we found that Tnfrsf23 is expressed in two alternative splice forms, and we determined that the smaller isoform is missing exon 4.
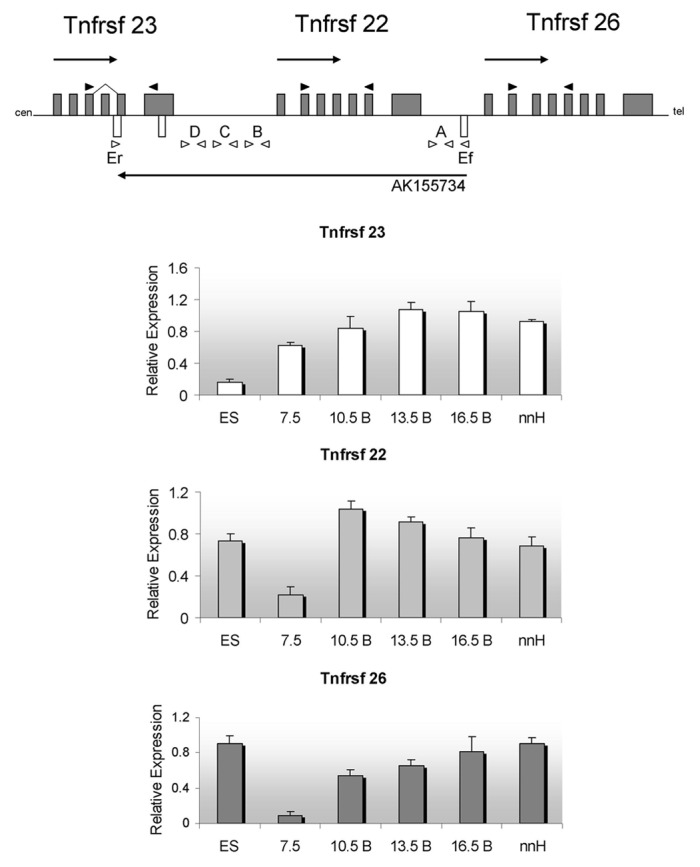
Figure 2. Expression profiles during development for the murine Tnfrsf genes. Top, schematic of the three Tnfrsf genes and their linear organization. Arrows above the line indicate transcriptional direction of Tnfrsf genes, arrow below the line, direction of transcription for AK155734. Cen, centromere; tel, telomere. The Kcnq1 and Kcnq1ot1 genes are telomeric to the Tnfrsf genes. Gray boxes, Tnfrsf exons, white boxes, predicted AK155734 exons. Dark arrows indicate the PCR primers used for the Tnfrsf genes, and white arrows indicate PCR primer sets (A, B, C, D, and Ef/Er) used to determine the presence and splicing of the antisense AK155734 (primer sequences in Table S1). Below, expression levels of each of the indicated Tnfrsf genes relative to Gapdh. ES, mouse embryonic stem cells, 7.5, whole embryos at 7.5 dpc; 10.5 B, 13.5 B, 16.5 B, bodies of embryos at 10.5, 13.5 and 16.5 dpc; nnH, neonatal hearts. Error bars represent standard deviations.
We investigated the allelic pattern for each of the genes in F1 hybrid 13.5 dpc embryos (Fig. 3A) and neonatal tissues (data not shown) from reciprocal crosses of C57BL/6J mice and C57BL/6J mice with a Castaneus chromosome 7, designated as B6(CAST7). Polymorphisms on chromosome 7 allow us to distinguish between the two parental alleles. When expressing RNA levels emanating from the paternal allele as percentages of the maternal RNA levels, all three paralogues showed a strong maternal bias in both crosses, indicating that they are possibly under the control of the KvDMR imprinting control element.
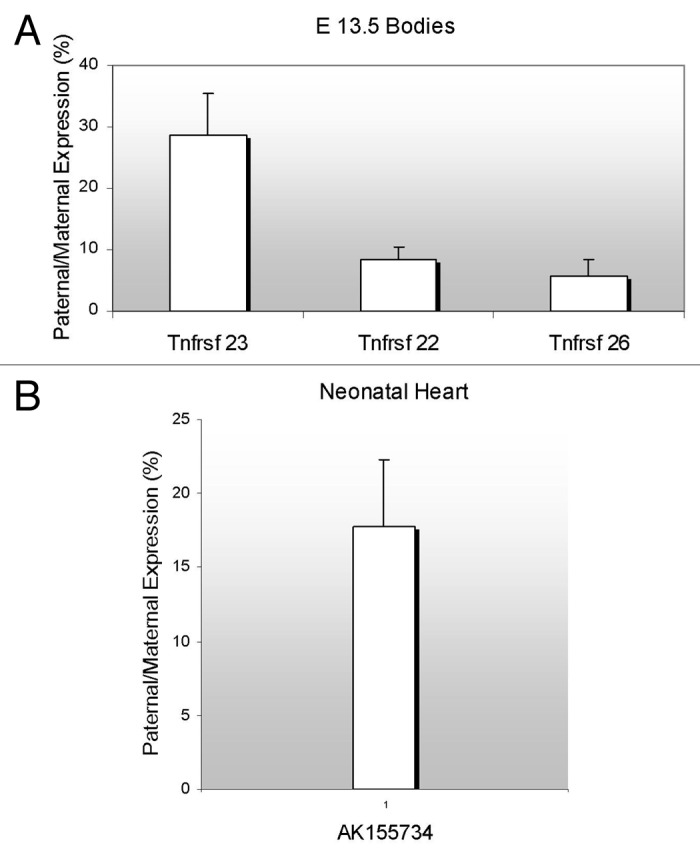
Figure 3. Parent-of-origin expression pattern of murine Tnfrsf genes and the antisense AK155734. (A) RNAs from F1 hybrid E13.5 were subjected to RT-PCR and restriction digests. Allele-specific bands were quantified and the ratio of paternal to maternal transcript was determined. (B) Neonatal heart RNAs from F1 hybrid mice were subjected to RT-PCR and restriction digests, as in A, and allelic ratios were determined. For both (A and B), the results of progeny of C57BL/6J x B6(CAST7) crosses are shown. The reciprocal crosses yielded similar results.
An antisense non-coding RNA (ncRNA) within the Tnfrsf region is initiated, but not transcribed full-length during development, and is imprinted.
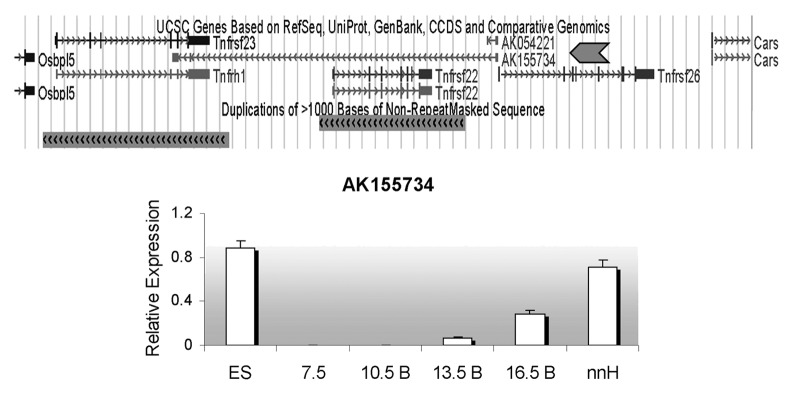
A 42 kb ncRNA, designated as AK155734, is annotated in the UCSC dbase with a start site less than 400 bp away from the Tnfrsf26 promoter and transcribed in antisense direction. AK155734 overlaps the Tnfrsf22 and 23 genes (Fig. 4). Three exons have been predicted to be spliced into a 1 kb mature transcript. To confirm the existence of the ncRNA, we designed primers along the length of the unspliced transcript in intergenic regions and tested RNAs from a range of developmental stages (primer sets A, B, C, D shown in Fig. 2), i.e., ES cells, embryos and neonatal tissues. Primer set A was positive for AK155734 in ES cells, but in embryos, the signal was only present after 13.5 dpc. In both ES cells, embryos and 16.5 dpc placenta, primer sets B, C and D did not produce signals, suggesting that the full-length RNA was not produced. The ncRNA is highly expressed in neonatal liver and heart, and is detectable with all primer sets (Fig. 4).
Figure 4. Detection of an antisense non-coding RNA at the Tnfrsf locus. Top, UCSC browser screen of the three Tnfrsf genes. Dark gray block arrow signals the reported antisense gene, AK155734. Below, expression relative to Gapdh of the AK155734 RNA, using primer set A (from Fig. 2) for embryonic stem cells (ES), whole embryos at E7.5, bodies from embryos (B) at E10.5, 13.5 and 16.5 and neonatal heart (nnH).
To determine if there was splicing of the transcript, we designed primers in exon 1 and 3, i.e., spanning introns 1 and 2 and exon 2 (primers Ef and Er in Fig. 2). A spliced transcript was detected only in neonatal liver, and this co-existed with the full unspliced form (data not shown).
We investigated if the ncRNA was imprinted and if so, whether it was maternally or paternally expressed. Taking advantage of polymorphisms between C57BL/6J and CAST/EiJ strains of mice, we performed RT-PCRs and restriction digests on RNA from neonatal hearts of F1 hybrid mice. We found that AK155734 is expressed with a strong maternal bias in reciprocal crosses, with the paternal expression reaching 17% of the total maternal level (Fig. 3B). Thus, the ncRNA is subjected to the same regulation as the sense Tnfrsf genes.
Phylogenetic distribution of Tnfrsf22, Tnfrsf23 and Tnfrsf26 orthologs in vertebrates
To date, no human homolog of the mouse Tnfrsf22, Tnfrsf23 and Tnfrsf26 genes has been identified in the KCNQ1 domain. Therefore, these genes might have arisen by an insertion of Tnfrsf sequences during the evolution of the mouse Kcnq1 domain, or might have degenerated or been deleted or relocated during the evolution of the human region. To discriminate between these two possibilities, we analyzed the distribution and evolution of Tnfrsf22, Tnfrsf23 and Tnfrsf26 orthologous sequences in multiple species. Although the Tnfrsf genes constitute a large family, we focused our study on those located within regions that are orthologous to the Kcnq1 domain (see Materials and Methods).
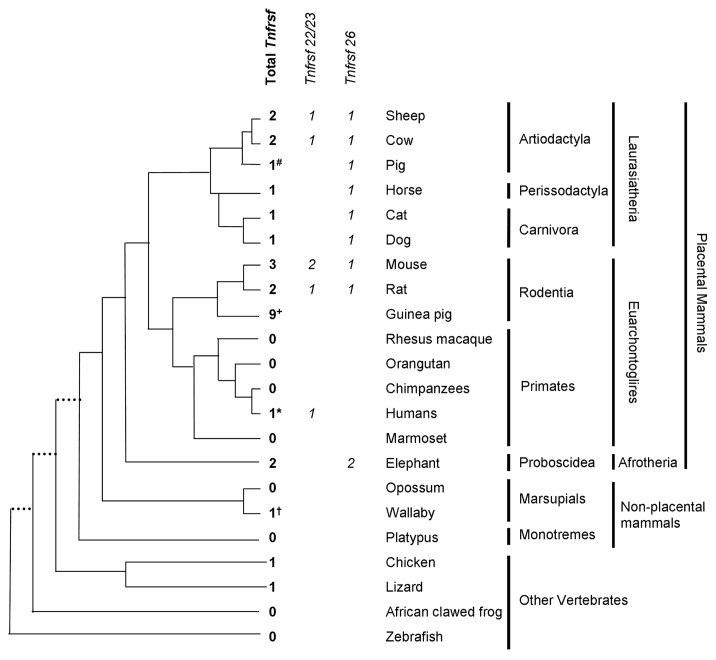
As summarized in Figure 5, we observed that TNFRSF sequences are present in orthologous regions in many vertebrates. Therefore, they are not exclusive of mice due to a rodent-specific insertion in the Kcnq1 domain.
Figure 5. Distribution of Tnfrsf homologous sequences in Kcnq1 orthologous regions. The topology is drawn according to www.tolweb.org/tree and Murphy et al.15 The total numbers of Tnfrsf sequences observed in each taxon are indicated, even in those in which no Tnfrsf homologs are detected, in order to reflect both gene gains and losses within the Kcnq1 region. The figure also illustrates how many of them correspond to orthologs of Tnfrsf22 or Tnfrsf23 and of Tnfrsf26 only for placental mammals, according to the phylograms depicted in Figures 6 and 7. Only mouse, human and frog Kcnq1 orthologous regions are devoid of sequencing gaps that result from incomplete sequence assembly. #In pig, three identical sequences were found; they could be the result of assembly errors or very recent duplications and, therefore, only one was analyzed in our phylogenetic trees. +In guinea pig, multiple sequences with significant similarity to mouse Tnfrsfs were found; we manually annotated a minimum of nine. *In humans, we only detected one sequence that is much shorter than any of the ones found in other species (Sup. Material). †In wallaby, we could not confirm the location of the Tnfrsf homologous sequences within the Kcnq1 orthologous region (see Materials and Methods).
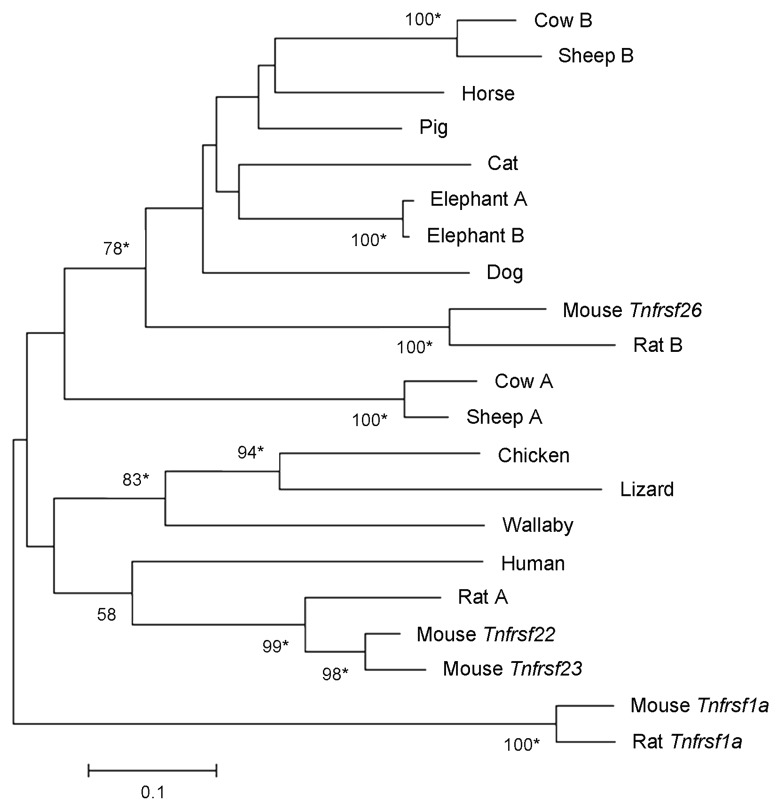
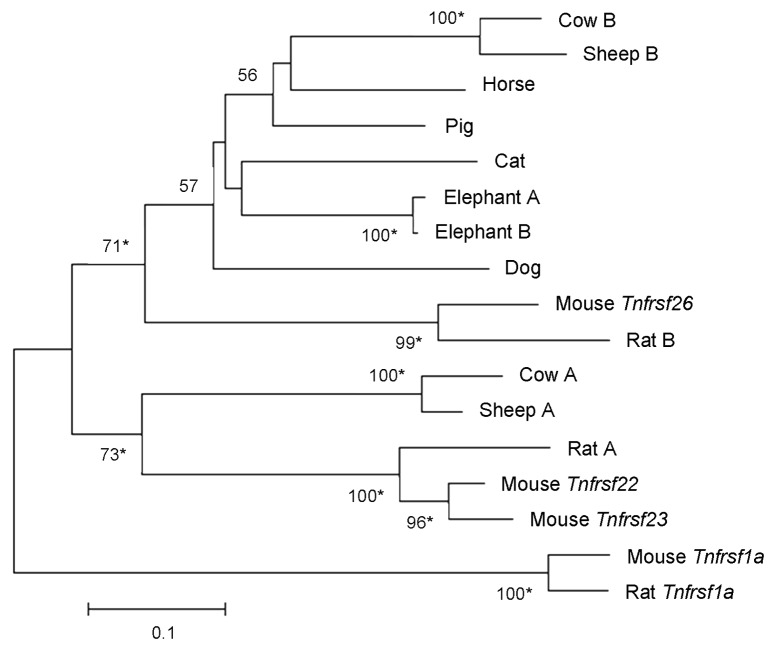
Although we found only one TNFRSF member in organisms other than mammals, there are variable numbers in mammalian species. A Neighbor-Joining phylogram of their nucleotide sequences (see Material and Methods) revealed multiple lineage-specific duplications (Fig. 6). For instance, several duplications have occurred in the rodent lineage, especially in guinea pig. Rats and mice have one Tnfrsf26 ortholog each, while a mouse lineage-specific duplication generated the Tnfrsf22 and Tnfrsf23 genes. Similar results are observed in a Neighbor-Joining tree of amino acid sequences of the same TNFRSF orthologs (data not shown).
Figure 6. Phylogeny of Tnfrsf homologous sequences within Kcnq1 orthologous regions. The evolutionary history was inferred using the Neighbor-Joining method. Nucleotide sequences that aligned unambiguously to mouse Tnfrsf22, Tnfrsf23 or Tnfrsf26 were analyzed (see Materials and Methods); nine guinea pig sequences were removed from this analysis for the sake of clarity, although similar results were obtained in trees that included them (data not shown). This is a rooted phylogram obtained using MEGA 5.05.13 The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. Numbers at nodes represent bootstrap support values based on 5000 pseudoreplicates. Values below 50% were removed and * indicates 70% or above bootstrap support.16
The phylogenetic tree in Figure 6 reveals one clade that includes mouse Tnfrsf26 and other mammalian sequences. When we restrict our study to the placental mammalian orthologs, the resulting phylogenetic tree also supports a clade including mouse Tnfrsf 26, as well as a second clade that contains the mouse Tnfrsf22 and Tnfrsf23 genes (Fig. 7). Both clades include very diverse mammalian species; moreover, divergent species [e.g., sheep and cow (laurasitheria) vs. mouse and rat (euarchontoglires)] have TNFRSF members in both clades (Fig. 5). These results suggest that the duplication that originated Tnfrsf26 and Tnfrsf22 (or Tnfrsf23) occurred no later than the split between euarchontoglires and laurasitherian mammals.
Figure 7. Phylogeny of Tnfrsf homologous sequences within Kcnq1 orthologous regions of placental mammals. The phylogeny was based on the Neighbor-Joining analysis of nucleotide sequences. Analysis was restricted to placental mammalian sequences that aligned unambiguously to mouse Tnfrsf22, Tnfrsf23 or Tnfrsf26, with the exception of human (due to the small size of the orthologous sequences) and guinea pig (see Materials and Methods). This rooted phylogram was obtained using the Maximum Composite Likelihood method implemented in MEGA 5.05.13 The tree is drawn at a scale that represents the number of base substitutions per site. Numbers at nodes represent bootstrap support values based on 5000 pseudoreplicates. Values below 50% were removed and * indicates 70% or above bootstrap support.16
In contrast, in some mammals, only TNFRSF members of one clade are present in the Kcnq1 orthologous region. Moreover, they appear to be absent in most primates (Fig. 5). With the exception of humans, these orthologous regions contain sequencing gaps due to incomplete assembling and, therefore, we cannot exclude the possible existence of additional TNFRSF sequences. Nevertheless, the data suggest that TNFRSF sequences degenerated or were relocated or deleted from the KCNQ1 region several times during the evolution of mammals.
Indeed, a detailed search allowed us to identify a short sequence (46 aminoacids) within the human KCNQ1 domain (Sup. Material). This sequence had 43% and 41% identities with mouse TNFRSF23 and TNFRSF22, respectively. No significant hits were found in the orthologous domains of other primate species by tblastn searches of the orthologous domains with either mouse (TNFRSF 22, TNFRSF 23 and TNFRSF 26) or the small human amino acid sequence queries. Blast search of NCBI RNA databases with this human sequence as a query showed no significant hits with either experimentally supported (clones and ESTs) or predicted human RNAs. This suggests that during the evolution of the KCNQ1 region in the human lineage, multiple mutations (point mutations and/or deletions) have accumulated on the ancestral TNFRSF, rendering it non-functional.
Discussion
We have found that three Tnfrsf genes present in the murine Kcnq1 domain, Tnfrsf23, 22 and 26, are expressed in embryos and are developmentally regulated. We also found that all three genes are expressed with a strong maternal bias. An antisense RNA, AK155734, is co-expressed and overlaps with Tnfrsf 22, and at least in neonatal heart, with Tnfrsf23. This non-coding RNA is imprinted in the same direction as the Tnfrsf genes.
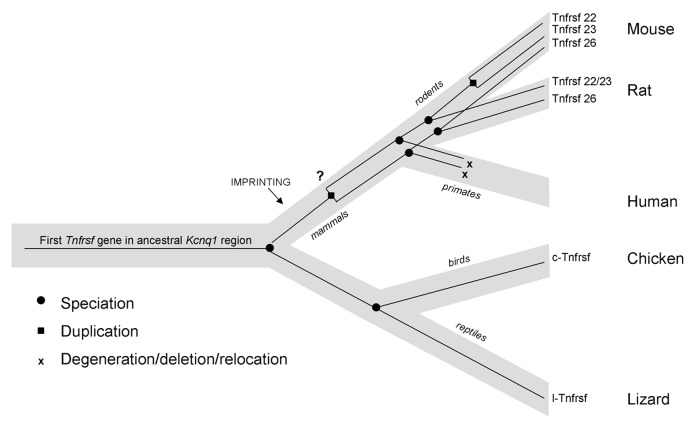
In analyzing the evolutionary origin of Tnfrsf22, 23 and 26, we find that one Tnfrsf gene is present within the Kcnq1 orthologous domain in non-mammalian vertebrates such as chicken and lizard. This suggests that one copy (at least) was also present in their common ancestor with mammals. Later in mammalian evolution, multiple duplications occurred. Although the scarcity of information in monotremes and marsupials does not allow us to determine when the first duplication took place, our data suggest that it occurred early during the evolution of mammals; we cannot, however, rule out that it occurred even earlier. What is clear is that after this initial duplication, Tnfrsf22/23 and Tnfrsf26 diverged. In some lineages, additional duplications occurred, while in others, (such as in primates) they appear to have been lost (Fig. 8). In mouse, the three Tnfrsf genes are the result of two duplication events: the one that occurred before the split of euarchontoglires from laurasitherian mammals and a second duplication after the split of the mouse and rat lineages (Fig. 8). In fact, the Tnfrsf22 and 23 genes are located on two segmental duplications, as annotated in the UCSC genome browser (www.ucsc.edu) (Fig. 4).
Figure 8. Model of the evolution of mouse and human Tnfrsf genes within the Kcnq1 region. Light gray line represents the species tree; thin black lines within represent the Tnfrsf gene tree. This figure represents only relevant lineages and species in order to summarize the major events in evolution of mouse and human Tnfrsf genes. Tnfrsf sequences were present in the Kcnq1 ancestral region before the first establishment of imprinting in mammals (notice that imprinting acquisition did not occur for all genes at the same time and it remains to be determined when parental bias was established in the Tnfrsf genes). Two duplications (one early or prior to mammalian evolution and one after the mouse and rat lineage split) originated the three Tnfrsf genes present in mouse. It has not been established whether the earlier duplication occurred before or after imprinting emerged in mammals (indicated with a question mark). In primates, Tnfrsf genes degenerated or were deleted or relocated, and in humans only a single short homologous sequence is observed.
We conclude that these genes have been present in the Kcnq1 orthologous region of diverse vertebrates since before the establishment of imprinted expression in mammals (Fig. 8);6 therefore, Tnfrsf genes did not initially acquire imprinting due to an insertion into a preexisting imprinted domain.
The Kcnq1 domain is regulated by a paternally expressed long non-coding RNA, Kcnq1ot1. Expression of Kcnq1ot1 leads to silencing of neighboring genes, with a range in the embryo that was assumed to be approximately half that of the placenta in the mouse (Fig. 1). The fact that the Tnfrsf genes exhibit imprinted expression suggests that they may be under the control of the Kcnq1ot1 RNA, although how the genes between Phlda2 and Tnfrsf26 escape repression will have to be investigated. There are other examples of escapees, such as Trpm5 and Tspan32 in the placenta, showing that silencing of genes is not uniform along the chromatin fiber. It is intriguing that the bias in expression is greatest in the Tnfrsf26 gene, the copy closest to the Kcnq1ot1 transcriptional unit. An alternative possibility is that there is an independent mechanism by which imprinting of the Tnfrsf genes is regulated. Several existing knockout mouse models will allow us to address this issue.
Neither of the Tnfrsf genes has a CG-rich promoter, so the mechanism of relative paternal repression may not be dependent on DNA methylation. Several of the genes in the domain have methylation-independent imprinted expression, but this is only true in the placenta. We cannot rule out that methylation marks on sequences that do not qualify as CG islands are important for imprinting at the Tnfrsf genes.
Interestingly, the antisense AK155734 gene has a similar expression pattern to the sense genes, albeit with a slightly later appearance. In addition, both have maternal bias, suggesting that either there is no transcriptional interference, or that they are expressed in distinct cells. Further experiments will be necessary to distinguish between these possibilities and to determine if AK155734 is functional.
No homolog of the murine Tnfrsf22, 23 and 26 genes had been identified in the human counterpart of the Kcnq1 domain to date, so we were interested in tracing the origin of this cluster. Our phylogenetic data showed that in fact, there are multiple Tnfrsf sequences in orthologous regions of many mammals, as well as in other vertebrates such as chicken and in lizard. In humans, there is a very short sequence with limited similarity to the murine Tnfrsf genes, which appears to have lost its function. Further studies will be needed to determine the selection regime (positive vs. purifying selection) operating during the evolution of this gene family within the Kcnq1 domain.
Duplicated loci are usually either maintained or lost during evolution, and if maintained, they can potentially serve as the raw material for neofunctionalization.7 New paralogs that are located in different genomic regions are more likely to have undergone adaptive evolution,8 and may acquire new regulatory signals and different expression patterns. Gene families that have rapidly expanded their copy number in mammals include those involved in immunity, such as the Tnf and Tnfr superfamilies, with rapid gene gain and loss. The Tnfrsf genes have been very dynamic during mammalian evolution with regards to species-specific gains and losses. For example, in rodents, the guinea pig lineage has undergone numerous expansions of Tnfrsf, and the mouse has had a duplication after the split with rat. On the other hand, primates may have lost the genes within the Kcnq1 region altogether, with only a trace remaining in humans (Fig. 8). It is interesting to note that the murine Tnfrsf genes lack cytoplasmic domains, suggesting they are snippets of original genes that were duplicated or relocated from other regions and can provide the substrate for expansion of their functions by adding different domains.
In conclusion, our results, in conjunction with the detailed biochemical studies previously reported,3 are suggestive of a developmental function for the Tnfrsf genes in the mouse embryo, possibly acting as decoy receptors. The allele-specific studies show parent-of-origin biases in expression, although further studies are required to determine if the Kcnq1 imprinting control region or the Kcnq1ot1 non-coding RNA are implicated in these patterns. Furthermore, our phylogenetic analysis shows that Tnfrsf genes were present within the Kcnq1 region before the establishment of imprinting.
Materials and Methods
RNA purification
ES cells (C57BL/6J from Jackson labs), embryos collected at appropriate days of gestation and neonatal tissues were dissected. To distinguish between parental alleles, reciprocal crosses between C57BL/6J and B6(CAST7)17 (mice with a CAST/EiJ chromosome 7 on a C57BL/6J background) were set up and F1 hybrid embryos and neonatal tissues were collected. RNA was extracted using TRIzol Reagent (Invitrogen, #15596–018) and following manufacturer's protocol for RNA extraction from tissues. All RNA samples were subjected to DNase treatment using Turbo DNA-free (Ambion, #AM1907) with the rigorous DNase treatment protocol. Three to five biological samples were collected for each embryo stage and neonatal tissue analyzed.
Reverse Transcription
Following the manufacturer's instructions, cDNA synthesis was performed on total RNA using SuperScript II Reverse Transcriptase (Invitrogen, 18064–014). A Reverse Transcriptase negative control was used to ensure there was no DNA contamination
Allele Specific RT-PCR and Quantification
The Tnfrsf23, 22 and 26 transcripts were amplified using Ruby Taq Master Mix (Affymetrix, 71191) in a reduced 15 µl reaction, in all cases with primers that spanned introns and contained a polymorphism in the coding regions (Table S1). PCR products were digested with restriction enzymes that distinguished between the C57BL/6J and CAST/EiJ alleles: for Tnfrsf 26, SfcI cuts the CAST/EiJ allele; for Tnfrsf22, NlaIII has 3 sites in the C57BL/6J and 2 in the CAST/EiJ allele; for Tnfrsf23, Hsp92II cuts the C57BL/6J and not the CAST/EiJ allele; and for AK155734, NlaIII cuts the C57BL/6J allele twice and the CAST/EiJ allele 3 times. PCR and digestion products were run on 7% polyacrylamide gels and quantified using the Kodak Gel Logic 2000 imaging system. Three independent biological samples from reciprocal crosses of C57BL/6J and B6(CAST7) mice were tested. 13.5 dpc embryos and neonatal hearts were analyzed for Tnfrsf imprinting, whereas AK155734 imprinting was analyzed in neonatal heart. The relative paternal to maternal band intensities were calculated and graphed. For quantification, RT-PCR products from Tnfrsf and AK155734 genes were graphed relative to Gapdh. Gapdh PCR was performed using the following primers: 5′-ATCACTGCCACCCAGAACAC-3′ and 5′-ATCCACGACGGACACATTGG-3′.
Identification of Tnfrsf22, Tnfrsf23 and Tnfrsf26 orthologous sequences in diverse species
The Tnfrsf gene family has many members, most of them located outside the Kcnq1 imprinted region. We restricted our study to the evolution of the orthologs of these three mouse genes within the imprinted domain, specifically within the region flanked by the Nap1l4 and Cars genes at the proximal boundary and by Osbpl5 and Nadsyn1 at the distal edge (Fig. 1). These genes were used as anchors to retrieve the sequences of orthologous regions in diverse organisms. First, the positions of the anchor orthologs were identified either by gene name or by blastp and tblastn search of NCBI databases,9 using mouse sequences as queries. We have restricted our study to those species* (1) in which we could identify at least one of the two genes at each boundary, (2) located on the same chromosome (syntenic); and (3) with deep sequencing coverage, so that we could retrieve the complete sequences of the orthologous region. The orthologous sequences within those boundaries were then retrieved from genome.ucsc.edu/cgi-bin/hgGateway. In some species, we located the anchor genes in SuperContig scaffolds and obtained the sequences within them. Finally, we searched these regions with mouse TNFRSF22, TNFRSF23 and TNFRSF26 amino acid sequences; these were restricted to the protein regions that are more conserved among the three mouse genes (Sup. Material). A search by tblastn9 allowed us to retrieve orthologous sequences with a threshold of e < 0.01. Both nucleotide and amino acid sequences of each ortholog were obtained, although some of them had to be edited in order to reconstruct ORFs (Sup. Material). This approach allowed us to identify novel Tnfrsf orthologous sequences that had not been previously described. In order to facilitate the interpretation of the phylogenetic analyses, we have identified them with the species name. When multiple paralogs were present within a species, we have distinguished them by adding a letter in alphabetic order (e.g., elephant A and elephant B), regardless of their phylogenetic origin.
*Wallaby constitutes the only exception: Cars and by Osbpl5 orthologous sequences were found on different scaffolds and we could not retrieve any Tnfrsf orthologs in them. However, genome-wide tblastn search with mouse TNFRSF22, TNFRSF23 and TNFRSF26 amino acid sequences allowed us to identify one DNA fragment containing wallaby Tnfrsf orthologous sequences; a similar approach did not reveal any Tnfrsf ortholog in platypus. The wallaby Tnfrsf orthologous sequences were used as a query to perform a tblastn search in the refseq_rna mouse sequence database, confirming they are more similar to Tnfrsf22 and Tnfrsf23 than to any other genes. In spite of not being able to verify their location in the wallaby Kcnq1 orthologous regions, they are placed in the same branch of as those of chicken and lizard in the phylogenetic tree depicted in Figure 6. Due to these reasons, and to the fact of being the only marsupial Tnfrsf ortholog found, we have included it in our study.
Phylogenetic analyses
Mouse TNFRSF22/23/26 and their orthologous sequences were aligned with ClustalW210 and edited with GeneDoc (www.psc.edu/biomed/genedoc). Regions of ambiguous alignment were excluded from the analyses.11 We obtained a Neighbor-Joining phylogram using MEGA 5.0512,13 and rooted the tree with two paralogous sequences (mouse and rat TNFRSF1a, which are the most closely related to TNFRSF22/23/26). We employed the Maximum Composite Likelihood model for nucleotide evolution, which is a likelihood-based implementation of the Tamura-Nei model that enhances the accuracy of calculating the pairwise distances.14 For amino acid evolution, the equal input model (which corrects for variation in amino acid frequency) was applied. Node support was assessed by conducting 5000 nonparametric bootstrap pseudoreplicates.
Supplementary Material
Acknowledgments
The authors woud like to thank Antonio Mas and Esther Betrán for helpful suggestions for the phylogenetic analysis. This study was supported by funding from the NIH R01GM093066 and NCI K22CA140361–3 (N.E.); E.D.E. and G.C. were supported by the Consejería de Educación y Ciencia de la Junta de Comunidades de Castilla-La Mancha (PPII10–0259–4347) and the European Social Fund.
Glossary
Abbreviations:
- TNFRSF
tumor necrosis factor receptor superfamily
- TNFSF
tumor necrosis factor superfamily
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/20243
References
- 1.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/S0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 2.Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/S0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 3.Schneider P, Olson D, Tardivel A, Browning B, Lugovskoy A, Gong D, et al. Identification of a new murine tumor necrosis factor receptor locus that contains two novel murine receptors for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) J Biol Chem. 2003;278:5444–54. doi: 10.1074/jbc.M210783200. [DOI] [PubMed] [Google Scholar]
- 4.Wiens GD, Glenney GW. Origin and evolution of TNF and TNF receptor superfamilies. Dev Comp Immunol. 2011;35:1324–35. doi: 10.1016/j.dci.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 5.Clark L, Wei M, Cattoretti G, Mendelsohn C, Tycko B. The Tnfrh1 (Tnfrsf23) gene is weakly imprinted in several organs and expressed at the trophoblast-decidua interface. BMC Genet. 2002;3:11–5. doi: 10.1186/1471-2156-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Killian JK, Nolan CM, Stewart N, Munday BL, Andersen NA, Nicol S, et al. Monotreme IGF2 expression and ancestral origin of genomic imprinting. J Exp Zool. 2001;291:205–12. doi: 10.1002/jez.1070. [DOI] [PubMed] [Google Scholar]
- 7.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20:1313–26. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–67. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 11.Bustos O, Naik S, Ayers G, Casola C, Perez-Lamigueiro MA, Chippindale PT, et al. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene. 2009;447:1–11. doi: 10.1016/j.gene.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–5. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–21. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillis DM, Allard MW, Miyamoto MM. Analysis of DNA sequence data: phylogenetic inference. Methods Enzymol. 1993;224:456–87. doi: 10.1016/0076-6879(93)24035-S. [DOI] [PubMed] [Google Scholar]
- 17.Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod. 2003;69:902–14. doi: 10.1095/biolreprod.103.017293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.