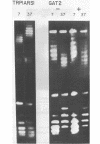
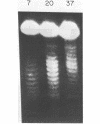
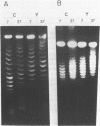
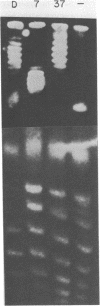
Abstract
Thermal untwisting of DNA is suppressed in vitro in nucleosomes formed with chicken or monkey histones. In contrast, results obtained for the 2 micron plasmid in Saccharomyces cerevisiae are consistent with only 30% of the DNA being constrained from thermal untwisting in vivo. In this paper, we examine thermal untwisting of several plasmids in yeast cells, nuclei, and nuclear extracts. All show the same quantitative degree of thermal untwisting, indicating that this phenomenon is independent of DNA sequence. Highly purified yeast plasmid chromatin also shows a large degree of thermal untwisting, whereas circular chromatin reconstituted using chicken histones is restrained from thermal untwisting in yeast nuclear extracts. Thus, the difference in thermal untwisting between yeast chromatin and that assembled with chicken histones is most likely due to differences in the constituent histone proteins.
Full text
PDF


















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrose C., McLaughlin R., Bina M. The flexibility and topology of simian virus 40 DNA in minichromosomes. Nucleic Acids Res. 1987 May 11;15(9):3703–3721. doi: 10.1093/nar/15.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel R., Feigelson P., Schutz G. Analysis of the complexity and diversity of mRNA from chicken liver and oviduct. Cell. 1976 Feb;7(2):247–254. doi: 10.1016/0092-8674(76)90024-6. [DOI] [PubMed] [Google Scholar]
- Bina-Stein M., Vogel T., Singer D. S., Singer M. F. H5 Histone and DNA-relaxing enzyme of chicken erythrocytes. Interaction with superhelical DNA. J Biol Chem. 1976 Dec 10;251(23):7363–7366. [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Brandt W. F., von Holt C. The primary structure of yeast histone H3. Eur J Biochem. 1982 Jan;121(3):501–510. doi: 10.1111/j.1432-1033.1982.tb05815.x. [DOI] [PubMed] [Google Scholar]
- Certa U., Colavito-Shepanski M., Grunstein M. Yeast may not contain histone H1: the only known 'histone H1-like' protein in Saccharomyces cerevisiae is a mitochondrial protein. Nucleic Acids Res. 1984 Nov 12;12(21):7975–7985. doi: 10.1093/nar/12.21.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R. Genetic information in oocyte RNA. J Mol Biol. 1971 Mar 28;56(3):491–506. doi: 10.1016/0022-2836(71)90396-2. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller F. B. The writhing number of a space curve. Proc Natl Acad Sci U S A. 1971 Apr;68(4):815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Felsenfeld G., O'Dea M. H., Gellert M. Effects of DNA supercoiling on the topological properties of nucleosomes. Proc Natl Acad Sci U S A. 1987 May;84(9):2620–2623. doi: 10.1073/pnas.84.9.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M. E., Rooney T. F., Austin R. H. Evidence for kinks in DNA folding in the nucleosome. Nature. 1987 Aug 6;328(6130):554–557. doi: 10.1038/328554a0. [DOI] [PubMed] [Google Scholar]
- Holm C., Meeks-Wagner D. W., Fangman W. L., Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42(2):169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Müller U., Eicken I., Wendel I., Zentgraf H. Biochemical and ultrastructural analysis of SV40 chromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):227–244. doi: 10.1101/sqb.1978.042.01.025. [DOI] [PubMed] [Google Scholar]
- Lee K. P., Baxter H. J., Guillemette J. G., Lawford H. G., Lewis P. N. Structural studies on yeast nucleosomes. Can J Biochem. 1982 Mar;60(3):379–388. doi: 10.1139/o82-045. [DOI] [PubMed] [Google Scholar]
- Lewis P. N., Chiu S. S. Effect of histone H3 sulfhydryl modifications on histone-histone interactions and nucleosome formation and structure. Eur J Biochem. 1980 Aug;109(2):369–376. doi: 10.1111/j.1432-1033.1980.tb04803.x. [DOI] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Yeast chromatin subunit structure. Science. 1975 Apr 11;188(4184):165–166. doi: 10.1126/science.1090006. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Morse R. H., Cantor C. R. Effect of trypsinization and histone H5 addition on DNA twist and topology in reconstituted minichromosomes. Nucleic Acids Res. 1986 Apr 25;14(8):3293–3310. doi: 10.1093/nar/14.8.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Cantor C. R. Nucleosome core particles suppress the thermal untwisting of core DNA and adjacent linker DNA. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4653–4657. doi: 10.1073/pnas.82.14.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson D. S., Thoma F., Simpson R. T. Core particle, fiber, and transcriptionally active chromatin structure. Annu Rev Cell Biol. 1986;2:117–147. doi: 10.1146/annurev.cb.02.110186.001001. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Venkatesan M., Thoma F., Simpson R. T. Isolation of an episomal yeast gene and replication origin as chromatin. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7206–7210. doi: 10.1073/pnas.83.19.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Littau V. C., Allfrey V. G. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983 Oct;34(3):1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Saavedra R. A., Huberman J. A. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell. 1986 Apr 11;45(1):65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- Sigurdson D. C., Gaarder M. E., Livingston D. M. Characterization of the transmission during cytoductant formation of the 2 micrometers DNA plasmid from Saccharomyces. Mol Gen Genet. 1981;183(1):59–65. doi: 10.1007/BF00270139. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Mechanism of a reversible, thermally induced conformational change in chromatin core particles. J Biol Chem. 1979 Oct 25;254(20):10123–10127. [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Tricoli J. V., Kowalski D. Topoisomerase I from chicken erythrocytes: purification, characterization, and detection by a deoxyribonucleic acid binding assay. Biochemistry. 1983 Apr 12;22(8):2025–2031. doi: 10.1021/bi00277a045. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. The path of DNA in the nucleosome. Cell. 1982 Jul;29(3):724–726. doi: 10.1016/0092-8674(82)90433-0. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D. E. Compilation analysis of histones and histone genes. Nucleic Acids Res. 1986;14 (Suppl):r119–r149. doi: 10.1093/nar/14.suppl.r119. [DOI] [PMC free article] [PubMed] [Google Scholar]