Summary
Assembly of microRNA Ribonucleoproteins (miRNPs) or RNA-Induced Silencing Complexes (RISCs) is essential for the function of miRNAs and initiates from processing of precursor miRNAs (pre-miRNAs) by Dicer or by Ago2. Here, we report an in-vitro miRNP/RISC assembly assay programmed by pre-miRNAs from mammalian cell lysates. Combining in-vivo studies in Dicer Knock-Out cells reconstituted with wild type or catalytically inactive Dicer, we find that the miRNA Loading Complex (miRLC) is the primary machinery linking pre-miRNA processing to miRNA loading. We show that a miRNA Precursor Deposit Complex (miPDC) plays a crucial role in Dicer-independent miRNA biogenesis and promotes miRNP assembly of certain Dicer-dependent miRNAs. Furthermore, we find that 5′-uridine, 3′-mid base pairing and 5′-mid mismatches within pre-miRNAs promote their assembly into miPDC. Our studies provide a comprehensive view of miRNP/RISC assembly pathways in mammals and our assay provides a versatile platform for further mechanistic dissection of such pathways in mammals.
Introduction
miRNAs are a class of ∼22 nucleotide (nt) RNAs that play important roles in almost every biological process through post-transcriptional gene regulation. Proper miRNA functioning requires its assembly into an RNA-induced Silencing Complex (RISC) or microRNP (miRNP), where the miRNA serves as the specificity guide for target RNA recognition (Hammond et al., 2000; Hutvagner and Zamore, 2002; Martinez et al., 2002; Mourelatos et al., 2002; Nykanen et al., 2001). At the core of every miRNP/RISC is an Argonaute (Ago) protein, which directly binds to a single-stranded miRNA and upon target RNA recognition, orchestrates translational repression and degradation of the targeted mRNA (Djuranovic et al., 2011; Hutvagner et al., 2001; Martinez et al., 2002; Mourelatos et al., 2002; Siomi and Siomi, 2009). Most miRNAs are excised from the stems of stem-loop precursors (pre-miRNAs) by Dicer to generate transient, double-stranded miRNA duplexes (Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001; Knight and Bass, 2001; Zhang et al., 2004). Loading of miRNA duplexes to Ago proteins is assisted by Hsp70 and Hsp90 chaperones (Iwasaki et al., 2010; Maniataki and Mourelatos, 2005; Yoda et al., 2010), and is followed by unwinding of the duplex and retention of the single stranded miRNA by Ago. The miRNA duplex is structurally asymmetric and the strand with a thermodynamically less stable 5′ end becomes the miRNA (guide strand) that is retained by Ago, while the other strand (passenger) is degraded (Khvorova et al., 2003; Schwarz et al., 2003). Recent studies have found that unwinding is facilitated by mismatches present in the seed (guide positions 2-8) or 3′-mid (guide positions 12-16) of miRNA duplexes (Kawamata et al., 2009; Matranga et al., 2005; Yoda et al., 2010).
Of the four mammalian Ago proteins, Ago2 is the only one with robust endonucleolytic activity towards RNA targets with extensive complementarity to the Ago2-bound miRNAs (Liu et al., 2004; Meister et al., 2004; Rivas et al., 2005). The nucleolytic activity of Ago2 is critical for cleavage of the passenger strand of small interfering RNA (siRNA) duplexes and assembly of the guide strand into RISC; it is also essential for the maturation of miR-451, a Dicer-independent miRNA, through cleavage of the 3′ arm of pre-miR-451 (Cheloufi et al., 2010; Cifuentes et al., 2010; Leuschner et al., 2006; Matranga et al., 2005; Miyoshi et al., 2005; Rand et al., 2005; Yang et al., 2010). However, in terms of miRNA-directed mRNA translational repression and mRNA decay, all four Ago proteins function in a similar manner (Pillai et al., 2004; Wu et al., 2008). Not surprisingly, mammalian Ago proteins share a similar repertoire of miRNAs (Azuma-Mukai et al., 2008), which are enriched in 5′-U (Hu et al., 2009; Seitz et al., 2011), and have similar structural preference (i.e. mismatches at guide positions 9-11) for binding to miRNA duplexes (Yoda et al., 2010). The structural basis of the 5′-U preference was revealed in the crystallographic structure of human Ago2 in complex with the 5′ nucleotide mimics, where a nucleotide specificity loop, conserved among all mammalian Ago proteins, was identified in the MID domain (Frank et al., 2010).
We and other groups have previously identified a miRNA loading complex (miRLC), which displays both pre-miRNA processing and miRNP/RISC cleavage activity when it is exposed sequentially to a miRNA precursor and its target (Maniataki and Mourelatos, 2005; Gregory et al., 2005; MacRae et al., 2008). This result, together with the finding that the miRLC is composed of Dicer, miRNA-free Ago, Hsp90 and TRBP, led to the conclusion that miRLC is the miRNA loading complex in mammals (Gregory et al., 2005; Maniataki and Mourelatos, 2005). Structural studies lend further support for the formation of an Ago-Dicer-TRBP miRLC tripartite complex (Wang et al., 2009). On the contrary, a recent study showed that mammalian RISC can be assembled with a miRNA duplex using cell lysates in vitro, raising the question of whether the miRLC previously identified represented the main or a bypass mechanism of miRNP/RISC assembly (Yoda et al., 2010).
RISC assembly assays from cell lysates have provided illuminating insights into the mechanisms of siRNA and miRNA biogenesis in Drosophila melanogaster and mammals (Kawamata et al., 2009; Miyoshi et al., 2009; Pham et al., 2004; Tomari et al., 2004a; Yoda et al., 2010). The majority of these studies have employed the Dicer processing product, siRNA or miRNA duplexes, as the substrate for RISC assembly. In Drosophila, free miRNA duplexes are released from the Dicer-1 and Loquacious complex before they are sorted by Dicer-2 and R2D2 into distinct Agos (Kawamata and Tomari, 2010). However, because mammalian Dicer binds directly to Agos (Tahbaz et al., 2004) through an Ago-binding site that is conserved only within vertebrate Dicers (Wakiyama et al., 2007), it remains a subject of debate whether there are free miRNA duplexes in mammals under physiological conditions. More importantly, in vivo, all miRNA duplexes are born from pre-miRNAs, yet so far reconstitution of miRNP assembly in mammalian lysates with pre-miRNAs, the authentic miRNA precursors, has not been achieved.
Recently, the Haber lab found that a subset of pre-miRNAs can be cleaved by Ago2 (Diederichs and Haber, 2007). The Kiriakidou lab showed that pre-miRNAs remain associated with Ago proteins, even in the absence of Dicer, in vivo, implying that Ago2 may play additional roles in steps upstream of miRNPs (Tan et al., 2009). Yet within which context Ago2 binds to these pre-miRNAs remains unknown. More recently, it was found that the precursor of miR-451 (pre-miR-451), which has an unusually short stem with and extensive complementarity, is not processed by Dicer, but rather first cleaved by Ago2 and then trimmed into the mature miR-451. The mechanistic details of the Dicer-independent miRNA biogenesis pathway remain unclear. As these questions are related to pre-miRNAs rather than miRNA duplexes, establishment of an in-vitro assembly assay from mammalian lysates with pre-miRNAs will be needed to address these questions.
Here, we report a miRNP/RISC assembly assay using mammalian lysates programmed by pre-miRNAs instead of miRNA duplexes, which allows us to define the molecular steps of miRNP assembly of both Dicer-dependent and -independent miRNAs. Parallel studies in the Dicer Knock-Out cells and those reconstituted with either wild type or catalytically inactive Dicer, confirm the operation of the reported pathways in vivo. We reaffirm that the miRLC is the primary machinery responsible for processing of Dicer-dependent miRNA precursors and for loading miRNAs to Ago. In addition, we show that a miRNA Precursor Deposit Complex (miPDC), whose assembly is promoted by 5′-uridine, 3′-mid base pairing and 5′-mid mismatches, enhances the expression of certain Dicer-dependent miRNAs and plays a crucial role in the maturation of the Dicer-independent miR-451.
Results
An miRNP/RISC Assembly Assay Programmed by pre-miRNAs from Mammalian Cell Lysates
To further dissect the miRNA biogenesis pathways in mammals, we decided to establish a pre-miRNA programmed miRNP/RISC assembly assay. To distinguish RNA-protein complexes that are related to the miRNP assembly from those that are unrelated, we assembled miRNP with the precursor of a Dicer-dependent miRNA in lysates from inducible Dicer Knock-Out (Dicerfl/fl) or -heterozygous (Dicer+/fl) mouse embryonic fibroblasts (MEFs) after deletion of the floxed allele(s) with Hydroxytamoxifen (4-OHT) (Figure 1A) (Harfe et al., 2005; Tan et al., 2009).
Figure 1. miRNP/RISC Assembly Assay Programmed by pre-miRNAs from Mammalian Cell Lysates.
A. Schematic of assay. S100 lysates from MEFs heterozygous (+/−) or homozygous Dicer (−/−) knock-out are mixed with S100 lysates from Dicer−/− MEFs overexpressing myc-Ago2 (Ago2+); end-radiolabeled synthetic pre-miRNAs are added and complexes are analyzed by native agarose gel electrophoresis (NAGE).
B. Secondary structure of pre-miR-28. Guide (miRNA) and passenger strand is depicted in red and blue respectively. The same notation applies to all figures.
C. NAGE of assembly reactions with pre-miR-28 in equal mixture of Dicer+/− or Dicer−/− lysates and lysates from Dicer−/− MEFs overexpressing Ago2 (Ago2+) or not (Ago2−). Dotted lines delineate the five sections that were sliced.
D. Native Polyacrylamide Gel Electrophoresis (PAGE) analysis of RNAs isolated from gel slices as delineated in (C).
E. Antibody supershift experiments. Reactions were assembled with pre-miR-28 in equal mixture of Ago2− overexpressing Dicer−/− MEF lysates with lysates from Dicer−/− MEFs reconstituted with an HA-tagged Dicer transgene (WT). The red line marks the supershifted complex while the blue line denotes the disappeared complex. The same symbol representations apply to all figures.
F. Native PAGE of RNAs isolated from complex E (shown in SFigure 1A) in reactions programmed with 5′-radiolabeled pre-miR-28 without “cold” RNA target (N/A), or with “cold” RNA target with bulged miRNA binding sites of mir-28 or mir-16. Complex E, containing miR-28 miRNP that was assembled from pre-miR-28, recognizes specifically the miR-28 RNA target as evidenced by the shifting of labeled miR-28, bound to its cognate RNA target, towards the top of the gel.
We first incubated 5′ radiolabeled pre-miR-28 (Figure 1B) with equal mixture of cytoplasmic S100 lysates from Dicer+/− and Dice−/− MEFs at 37 °C for 60 min and resolved the assembled complexes by native agarose gel electrophoresis (NAGE). One prominent complex, which we term complex D was detected (Figure 1C, reaction 1). We then sectioned the gel into five equal slices from the top and eluted RNAs from each slice (Figure 1C). Native polyacrylamide gel electrophoresis (PAGE) of the eluted RNAs showed that slice IV (complex D) contained predominantly pre-miR-28, with some miR-28 duplex but little single-stranded miR-28 (Figure 1D, reaction 1). This implies that pre-miR-28 is processed by Dicer within complex D. Interestingly, slices III and II above complex D contained traces of pre-miR-28, some miR-28 duplex and little single-stranded miR-28, with slice II containing more single-stranded miR-28 and less miR-28 duplex than slice III (Figure 1D, reaction 1). This finding is reminiscent of the structural change of an siRNA duplex during the RLC to RISC transition in the RISC assembly assay in Drosophila (Tomari et al, 2004b). Thus, we reasoned that a similar transition also occurred in our assay and hypothesized that complex D contains miRLC while the region above complex D contains miRNP, even though its assembly was so inefficient that it was only faintly detected.
Further attempts to enhance the miRNP assembly efficiency led us to find that we could supplement ectopic Ago2 to stimulate miRNP assembly by replacing the Dicer−/− lysate with the lysate from a Dicer −/− MEF line that we engineered to overexpress myc-Ago2 from a stably transduced lentivirus (Ago2+) (Figure 1A). Now, another complex (E), whose assembly strictly depended on Dicer (Figure 1C, D reaction 3), was clearly detected in slices II and III (Figure 1C, reaction 2). Complex E contained single-stranded miR-28 and miR-28 duplex with very little pre-miR-28 (Figure 1D, reaction 2), suggesting that complex E is the miRNP. As every miRNP/RISC is composed of an Ago protein with a miRNA, stimulation of the miRNP assembly by supplementation of Ago2 also suggest s that Ago2 is the core constituent of complex E.
A signature function of miRNP/RISC is its ability to directly bind to cognate RNA targets, which leads to either mRNA cleavage or translational repression and mRNA degradation (Djuranovic et al., 2011; Siomi and Siomi, 2009). As shown in Figure 1F, miR-28 directs the miRNP in complex E (SFigure 1A) to bind specifically to a “cold” RNA target with miRNA recognition elements (MREs) for miR-28 but not miR-16. In addition, miR-28 can guide Ago2 to cleave a cognate target when a short perfectly complementary target is added to the immunoprecipitated miRNP after pre-incubation of the assembly reaction with 5′ phosphorylated, “cold” pre-miR-28 (SFigure 1B). Taken together, these findings indicate that complex E is indeed a functional miRNP/RISC.
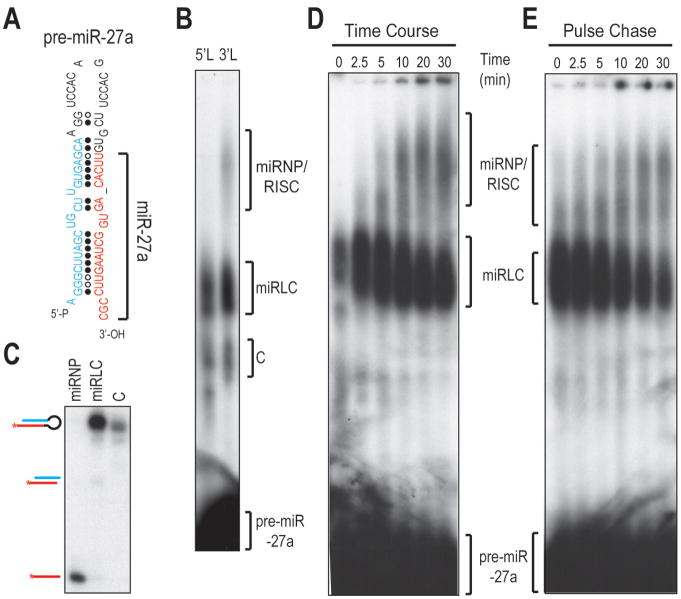
We next tested whether our assay was capable of assembling miRNP/RISC from pre-miR-27a, whose miRNA originates from the 3′-arm instead of the 5′-arm (Figure 2A). When 5′ (5′L) or 3′ (3′L) radiolabeled pre-miR-27a was used to program assembly reactions, only miR-27a emanating from the 3′ arm assembled into miRNP (Figure 2B, C and SFigure 2A, B). This confirms that our assay is capable of recapitulating the asymmetric incorporation of miRNAs in miRNP/RISC (Figure 2B).
Figure 2. Asymmetric miRNA loading in miRNP/RISC is faithfully recapitulated with a pre-miRNA substrate.
A. Schematic of pre-miR-27a.
B. NAGE of assembly reactions with 5′- (5′L) or 3′- radiolabeled (3′L) pre-miR-27a.
C. Native PAGE analysis of RNAs isolated from complexes assembled with pre-miR-27a-3′L in (B).
D. Kinetics of the miRNP/RISC assembly with pre-miR-27a-3′L.
E. Pulse-chase experiment. miRLC was pulsed in equal mixture of Ago2+ lysates with WT lysates at 37 °C for 5 min with 3′-radiolabeled pre-miR-27a before 20-fold unlabeled pre-miR-27a was added; chase of complex miRLC into miRNP was monitored at 30°C.
Kinetic analysis of RNA-protein complex formation with pre-miR-27a shows that the amount of complex E is in reverse proportion to that of complex D, suggesting that complex D is the precursor to complex E (Figure 2D). Furthermore, complex D can be “chased” into complex E (Figure 2E). Thus, complex D is the precursor to miRNP (complex E). Complex D is composed of Dicer, Ago and TRBP as antibodies against Dicer, Ago and TRBP, but not KSRP, an RNA binding protein not involved in miR-28 biogenesis (Trabucchi et al., 2009), could supershift complex D (Figure 1E and SFigure 2B). Thus, complex D is the miRLC complex identified previously (Gregory et al., 2005; MacRae et al., 2008; Maniataki and Mourelatos, 2005).
miRLC is the Primary miRNA Loading Machinery
miRLC contains not only pre-miRNAs and miRNA duplexes but also single-stranded miRNAs (Figures 1D), indicating that both pre-miRNA processing and miRNA loading occur within miRLC. miRNA duplexes are loaded into Agos and their unwinding by Agos is facilitated by mismatches and G/U wobble base pairs present in the seed and the 3′ mid of miRNAs (Kawamata et al., 2009; Yoda et al., 2010). If miRNA loading occurs within miRLC, the miRNA duplex should remain in the miRLC when its unwinding is blocked. To test this hypothesis, we programmed assembly reactions with a defective unwinding mutant of pre-miR-28 (pre-miR-28-dum) in the Dicerfl/fl ;myc-Ago2 lysate (SFigure 1C, D). pre-miR-28-dum was processed by Dicer into the miRNA duplex within the miRLC, but miRNP did not assemble as evidenced by the absence of single-stranded miR-28 (SFigure 1E). Instead, a complex that contained pre-miR-28-dum and a faster migrating RNA whose mobility corresponded to Ago2-cleaved pre-miR-28-dum (ac-pre-miR-28-dum), appeared at the same position as miRNP (complex E) (SFigure 1D). As ac-pre-miR-28-dum disappeared upon incubation of pre-miR-28-dum in the Dicerfl/fl lysate that overexpresses the catalytically-inactive myc-Ago2 (SFigure 1D, E), this suggested that Ago2 bound to and cleaved pre-miR-28-dum within complex E. Thus, the complex E formed by pre-miR-28-dum does not correspond to miRNP but to an Ago2-pre-miRNA containing complex and will be discussed further below. Notably, the miRNA duplex processed from pre-miR-28-dum resides mainly in the miRLC, confirming that miRNA loading primarily occurs within the miRLC (SFigure 1D, E).
miRLC is the Primary Machinery of Pre-miRNA Processing
The presence of pre-miRNAs, miRNA duplexes and single-stranded miRNAs in the miRLC strongly indicates that pre-miRNA processing and miRNA loading initiate in and are streamlined by the miRLC. The assembly of complex D strictly depends on Dicer (Figure 1C and SFigure 2A), indicating that Dicer is a core component of complex D. In that case, one would expect that the miRLC would accumulate when the catalytic activity of Dicer is inactivated as catalytically-inactive Dicer would trap the protein and RNA (pre-miRNA) components of miRLC. To test this hypothesis, we generated inducible Dicer Knock-Out MEFs that express HA-tagged wildtype (WT) or catalytically inactive mutant (D2A) Dicer transgene in a Doxycycline (Dox) responsive manner (SFigure 3A). We confirmed that only the introduction of the WT, but not the D2A mutant, could rescue the miRNA biogenesis defect in the Dicer−/− MEFs (SFigure 3). Consistent with our hypothesis, we observed a dramatic accumulation of miRLC in assembly reactions with the D2A lysate (Figure 3A), indicating that miRLC is the primary machinery of pre-miRNA processing in vitro.
Figure 3. pre-miRNA processing and miRNA loading initiate in and are streamlined by the miRLC.
A. NAGE of assembly reactions with pre-miR-28 in equal mixture of Ago2-overexpressing Dicer−/− MEF, lysates and lysates from Dice−/− MEFs expressing wild-type (WT) or catalytically inactive (D2A) Dicer transgenes.
B,C. Western Blots of whole cell lysates and Ago (B) or Dicer (C) immunoprecipitates (IP). NMS: Nonimmune mouse serum.
D. Northern Blots of RNAs isolated from Dicer immunoprecipitates.
Pre-miRNA is also processed by miRLC in vivo. As shown in Figure 3B, C, the amount of Dicer that associates with Agos and TRBP is increased in the D2A MEFs. In addition, increased amounts of pre-miRNAs were immunoprecipitated with Dicer from the D2A MEFs (Figure 3D), suggesting that pre-miRNAs are also processed in vivo by the miRLC. Collectively, all the results presented above indicate that miRLC is the primary machinery of pre-miRNA processing.
miRNP assembly with the precursor of a Dicer-independent miRNA via an Ago-containing, miRNA Precursor Deposit Complex (miPDC)
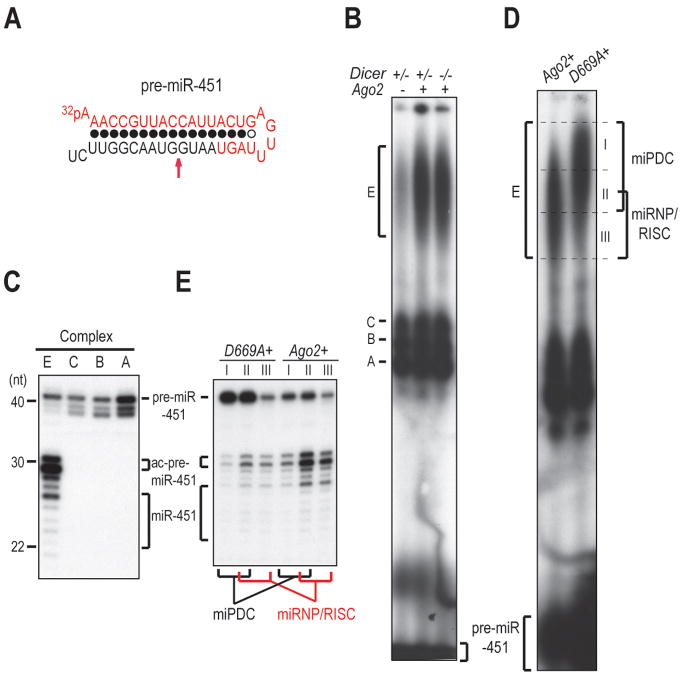
The biogenesis of miR-451 is independent of Dicer but is dependent upon the endonuclease activity of Ago2 (Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010). Though the primary transcript of miR-451 (pri-miR-451) is not expressed in MEFs, the biogenesis of miR-451 can be recapitulated when pri-miR-451 is ectopically expressed in MEFs (Yang et al., 2010). To test whether our assembly assay is capable of assembling miRNP with a Dicer-independent miRNA precursor, we incubated 5′ radiolabeled pre-miR-451 (Figure 4A) with equal mixture of Dicer+/− lysates with lysates from Dicer−/− MEFs that were overexpressing Ago2 (Ago2+). Four complexes assembled (Figure 4B), yet only complex E contained miR-451 (Figure 4C), suggesting complex E contains miRNP. In addition, complex E contained Ago2-cleaved pre-miR-451 (ac-pre-miR-451) and pre-miR-451 (Figure 4C), indicating that pre-miR-451 is directly bound and cleaved by Ago2 within complex E. The assembly of complex E is stimulated by supplementation of Ago2 but is independent of Dicer (Figure 4B and SFigure 4A), consistent with the independent role of Dicer in the maturation of miR-451 in vivo (Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010). However, “pulsing” of complex E failed to “chase” it into any other complex (SFigure 4B) and kinetic analysis of the assembled RNA-protein complexes with pre-miR-451 also failed to identify any precursor complex to complex E (SFigure 4C), implying that the mature miR-451 resides in the miRNP while pre-miR-451 and ac-pre-miR-451 are probably retained in another complex that we name “miRNA Precursor Deposit Complex” (miPDC).
Figure 4. miRNP/RISC assembly with the precursor of a Dicer-independent miRNA via a miRNA Precursor Deposit Complex (miPDC).
A. Secondary structure of pre-miR-451. The red arrow denotes the Ago2 cleavage site.
B. NAGE of assembly reactions with pre-miR-451 in equal mixture of Dicer+/− or Dicer−/− lysates and lysates from Dicer−/− MEFs overexpressing Ago2 (Ago2+) or not (Ago2−).
C. Denaturing PAGE of RNAs isolated from indicated RNA-protein complexes assembled in equal mixture of Dicer+/− lysates and Ago2+ lysates.
D. NAGE of assembly reactions with pre-miR-451 in equal mixture of Dicer+/− lysates and lysates from Dicer−/− MEFs overexpressing wild-type (Ago2+) or catalytically inactive (D669A+) Ago2 transgenes.
E. Denaturing PAGE of RNAs isolated from the sliced sections of NAGE shown in (C). Trace amounts of ac-pre-miR-451 in reactions assembled with catalytically-inactive D669A Ago2, are generated by small amount of endogenous Ago2 in the lysate.
To test this hypothesis, assembly reactions were set up in lysates from Dicer−/− MEFs overexpressing a catalytically inactive Ago2 mutant (D669A+) instead of the wild-type Ago2. Interestingly, assembly in the D669A+ lysate retarded the migration of complex E (Figure 4D). This is likely the result of blocking the pre-miR-451 cleavage, as a similar retardation is also observed when a pre-miR-451 mutant that is resistant to Ago2 cleavage (m10) is assembled in the Ago2+ lysate (SFigure 4D). Next, we asked which portion of complex E contained miPDC and which contained miRNP by sectioning the gel region corresponding to complex E into three slices (Figure 4D). Slice I, enriched in the reaction with the D669A+ lysate (Figure 4D), contains predominantly pre-miR-451, (Figure 4E) and thus corresponds to miPDC. Slice II, common to both reactions assembled with the Ago2+ and the D669A+ lysate (Figure 4D), contains a mixture of pre-miR-451, ac-pre-miR-451 and mature miR-451 (Figure 4E) and thus is composed of both miPDC and miRNP. Slice III, enriched in the reaction with the Ago2+ lysate (Figure 4D), contains primarily mature miR-451 and ac-pre-miR-451, (Figure 4E) and thus consists of miRNP. Taken together, we conclude that complex E is composed of both miPDC and miRNP.
The Hsp70 inhibitor 2-phenylethynesulfonamide (PES) inhibited the assembly of complex E (SFigure 4E), suggesting that pre-miR-451 is loaded into Ago2 by a similar PES-sensitive machinery that loads the miRNA duplex into Ago proteins (Iki et al., 2010; Iwasaki et al., 2010; Miyoshi et al., 2010). Thus the loading of pre-miR-451 into Ago2 is crucial for miPDC and miRNP assembly.
Assembly of certain Dicer-dependent pre-miRNAs into miRLC and miPDC
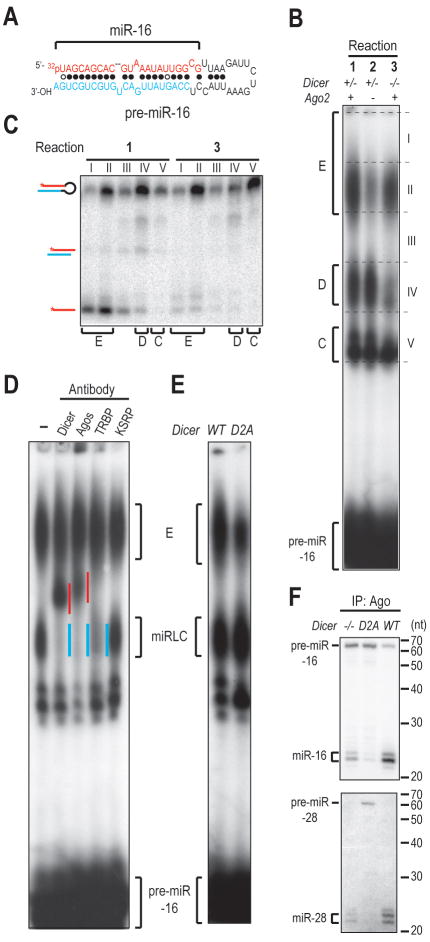
The assembly of miPDC with pre-miR-451 and with the defective unwinding mutant of pre-miR-28 pre-miR-28-dum (SFigure 1C, D, E) prompted us to test other pre-miRNAs for their ability to assemble miPDC. One such pre-miRNA is pre-miR-16 (Figure 5A). Similar to pre-miR-28 and pre-miR-27a, the miRLC for pre-miR-16 resides in complex D (Figure 5B, C, D, E) and the assembly of miRNP from pre-miR-16 is strongly stimulated by supplementation of Ago2 (Figure 5B, C). However, in assembly reactions with pre-miR-16, complex E contains in addition to miRNPs, significant amounts of pre-miR-16 (Figure 5B, C, reaction 1), in contrast to the E complexes formed with pre-miR-28 and pre-miR-27a, which are composed primarily of miRNPs. In addition, complex E containing pre-miR-16 still formed in the absence of Dicer (Figure 5B, C, reaction 3). The assembly of pre-miR16 into complex E is not an artifact due to the incorporation of “unstructured” single-stranded pre-miR-16 into Ago2, as a pre-miR-16 mutant with a 2-nt 5′ overhang instead of the native 3′ overhang, does not assemble complex E (SFigure 5A). Single-stranded miR-16 also resides in complex E (Figure 5B, C, reaction 1), suggesting that similar to pre-miR-451, the complex E formed in assembly reactions with pre-miR-16 is also composed of two co-migrating complexes-- miRNP with single-stranded miR-16 and another complex with pre-miR-16. Three lines of evidence support that the complex that co-migrates with miRNP is miPDC. First, this complex is supershifted by an anti-Myc antibody, suggesting that pre-miR-16 is bound by myc-Ago2 (SFigure 5E). In addition, an Ago2-cleavable pre-miR-16 mutant (cm), which has complementary mutations introduced into the 3′-mid region surrounding the Ago2-cleavage site (SFigure 5B), is cleaved by Ago2 (SFigure 5D), indicating that pre-miR-16 is bound to Ago2 by anchoring its 5′ phosphate in the MID domain of Ago2 and uses the 5′ arm as a guide to cleave the 3′ arm. Interestingly, assembly of miPDCs with pre-miR-28-dum and pre-miR-16-cm in the D669A+ lysate does not cause retardation of complex E (SFigure 1D and SFigure 5C), which is unlike the retardation seen when pre-miR-451's miPDC is formed in the D669A+ lysate. This suggests that the retardation observed by blockage of Ago2 cleavage is specific to pre-miR-451, which is probably resulting from the unusually short length of pre-miR-451 and the extensive complementarity of its stem. Finally, the assembly of pre-miR-16 into complex E is also strongly inhibited by the Hsp70 inhibitor PES (SFigure 5F, G). Collectively, these results suggest that complex E formed in pre-miR-16 programmed assembly reactions is composed of both miRNP and miPDC.
Figure 5. Assembly of the Precursor of a Dicer-dependent miRNA into miRLC and miPDC.
A. Secondary structure of pre-miR-16-1., B. NAGE of assembly reactions with 5′-radiolabeled pre-miR-16 in equal mixture of Dicer+/− or Dicer−/− lysates and lysates from Dicer−/− MEFs overexpressing Ago2 (Ago2+) or not (Ago2−)., C. Native PAGE of RNAs isolated from gel slices as delineated in (B)., D. Antibody supershift experiments in equal mixture of Ago2-overexpressing Dicer−/− lysates and lysates from Dicer−/− MEFs reconstituted with HA-tagged Dicer transgene., E. NAGE of assembly reactions in equal mixture of Ago2-overexpressing Dicer−/− lysates and lysates from Dicer Knock-Out MEFs expressing wild-type (WT) or catalytically inactive (D2A) Dicer transgene., F. Northern blots of RNAs isolated from Ago immunoprecipitates.
We next tested whether the complexes that we identified in our in vitro assembly assays are also found in vivo. Ago is the core component of both miRLC and miPDC. In the Dicer−/− MEFs, assembly of miRLC is greatly reduced while that of miPDC is not affected (Figure 5B, reaction 3). In contrast, in the D2A MEFs, miRNP is reduced but neither miRLC nor miPDC assembly is affected (Figure 5E, SFigure 5H). If a pre-miRNA can remain associated with Agos in the Dicer−/− MEFs, where miRLC assembly does not take place, this implies that the pre-miRNA interacts with Ago through miPDC. Indeed, Figure 5F and SFigure 5J show that pre-miR-16 and pre-miR-21, which assemble both miRLC and miPDC (Figure 5B and SFigure 5I), co-immunoprecipitate with Agos in both the Dicer−/− and the D2A MEFs. In contrast, pre-miR-28 and pre-miR-27a, which assemble only miRLC, co-immunoprecipitate with Agos only in the D2A but not the Dicer−/− MEFs (Figure 5F and SFigure 5J). Notably, the interaction of pre-miR-16 and pre-miR-21 with Agos is observed not only when miRNA biogenesis is blocked in the Dicer−/− and the D2A MEFs, but also in the WT MEFs (Figure 5F and SFigure 5J), arguing that their interaction with Ago is not a simple result of the availability of more pre-miRNAs in the Dicer−/− and the D2A MEFs but is physiologically relevant. Taken together, these results suggest that some pre-miRNAs (e.g. pre-miR-16 and pre-miR-21) assemble both miRLC and miPDC whereas others (e.g. pre-miR-28 and pre-miR27a) assemble only miRLC in vivo.
Processing of pre-miR-16 by Dicer occurs in the miRLC as photocrosslinking experiment with pre-miR-16 containing a single 4-thiouridine adjacent to Dicer's cleavage site (pre-miR-16-s21U) showed that pre-miR-16 was crosslinked to Dicer only in the miRLC but not in the miPDC (SFigure 5K, L, M). A previous study found that certain Dicer-dependent miRNA precursors, such as the pre-let-7 family members, were cleaved by Ago2 before Dicer-processing to facilitate removal of the passenger strand during unwinding (Diederichs and Haber, 2007). Our results suggest that Dicer-dependent miRNA precursors are cleaved by Ago2 primarily in the miPDC before they are processed by Dicer in the miRLC (SFigure 5D). Indeed, a small amount of ac-pre-miR-16-cm, which was produced by Ago2 cleavage within the miPDC, was also recovered in the miRLC (SFigure 5D), suggesting that miPDC may be a precursor to miRLC. In that scenario, one would expect accumulation of miPDC when the assembly of miRLC and miRNP is blocked in the Dicer−/− lysate. As shown in Figure 5B, reaction 3 and SFigure 5I, that is exactly what occurs. On the contrary, miPDC dis not accumulate as miRLC accumulated in the D2A lysate (Figure 5E and SFigure 5H) suggesting that the accumulation of miPDC in the Dicer−/− lysate is not simply due to availability of more “free” pre-miRNAs in the Dicer−/− lysate. Hence, Ago stimulates miRNP assembly not only by serving as the acceptor of miRNAs but also by promoting the assembly of certain pre-miRNAs into the miPDC.
5′ Nucleotide Identity and Structural Features Important for miPDC Assembly
We have demonstrated that pre-miRNAs containing extensive base pairing around the 10th nucleotide from the 5′ end are cleaved by Ago2 within the miPDC in a similar way that miRNAs cleave their targets. This indicates that the 5′ ends of such pre-miRNAs are anchored in the MID domain of Ago2, which was shown to mediate the interaction of Ago2 with the 5′ phosphate of RNA guides (Ma et al., 2005; Parker et al., 2005). A recent structural study identified a nucleotide specificity loop in human Ago2 that plays a critical role in determining the nucleotide binding preference (U > A ≫ G ≈ C) at the 5′ end of miRNAs (Frank et al., 2010). We notice that both pre-miR-27a and pre-miR-28, which do not assemble miPDC (Figure 1B, C and Figure 2A, B), begin with an A. In contrast, pre-miR-16 and pre-miR-21, which assemble miPDC (Figure 5B and SFigure 5I), begin with a U. However, the first nucleotide is not the only determinant for miPDC assembly because pre-miR-451, which assembles miPDC, begins with an A. In addition, the defective unwinding mutant of pre-miR-28 (pre-miR-28-dum), which has two complementary mutations introduced into both the seed and the 3′-mid, can also assemble miPDC although it begins with an A (SFigure 1C, D, E), indicating that the secondary structure of a pre-miRNA also plays a role in the miPDC assembly.
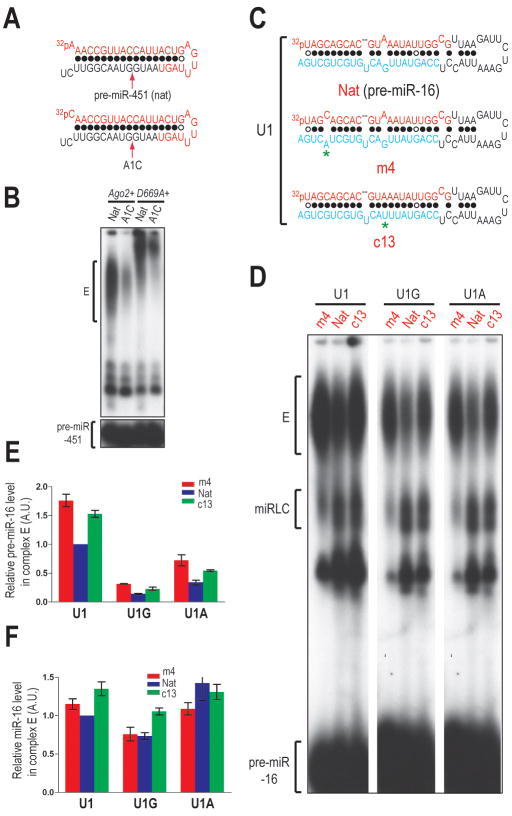
To identify the 5′ nucleotide and structural features of pre-miR-16 important for miPDC assembly, we first changed its first nucleotide into an A (U1A) or a G (U1G). Mutation to a C was not attempted as such a mutation would allow the first nucleotide to base pair with the 3′ penultimate nucleotide, which could affect guide strand (mature miRNA) selection. However, changing the first nucleotide of pre-miR-451 to a C (A1C) led to marked inhibition of miPDC assembly and as a result miRNP formation, verifying that miPDC assembly is critical for generation of miR-451 miRNP (Figure 6A, B). Mutation of the first nucleotide of pre-miR-16 to an A or a G resulted in 2- and 4-fold reduction in the efficiency of miPDC assembly, respectively (Figure 6D, E), indicating that the identity of the first nucleotide of pre-miR-16 plays a major role in guiding miPDC assembly. In addition, the amount of miR-16 was reduced when the first nucleotide of pre-miR-16 was mutated to a G (Figure 6F). Next, we introduced point mutations that alter base pairing properties of nucleotide 4 (m4) or 13 (c13) of pre-miR-16 (Figure 6C). In addition, we introduced these secondary structure mutations into the 5′ nucleotide mutants (U1A or U1G) to assess their relative contribution to the miPDC assembly. As shown in Figure 6D, E, F and SFigure 6A, mismatch in the 4th position and base pairing in the 13th position promoted both miPDC and miRNP assembly. However, structural mutants with 5′-G and 5′-A did not assemble more miPDC than their 5′-A and 5′-U counterparts with the native structure respectively (Figure 6D, E), suggesting that the secondary structure plays an accessory role in the miPDC assembly. when a pre-miR-28 point mutant with base pairing at the 13th nucleotide (pre-miR-28-c13) assembled more miPDC and miRNP than its native counterpart (SFigure 6B, C, D).
Figure 6. 5′ Nucleotide Identity and Structural Features Important for miPDC Assembly.
A. Secondary structures of the native (Nat) and the 5′-cytidine mutant (A1C) of pre-miR-451.
B. NAGE of assembly reactions in lysates from Dicerfl/fl MEFs overexpressing wild-type (Ago2+) or catalytically inactive (D669A+) Ago2 transgenes.
C. Secondary structures of pre-miR-16, m4 or c13 mutants. Mutated nucleotides are marked with asterisks.
D. NAGE of assembly reactions programmed with pre-miR-16 containing native (Nat) or altered secondary structures (m4 or c13) in the 5′ wild-type (U1) or 5′ mutant (U1G or U1A) background.
E, F. Relative amounts of pre-miR-16 (E) and miR-16 (F) in complex E. The standard error of the mean (SEM) is from two different experiments.
Next, we introduced single mismatch mutations, which disrupt base pairing of the seed (m2-m8) or 3′ mid (m12-m16) region of miR-451 without changing the sequence of mature miR-451, into the 3′ arm of pre-miR-451 (SFigure 6E). Consistent with the findings with pre-miR-16, mismatches at the 3′ mid (12th-16th) region attenuated miPDC assembly whereas mismatches at the 5′-mid (5th-7th) positions enhanced miPDC assembly (SFigure 6F, G). The inhibition of miPDC assembly by mismatches at the 2nd and 3rd positions (SFigure 6F, G) are specific to pre-miR-451 probably because of its extremely short stem, as we did not observe inhibition of miPDC assembly when a similar mismatch was introduced into the 3rd position of pre-miR-16 (our unpublished data).
Taken together, the findings presented above indicate that the 5′ nucleotide identity and the secondary structure of a pre-miRNA play major roles in miPDC assembly: 5′-U and base pairing at the 3′ mid region promotes miPDC assembly while base pairing at the 5′ mid region attenuates miPDC assembly.
Discussion
The paucity of in-vitro systems that program assembly of miRNP/RISC with pre-miRNAs has been a stumbling block for dissection of mechanistic details of mammalian miRNP/RISC assembly pathways. With supplementation of Ago2 protein, we have established a miRNP/RISC assembly assay in mammals, which can be programmed with both Dicer-dependent and Dicer-independent miRNA precursors and which faithfuly recapitulates many key processes during miRNA biogenesis in vivo. Our studies reaffirm that miRLC is the primary machinery that carries both pre-miRNA processing and miRNA loading for Dicer-dependent miRNA precursors in mammals. Hence, contrary to a previous report that used miRNA duplexes instead of pre-miRNAs to study miRNP assembly (Yoda et al., 2010) but in agreement with a recent report (Noland et al., 2011), miRLC is the canonical miRNA loading complex in mammals and plays a key role in pre-miRNA processing, in loading of the Dicer products (miRNA duplexes) to Ago protein and in initiating miRNP formation.
We have found that miPDC, whose core is composed of a pre-miRNA directly binding to Ago2, is assembled with certain miRNA precursors beginning with 5′-U or 5′-A in the context of preferable secondary structure. Our studies confirm and extend the original discovery of Ago2-pre-miRNA complexes in vivo (Tan et al., 2009) and place them at the center of mammalian miRNP assembly pathways. Assembly of pre-miR-451 into miPDC is crucial for the maturation of miR-451 as miPDC couples Ago2 cleavage with the trimming of ac-pre-miR-451 to generate mature miR-45. The assembly of Dicer-dependent miRNA precursors into miPDC suggests that the Dicer-independent miR-451 maturation pathway probably evolved by using the pre-existing miPDC in the Dicer-dependent miRNA biogenesis pathway and diverting it into an Dicer-independent pathway.
The assembly of Dicer-dependent miRNA precursors into miPDC promotes their expression by escorting them to the miRLC for processing by Dicer. Notably, preferable features of pre-miRNAs that promote miPDC assembly resemble the characteristics of an authentic pre-miRNA and those of a productive miRNA duplex: a 5′-U and base pairing in the 3′-mid region for efficient miRNA loading (Kawamata et al., 2011; Seitz et al., 2011); and mismatches in the 5′-mid region for efficient unwinding (Kawamata et al., 2009; Yoda et al., 2010). However, although many of the features for miPDC assembly are shared with those for efficient loading or unwinding of Dicer products (miRNA duplexes), they are not identical. Importantly, some features present in natural mouse miRNA duplexes (Seitz et al., 2011), which cannot be explained by having a role in miRNA loading, make good sense when miPDC assembly is taken into account. For example, the Seitz lab has found that although miRNAs in mouse, D. melanogaster and C. elegans are enriched in 5′-U, regardless of their origin from the 5′ or 3′ arm of pre-miRNAs, the 5′-arm-derived miRNA*s (passenger strands) in mouse are enriched significantly in 5′-A compared to their 3′-arm-derived counterparts (Seitz et al., 2011). In theory, as the passenger (miRNA*) strand is not loaded into Ago, as long as it does not begin with a 5′-U to compete for guide (miRNA) strand selection, its first nucleotide does not need to be enriched in adenosine. Instead, we speculate that the preferential enrichment of 5′-A in the 5′-arm-derived miRNA*s, reflects their role in promoting miPDC assembly. It is also intriguing to note that the first nucleotide of pre-miR-451, which is strictly conserved in all organisms encoding miR-451, is an adenosine instead of a uridine. This does not seem to reflect a constraint for efficient Drosha processing, as neither A nor U base pairs with the opposite C. In addition, it is outside of the seed and thus should have no impact on mRNA targeting specificity. Instead, it is possible that the conservation of a 5′-A instead of a 5′-U in pre-miR-451 reflects a need for the cell to restrict excessive loading of pre-miR-451 to miPDC, which might interfere with the assembly of other pre-miRNAs into miPDC and lower the availability of Agos for miRLC assembly.
Many studies have revealed that processing of certain pri-miRNAs by Drosha is stimulated in response to various intrinsic and extrinsic stimuli (Krol et al., 2010). Interestingly, a recent study found that there is competition competition between Dicer mRNA and pre-miRNAs for export by Exportin-5 and expression of excessive pre-miRNAs results in downregulation of Dicer (Bennasser et al., 2011). Thus, it is tempting to speculate that the miPDC may also serve as a temporary storage site of pre-miRNAs during fluctuation of Dicer's level.
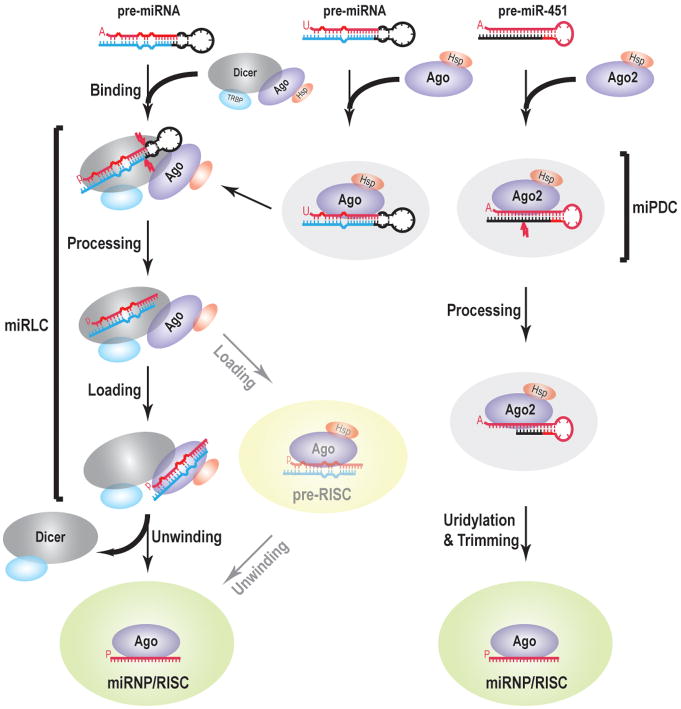
In summary, the pre-miRNA programmed miRNP/RISC assembly pathways revealed by our studies have allowed us to build a coherent model of how such pathways operate in mammals to form miRNPs/RISCs, the functional units for miRNAs (Figure 7). Furthermore, our assay provides a platform for studying and manipulating every step of the miRNP/RISC assembly pathway in vitro, which will facilitate elucidation of mechanistic details of miRNP biogenesis and its regulation.
Figure 7. A model for pre-miRNA programmed silencing complex assembly pathways in mammals.
Experimental Procedures
Preparation of Immortalized MEF Cell Lines and Cell Culture
The immortalized Dicer+/fl, Dicerfl/fl MEF cell lines and the induction of loxP recombination has been previously described (Tan et al., 2009). The expression of HA-Dicer or myc-Ago transgene was initiated by addition of 0.5-1 μg/ml of Doxycycline (Dox) to the medium. All transfections were performed with Lipofectamine 2000 (Invitrogen).
miRNP/RISC Assembly Assay
40 μg of two cytoplasmic S100 lysates from the indicated genotyped MEFs was mixed at 1:1 ratio and incubated with 50-100 fmole of 5′ 32P-radiolabeled pre-miRNAs with or without miRNA targets in 40 mM KOAc (pH 7.4), 3 mM Mg(OAc)2, 1 mM DTT, 1 mM ATP, 0.2 mM GTP and 1 U/μl of recombinant RNasin (Promega) at 37 °C for the indicated time. RNA products were isolated with TRIzol (Invitrogen) and resolved through a 15% denaturing polyacrylamide gel. To detect assembled RNA-protein complexes, native agarose gel electrophoresis was performed.
Native Agarose Gel Electrophoresis
Native agarose gel electrophoresis (NAGE) is modified from the protocol described in (Tomari et al., 2004a). In brief, a native agarose gel made of 1.5% of MetaPhor agarose (Lonza), 0.5% of SeaKem Gold agarose (Lonza) and 0.5×TBE with 1.5 mM MgCl2, was cast into a glass plate sandwich with a Gelbond film (Lonza) attached on one plate. After RISC assembly assay, Ficoll-400 was added to 3% and the RNA-protein complexes were resolved on a native agarose gel at 5 W for 3.5-4 hr at 4 °C.
Identification of RNA Components in RNA-Protein Complexes
To identify the RNA components in the RNA-Protein complexes, native agarose gels were sliced at 2-2.5 cm intervals from the top (excluding the well) to the last complex or gel slices containing individual complexes was recovered. The RNAs were eluted in PK buffer (100 mM Tris-HCl (pH 7.5), 150 mM NaCl, 12.5 mM EDTA, 0.5% SDS), extracted with acid phenol-chloroform and precipitated with ethanol. For denaturing PAGE analysis, the RNA pellets were redissolved in Gel Loading Buffer II (Ambion) and resolved on a 15% denaturing PAGE gel. Native PAGE analysis is adapted from (Tomari et al., 2004a). Specifically, RNA pellets were redissolved in native loading buffer and resolved on a 10% native polyacrylamide (19:1) gel at 4 °C at 7 W, using 0.5×TBE with 1 mM MgCl2 as the running buffer.
Supplementary Material
Highlights.
Development of in-vitro miRNP/RISC assembly assay programmed by pre-miRNAs.
miRLC is the primary machinery that links pre-miRNA processing to miRNA loading.
Roles of miPDC in miRNA biogenesis.
5′-U, 3′-mid base pairs, 5′-mid mismatches in pre-miRNAs promote miPDC assembly.
Acknowledgments
We are grateful to M. Simon and L. Fraser for sharing the pEN-Tmcs and pSLIK-Hyg plasmids, DD. Trono for the lentivirus packaging plasmids, Dr. V. Kim for the anti-Dicer antibody; E. Prak and A. Panigrahi for their help with real-time PCR quantitation and Developmental Hybridoma Studies Bank (DHSB) for the 9E10 antibody. We thank all members of our lab for helpful discussions. Supported by NIH grant GM072007 to ZM.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci U S A. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Chable-Bessia C, Triboulet R, Gibbings D, Gwizdek C, Dargemont C, Kremer EJ, Voinnet O, Benkirane M. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat Struct Mol Biol. 2011;18:323–327. doi: 10.1038/nsmb.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Zhou YH, Chen W, Khaitovich P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 2009;10:413. doi: 10.1186/1471-2164-10-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Yoda M, Tomari Y. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep. 2011;12:944–949. doi: 10.1038/embor.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Okada TN, Siomi H, Siomi MC. Characterization of the miRNA-RISC loading complex and miRNA-RISC formed in the Drosophila miRNA pathway. RNA. 2009;15:1282–1291. doi: 10.1261/rna.1541209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol. 2010;17:1024–1026. doi: 10.1038/nsmb.1875. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell. 2011;43:110–121. doi: 10.1016/j.molcel.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Seitz H, Tushir JS, Zamore PD. A 5′-uridine amplifies miRNA/miRNA* asymmetry in Drosophila by promoting RNA-induced silencing complex formation. Silence. 2011;2:4. doi: 10.1186/1758-907X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, Garchow BG, Liu X, Yeung J, Morris JPt, Cuellar TL, McManus MT, Kiriakidou M. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004a;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004b;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr Biol. 2008;18:1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.