Parvoviruses are unique among all known organisms in having DNA genomes that are both single stranded and linear; however, they manage to inveigle their host cell into replicating these apparently alien molecules with considerable efficiency. Members of this family that infect mammals can be divided into two functional subgroups, the adeno-associated viruses (AAV), which require coinfection with an adenovirus or herpesvirus for productive growth, and the autonomously replicating viruses, such as Minute Virus of Mice (MVM), which have no such requirement. In the absence of helper, wild-type AAV adopts a latent lifestyle during which it integrates into the host genome, predominantly at a single specific site. Because of this property, AAV has attracted much interest as a vector for gene therapy, despite being restricted in the size of insert it can transfer. Interestingly, although capable of efficient transduction into many cell types, none of the current AAV gene therapy vectors contain the viral gene responsible for site-specific integration, although they still integrate efficiently into the host genome in a sequence-independent manner. Studies over the last few years aimed at understanding transduction by these vectors into intact tissue targets, such as skeletal muscle, have now revealed a hitherto unsuspected twist to the generally accepted replication mechanism. This alternate pathway of parvoviral DNA metabolism has been exploited by Yan and colleagues (1) in an approach that uses a binary delivery system to overcome the limited genetic capacity of AAV-based vectors.
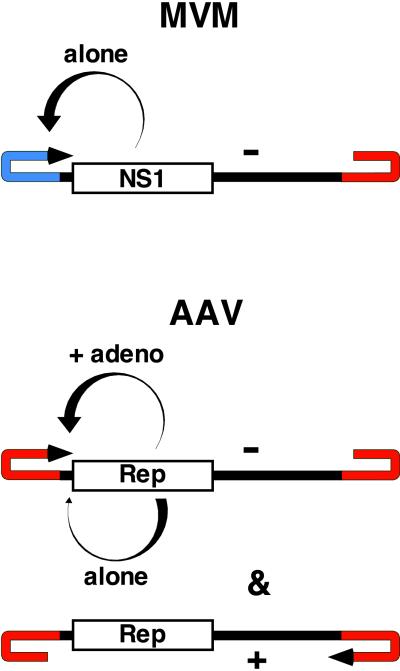
The typical parvoviral genome is between 4,500 and 5,000 nt long with a small terminal palindrome, capable of folding into a hairpin structure, at each end. In most autonomously replicating parvoviruses, these hairpins are different in sequence and structure at both ends of the molecule, as shown for MVM in Fig. 1, whereas for AAV, the palindromic sequences are part of a terminal repeat (TR) some 20 nt longer than the hairpin itself. AAV packages equal numbers of the transcriptionally positive and negative DNA strands, in separate virions, most likely because all of the cis-acting signals for both replication and packaging lie within the identical AAV TRs (2). In contrast, MVM packages almost exclusively the negative strand.
Figure 1.
Comparison of the genomes of a typical autonomous parvovirus (MVM) with that of AAV. Terminal hairpins are in red or blue. Arrowheads denote the 3′ ends of positive (+) or negative (−) sense strands. Curved tapered arrows denote the mode of regulation of NS1 or Rep of its own synthesis.
During productive infection, both types of virus replicate their genomes by using the 3′ hairpin to prime synthesis of a complementary strand, creating a monomer-length duplex intermediate. Continued DNA synthesis displaces and copies the hairpin at the opposite end, to produce an extended terminal palindrome, which can then be melted out and rearranged into 5′ and 3′ hairpin structures. The hairpin containing the new 3′ end then primes synthesis of another copy of the genome, creating a dimer intermediate in which the two genomic strands are arranged as a palindromic concatamer. Further replication by this “rolling hairpin” mechanism produces tetramers in which genomes are joined in alternating head-to-head and tail-to-tail arrangements (3). The hairpin sequences in these concatamers are regenerated and the individual monomer-length genome sequences released for packaging by the action of a virally encoded multifunctional protein called NS1 for MVM and Rep for AAV.
NS1 and Rep are site-specific DNA-binding proteins with nickase and helicase activity, which also regulate transcription of their own mRNAs. When the uncoated MVM genome is converted to a duplex form, early in S-phase, NS1-specific transcripts are expressed from the P4 promoter, leading to accumulation of NS1 protein in the nucleus. As indicated in Fig. 1, one function of NS1 is to up-regulate the P4 promoter itself, and this positive feedback loop appears to be a part of the “hard wiring” of infection that ensures rapid viral takeover of the cell. A similar enhancement of Rep synthesis occurs in AAV infection in the presence of helper virus, but when AAV infects on its own, the feedback effect of Rep is reversed, and Rep production shuts down. However, enough Rep is produced—between 1,000 and 4,000 molecules per cell (4)—to catalyze the integration of the viral genome into a specific site at chromosome 19q13.3-qter in the human genome (5, 6). Specific integration at this chromosomal locus requires the interaction of Rep with a target DNA sequence that has key features in common with the origin of AAV DNA replication embedded in each viral TR (7). When placed in a plasmid, the chromosome 19 sequence initiates Rep-dependent DNA replication in an in vitro system (8) and, as a “knock-in” transgene in the mouse X-chromosome, will direct site-specific integration of AAV in murine cells (4). When cloned into an EBV episome, the sequence also supports site-specific AAV integration in vivo, providing a system that can be genetically manipulated. Such studies have confirmed that this sequence must be able to function as an AAV replication origin to serve as an integration site (9). This “shuttle” episome system allows integrants to be recovered from the target cell by transformation into Escherichia coli and has been used to show that the trick of replicative integration is not unique to AAV. MVM can also integrate into a functional copy of one of its DNA replication origins (10), but the mammalian host cells harboring such MVM integrants cannot be cloned out because the virus is constitutively lytic.
Because Rep is required for site-specific integration, the vast majority of AAV-based vectors, which have been stripped of Rep because of its potential cytotoxicity and to increase the genetic capacity of the vector, are incapable of inserting specifically at the chromosome 19 locus. Nevertheless, transgenes packaged in recombinant AAV (rAAV) particles have shown a significant ability to integrate at numerous other sites in the human, simian, and mouse genomes and to persist, sometimes with high levels of expression, in many different tissues, although concordance between these phenomena is by no means complete.
The last 4 years have seen increasing interest in the use of rAAV vectors to target therapeutic genes to smooth muscle (11–17), a tissue that seems to be highly susceptible to rAAV-mediated transduction and that supports sustained high-level transgene expression. Vector DNA persists in muscle as concatamers (12), which were initially thought to be wholly integrated (18, 19), but subsequent analysis has suggested that a fraction of these transgenes are present as complex high molecular-weight episomes, apparently derived over time from monomer circles (20). Use of a shuttle version of rAAV, which allows such forms of persisting vector DNA to be recovered in bacteria, has permitted the isolation and characterization of such circular forms. These studies indicate that circular forms arise from a recombinatorial interaction between AAV TRs, with at least 1 in 400 of the input-transducing genomes becoming circularized (21). It also appears that the majority of circular concatamers contain transgenes organized in a head-to-tail manner, confirming that they are not derived from normal viral replication intermediates.
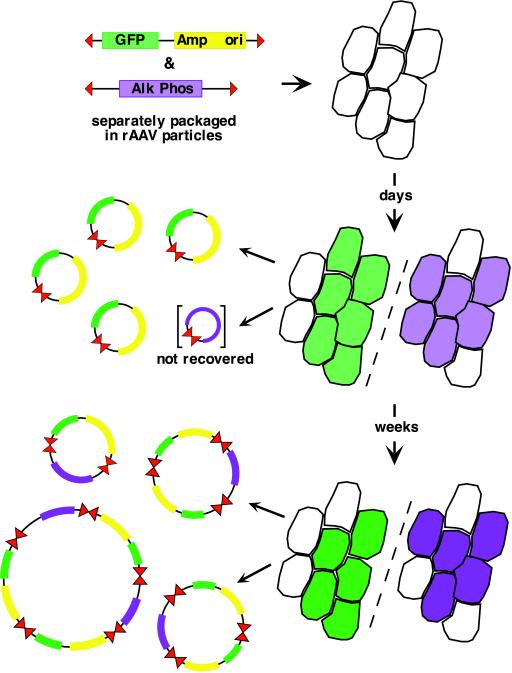
Suspecting that concatamer formation was a result of intermolecular recombination rather than de novo replication, Yang et al. (22) cotransduced mouse tibialis muscles with a pair of distinguishable rAAV vectors, one of which was a shuttle vector, as diagrammed in Fig. 2. In the first 2 weeks, the only circular forms recovered from coinfected muscle contained sequences exclusively derived from the shuttle vector, but from the end of the first month, and continuing thereafter, they observed an increase in transgene expression levels in individual myofibers and a corresponding steady increase in the isolation of chimeric plasmids. On analysis, these were found to be mostly head-to-tail arrangements of sequences from both the shuttle and the nonselected vector, separated by TR sequences. Because the TR is the only homologous sequence shared by both vectors, they concluded that these chimeric concatamers had arisen by recombination between the TRs of different circular intermediates. Because there is clearly much greater opportunity for reciprocal recombination to occur between homologous circular forms of each vector, the high level of chimera isolation, up to a third of all forms isolated after 4 months, suggests that interTR recombination is by far the more favored event. These results provide good evidence that the sequence and/or structure of the AAV TR is hyperrecombinogenic in vivo, and that the production of recombinant circular forms from different input rAAV-vectored sequences can be readily achieved.
Figure 2.
Diagram of the experiments reported in Yang et al. (22). GFP indicates the green fluorescent protein coding sequence (green), whereas Alk Phos denotes that of alkaline phosphatase (purple). The selectable marker and bacterial plasmid origin of the shuttle vector, Amp and ori, are shown in yellow. The vector TRs are shown as red triangles. The cells (Right) represent myofibers, cut in cross section, displaying transgene expression levels for GFP on the left and Alk Phos on the right.
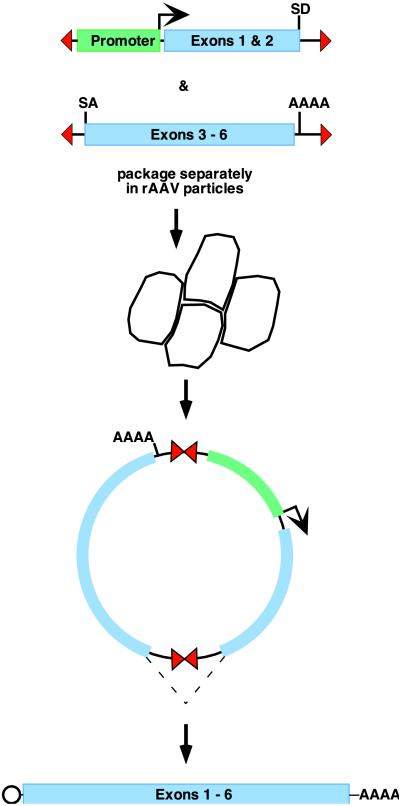
Yan and colleagues (1) have now followed up on this observation by constructing two rAAV vectors that each contain a different part of the murine erythropoietin (EPO) genomic sequence, arranged so that intron 3 spans the 3′ TR of one and the 5′ TR of the other. They find that such vectors also interact in vivo, some forming head-to-tail chimeric dimer circles that reassemble the native configuration of the gene. They confirm that rescued chimeras of this type are able to express EPO after transfection into cultured cells. As these chimeras first become demonstrable in the muscle tissue of transduced mice, the animals experience a significant rise in hematocrit. At the same time, they become resistant to anemia resulting from adenine treatment, which abrogates endogenous EPO production in the kidney. Thus the report offers convincing proof of concept for this binary vector approach. Because the EPO gene used is relatively small, each of the four introns was included, but this is probably not necessary, and binary vectors comprising long control regions and/or large cDNAs split by a single intron, as diagrammed in Fig. 3, become exciting possibilities for the future of AAV gene therapy.
Figure 3.
Putative vector pair for effectively doubling the capacity of rAAV, as suggested by the report of Yan et al. (1). A cDNA encoding a fictitious therapeutic gene of ≈6 kb, requiring an upstream control region of ≈3 kb, is split between two rAAV vectors across an intron, such that the 3′ end of the first gene segment contains a splice donor site (SD), and the 5′ end of the second gene segment contains a splice acceptor (SA) site. The black arrow indicates the start of transcription, and the polyadenylation site is denoted AAAA. The dotted line indicates the splicing event, occurring on transcription of the dimeric chimera, that creates an intact mRNA encoding the full-length gene product.
The coinfection results presented by Yan and colleagues (1) may also shed some light on a further puzzling feature of rAAV transduction, namely that it is so efficient in postmitotic cells. This appears counterintuitive from the perspective of the autonomous parvoviruses, which can be replicated only when their host cell enters S-phase of its own volition. As yet, there are no reports of definitive experiments, such as density transfer analyses, which would prove that rAAV DNA is ever replicated semiconservatively on entry into terminally differentiated cells. Given the demonstration by Yan et al. that multiple functional coinfections of the same cell can occur readily, it seems possible that viral DNA replication is irrelevant to this type of persistence, and that incoming parental plus and minus strands, derived from separate virions, simply anneal after uncoating. The hyperrecombinogenic nature of their terminal structures could then catalyze the assembly of these duplexes into circular and then concatameric forms, in the absence of either Rep or the cellular DNA replication machinery.
Acknowledgments
I thank Susan Cotmore for stimulating and profitable discussion.
Footnotes
See companion article on page 6716.
References
- 1.Yan Z, Zhang Y, Duan D, Engelhardt J F. Proc Natl Acad Sci USA. 2000;97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao X, Xiao W, Li J, Samulski R J. J Virol. 1997;71:941–948. doi: 10.1128/jvi.71.2.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cotmore S F, Tattersall P. In: DNA Replication in Eukaryotic Cells. DePamphilis M, editor. Plainview, New York: Cold Spring Harbor Lab. Press; 1996. pp. 799–813. [Google Scholar]
- 4.Young S M, McCarty D M, Degtyareva N, Samulski R J. J Virol. 2000;74:3953–3966. doi: 10.1128/jvi.74.9.3953-3966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotin R M, Menninger J C, Ward D C, Berns K I. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 7.Kotin R M, Linden R M, Berns K I. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R M. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linden R M, Winocour E, Berns K I. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corsini J, Tal J, Winocour E. J Virol. 1997;71:9008–9015. doi: 10.1128/jvi.71.12.9008-9015.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kessler P D, Podsakoff G M, Chen X, McQuiston S A, Colosi P C, Matelis L A, Kurtzman G J, Byrne B J. Proc Natl Acad Sci USA. 1996;93:14082–14087. doi: 10.1073/pnas.93.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark K R, Sferra T J, Johnson P R. Hum Gene Ther. 1997;8:659–669. doi: 10.1089/hum.1997.8.6-659. [DOI] [PubMed] [Google Scholar]
- 13.Qing K, Khuntirat B, Mah C, Kube D M, Wang X S, Ponnazhagan S, Zhou S, Dwarki V J, Yoder M C, Srivastava A. J Virol. 1998;72:1593–1599. doi: 10.1128/jvi.72.2.1593-1599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rendahl K G, Leff S E, Otten G R, Spratt S K, Bohl D, Van Roey M, Donahue B A, Cohen L K, Mandel R J, Danos O, et al. Nat Biotechnol. 1998;16:757–761. doi: 10.1038/nbt0898-757. [DOI] [PubMed] [Google Scholar]
- 15.Monahan P E, Samulski R J, Tazelaar J, Xiao X, Nichols T C, Bellinger D A, Read M S, Walsh C E. Gene Ther. 1998;5:40–49. doi: 10.1038/sj.gt.3300548. [DOI] [PubMed] [Google Scholar]
- 16.Xiao W, Chirmule N, Berta S C, McCullough B, Gao G, Wilson J M. J Virol. 1999;73:3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song S, Morgan M, Ellis T, Poirier A, Chesnut K, Wang J, Brantly M, Muzyczka N, Byrne B J, Atkinson M, et al. Proc Natl Acad Sci USA. 1998;95:14384–14388. doi: 10.1073/pnas.95.24.14384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao X, Li J, Samulski R J. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher K J, Jooss K, Alston J, Yang Y, Haecker S E, High K, Pathak R, Raper S E, Wilson J M. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 20.Vincent-Lacaze N, Snyder R O, Gluzman R, Bohl D, Lagarde C, Danos O. J Virol. 1999;73:1949–1955. doi: 10.1128/jvi.73.3.1949-1955.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, Fisher K J, Engelhardt J F. J Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Zhou W, Zhang Y, Zidon T, Ritchie T, Engelhardt J F. J Virol. 1999;73:9468–9477. doi: 10.1128/jvi.73.11.9468-9477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]