Abstract
The Latvian HIV-1 outbreak among intravenous drug users (IDUs) in 1997–1998 involved subtype A1. To obtain a more complete picture of the Latvian HIV-1 epidemic, 315 HIV-1-infected patients diagnosed in 1990–2005 representing different transmission groups and geographic regions were phylogenetically characterized using env V3 and gag p17 sequences. Subtypes A1 and B infections were found in 76% and 22% of the patients, respectively. The subtype A1 sequences formed one large cluster, which also included sequences from other parts of the former Soviet Union (FSU), whereas most subtype B sequences formed three distinct clusters. We estimated that subtype A1 was introduced from FSU around 1997 and initially spread explosively among IDUs in Riga. A recent increase of heterosexually infected persons did not form a separate subepidemic, but had multiple interactions with the IDU epidemic. Subtype B was introduced before the collapse of the Soviet Union and primarily has spread among men who have sex with men.
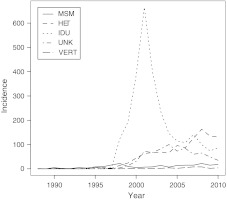
The first HIV-1-infected individual in Latvia was registered in 1987 and by the end of 2009 a total of 4614 cases had been identified.1 Of these, 825 patients had developed AIDS and 494 had died either from AIDS or from other causes. In the first 10 years of the Latvian epidemic transmissions occurred mainly through sexual contacts. However, in 1998 an explosive outbreak of HIV-1 infections started among intravenous drug users (IDUs). The outbreak culminated in 2001 when 807 new cases were reported of which 665 were IDUs (Fig. 1). In recent years the proportion of heterosexually acquired infections among newly diagnosed has increased from 12 % in 2002 to 48 % in 2009. Similar patterns have been reported from several other countries in the former Soviet Union (FSU) and Eastern Europe.2 We have previously molecularly characterized HIV-1 variants from 97 Latvian patients, who were newly diagnosed between 1995 and 2001 and of whom 89 were IDUs.3 We found that Latvian IDUs were infected with a subtype A1 virus variant that was similar to the A1 variant found in many other parts of the FSU and that was first described from the Ukraine.4,5 However, little is known about the transmission patterns in the early phase of the Latvian HIV-1 epidemic and in other transmission groups than IDUs. The aim of this study was to obtain a better understanding of the Latvian HIV-1 epidemic by molecularly characterizing HIV-1 in patients sampled over the entire time span of the epidemic and representing all transmission groups and geographic regions of Latvia.
FIG. 1.
Number of newly HIV-1 diagnosed individuals in different transmission groups in Latvia from 1987 to 2008. HET, heterosexual; MSM, men who have sex with men; VERT, mother-to-child transmission; IDU, intravenous drug use; Unkn, unknown transmission route.
The study included 315 Latvian patients with a known transmission route who were diagnosed as seropositive for HIV-1 between 1990 and 2005 and from whom frozen plasma samples with known time for sampling were available at the Infectology Center of Latvia (Table 1). We especially targeted samples from patients diagnosed in the early phase of the Latvian HIV-1 epidemic and samples from transmission groups other than IDUs. From IDUs we included a proportion of all available samples because our earlier study had shown that most IDUs were infected with closely related subtype A1 viruses.3 The samples had been collected between January 1998 and October 2005. A majority of the samples (n=218) were newly sequenced, but 97 sequences were from our previous study.3 The study population included 198 males, 111 females, and for 6 patients the gender was unknown. A majority of the patients (n=246) were from the capital Riga, which has the largest number of HIV-1 cases in the country, 52 patients were from other regions, and for 17 patients the place of residence was unknown. The reported route of infection was as follows: 135 IDUs, 87 heterosexuals, 56 men who have sex with men (MSM), 3 mother-to-child transmissions, and 34 unknowns (Table 1). The study was approved by the Latvian Ethical Committee (19.06.1998. Nr.2 and 24 March, 2005, A-4, decision nr.4) and the Regional Medical Ethics Board in Stockholm (Dnr. 2004/4:6 and Dnr. 2008/4:1).
Table 1.
Subtype Distribution and Route of Transmission in the 315 Latvian HIV-1 Patients Included in the Study
| |
Subtype |
||||||
|---|---|---|---|---|---|---|---|
| Route of transmission | A1 | B | C | G | CRF02_AG | CRF03_AB | Totala |
| Intravenous drug use | 132 | 2 | — | — | — | 1 | 135 (6%) |
| Heterosexual | 70 | 10 | 2 | 1 | 4 | — | 87 (19%) |
| Men who have sex with men | 5 | 51 | — | — | — | — | 56 (41%) |
| Mother-to-child | 3 | — | — | — | — | — | 3 (25%) |
| Unknown | 29 | 5 | — | — | — | — | 34 (7%) |
| Total | 239 | 68 | 2 | 1 | 4 | 1 | 315 (9%) |
In parentheses is the coverage in percent of HIV-1 registered patients by 2005.
Virus RNA was extracted from plasma and the env variable region V3 (V3) and gag p17 (p17) regions were amplified using nested PCR primers and sequenced as previously described.6–8 For subtype assignments, the sequences were manually aligned with the subtype reference dataset available at the Los Alamos HIV sequence Database5 using BioEdit.9 The alignments were used to construct neighbor-joining (NJ) phylogenetic trees with the maximum composite likelihood substitution model in Mega v4.1.10 Sequences in the Los Alamos Database5 that were closely related to our Latvian HIV-1 sequences were selected by a two-step procedure involving preliminary identification by HIV BLAST5 searches and verification using NJ tree analyses. Based on these analyses 279 subtype A1 and 125 subtype B V3 database sequences were selected for further phylogenetic analyses. PhyML v.3.011 was used to construct maximum likelihood (ML) phylogenetic trees with a generalized time reversible substitution model with gamma distributed rate variation among sites and invariant sites (GTR+G+I), which was identified as the best model by the FindModel tool.5 PhvML was also used to test the phylogenetic robustness using nonparametric bootstrap resampling (100 replicates). Bayesian coalescent methods as implemented in BEAST v.1.5.412 was used to estimate the time for the most recent common ancestor (MRCA) for certain sequence clusters as well as to construct maximum clade credibility phylogenetic trees for HIV-1 subtypes A1 and B in Latvia. For subtype B all Latvian sequences were included in the BEAST analyses. For subtype A1 the analyses were carried out using three subsets of 60 randomly selected Latvian sequences (for both p17 and V3) because the large number of subtype A1 sequences prohibited BEAST analyses using all sequences. We selected a relaxed clock with lognormal distributed rates and the Bayesian skyline demographic model (with 10 population size categories) based on our prior knowledge about the evolution of HIV-113,14 and the HIV-1 population dynamics in Latvia (Fig. 1), but other demographic models were also tried. The Bayesian search was run with a Markov Chain Monte Carlo (MCMC) chain for 108 generations with sampling every 105 generations. Convergence of the MCMC chain was assumed when the likelihood score showed an effective sampling size ≥300.
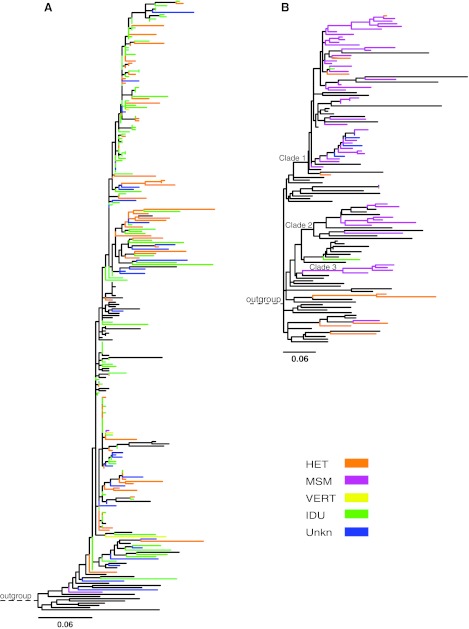
Out of the 315 Latvian HIV-1-infected patients included in the study, 296 patients with data from both V3 and p17 were consistent in the subtype assignments. For 6 and 12 patients, respectively, subtyping was successful only in p17 or V3. Subtypes A1 and B dominated and were found in 239 (76%) and 68 (22%) patients, respectively. A few other subtypes (C, G, CRF02_AG, and CRF03_AB) were also observed (Table 1). Most Latvian subtype A1 sequences (132 of 239) were obtained from IDUs, but 70 were obtained from heterosexually infected individuals and three from vertically infected children. Figure 2A shows the ML tree based on our V3 sequences of subtype A1 from Latvia and closely related sequences originating from other parts of the FSU that were selected based on HIV BLAST searches and NJ tree analyses. The subtype A1 sequences from IDUs and heterosexuals were intermingled in the tree indicating that they do not represent separate subepidemics. Likewise, sequences from other parts of the FSU were also intermingled with the Latvian subtype A1 sequences. The bootstrap support for the internal branches in this Latvian/FSU cluster was generally low (<70%) and no significant subclusters were identified, which prevented us from estimating the number of subtype A1 introductions in Latvia. Similar results, i.e., that the subtype A1 sequences from Latvia and the FSU formed one cluster with no identifiable substructure, were obtained when ML phylogenetic trees were constructed using p17 sequences as well as when phylogenetic trees were constructed with Bayesian and NJ methods (data not shown). A majority of the subtype A1 sequences were obtained from patients from Riga. Epidemiological data suggested that subtype A1 initially spread explosively among IDUs in Riga and later has spread to other transmission groups and to other regions of the country (Fig. 1).1 Our Bayesian inference indicated that the MRCA of the Latvian subtype A1 sequences existed around 1997. This estimate was robust to subsampling of sequences as well as to choice of demographic model and genetic region (data not shown). Because sequences from the FSU were intermixed with the Latvian sequences, the MRCA may slightly predate the time for the introduction of subtype A1 in Latvia, but the first subtype A1 sequence from Latvia was obtained in 1998.
FIG. 2.
Maximum likelihood trees of V3 sequences from Latvia and closely related database sequences for subtype A1 (A) and subtype B (B). The color of each branch represents the transmission route of Latvian sequences, while sequences from other countries are colored black. The scale at the bottom indicates nucleotide substitutions per site according to a GTR+G+I model. The subtype A1 tree was rooted using subtype A1 sequences SE8538 (AF069669), SE7253 (AF069670), and SE7535 (AF069671) and the subtype B tree was rooted using the subtype D sequence D.CD.84.84ZR085 (U88822). Three sequence clusters (1–3) in the subtype B tree are indicated. HET, heterosexual; MSM, men who have sex with men; VERT, mother-to-child transmission; IDU, intravenous drug use; Unkn, unknown transmission route.
A majority (51 of 68) of the Latvian subtype B sequences were obtained from MSM (Table 1). Figure 2B shows a ML tree based on Latvian subtype B V3 sequences, which were available from 64 Latvian patients, and 58 closely related database sequences, of which a majority were from other parts of the FSU and a few were from Western Europe, the United States, and Australia. A majority (55 of 64) of the Latvian subtype B sequences formed three separate sequence clusters, which are denoted Clade 1, 2, and 3 in Fig. 2B. These clusters were recovered in analyses of both V3 and p17 sequences as well as by different phylogenetic methods, i.e., ML, NJ, and the Bayesian inferences. Clade 1 was the largest cluster and contained 45 Latvian sequences as well as 11 sequences from the FSU and 1 sequence from the United States, Great Britain, and Australia, respectively. Clade 2 also included sequences from other parts of the FSU. The clusters included samples from the entire study period (patients diagnosed from 1990 to 2003), which indicates that these variants have spread in Latvia during many years. In agreement with this, our Bayesian inference indicated that the MRCA of Clades 1, 2, and 3 existed around 1985, 1985, and 1990, respectively. A majority of the subtype B sequences were obtained from patients from Riga.
In this study we have for the first time provided a comprehensive picture of HIV-1 spread in Latvia. Our study revealed the following main characteristics of the Latvian HIV-1 epidemic: (1) subtypes A1 and B dominated and were found in a majority of the patients; (2) subtype A1 was most likely imported from the FSU around 1997 and has been involved in the explosive outbreak among IDUs in Riga and has also started to spread heterosexually; (3) subtype B has been introduced several times and has mainly spread among MSM; and (4) other subtypes and CRFs have been introduced via intravenous drug use and through heterosexual contacts, but have so far shown no or very limited spread within Latvia. Thus, many HIV-1 introductions into Latvia have caused no or a few secondary cases, but a few (especially the subtype A1 variant) have been epidemiologically successful and currently account for most of the HIV-1 infections in the country. We found that HIV-1 subtype A1 entered Latvia at least once around 1997 and then gave rise to the explosive outbreak among IDUs. In agreement with our previous study,3 a majority of subtype A1 infections were found among IDUs in Riga. Here we show that this variant has also spread to other geographic regions of Latvia and has also started to spread heterosexually in the country. However, it cannot be excluded that some IDUs have chosen to report a heterosexual transmission route because intravenous drug use is associated with social stigma. In recent years the number of newly diagnosed IDUs has declined, but the increase in heterosexually acquired HIV-1 infections is worrying since it may create a foundation for a self-sustained heterosexual epidemic. Most subtype B variants were obtained from MSM and could be grouped into three clusters that appeared to have been imported from the FSU before the collapse of the Soviet Union in 1991.
Sequence Data
The sequences have been deposited in GenBank under accession numbers AY290866–AY290962, EU702489–EU702618 and FJ429476–FJ429551 (p17), and AY290963–AY291059, EU702619–EU702748, and FJ429395–FJ429475 (V3).
Acknowledgments
We thank Laura Selakova, Anda Lazdina, and Inga Upmace at the former Public Health Agency, AIDS and STI Prevention Centre, now HIV/AIDS Program unit of the Infectology Centre of Latvia, for epidemiological information, and Dr. Pauls Aldins, Dr. Diana Dusacka, and Dr. Ludmila Guseva at the Infectology Center of Latvia for preparing the samples and for epidemiological information. We thank Afsaneh Heidarian for excellent technical assistance.
The research leading to these results has received funding from the Swedish Research Council, the Swedish Baltic Sea Grant, the Latvian Council of Science, the National Institutes of Health (NIH) [Grant 5R01AI087520-02], and EU projects: SPREAD (QLK2-CT-2001-01344), EHR (LSHP-CT-2006-518211), and CHAIN (FP7/2007-2013) “Collaborative HIV and Anti-HIV Drug Resistance Network” Grant agreement no. 223131.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Public Health Agency, AIDS and STI, Riga, Latvia. sva.vi.gov.lv/lv/aidsunsti/statistika/hivaids/hivinfekcija/ sva.vi.gov.lv/lv/aidsunsti/statistika/hivaids/hivinfekcija/
- 2.UNAIDS Report on the global AIDS epidemic. 2010. www.unaids.org/globalreport/Global_report.htm www.unaids.org/globalreport/Global_report.htm
- 3.Balode D. Ferdats A. Dievberna I, et al. Rapid epidemic spread of HIV type 1 subtype A1 among intravenous drug users in Latvia and slower spread of subtype B among other risk groups. AIDS Res Hum Retroviruses. 2004;20:245–249. doi: 10.1089/088922204773004978. [DOI] [PubMed] [Google Scholar]
- 4.Liitsola K. Holm K. Bobkov A, et al. An AB recombinant and its parental HIV type 1 strains in the area of the former Soviet Union: Low requirements for sequence identity in recombination. UNAIDS Virus Isolation Network. AIDS Res Hum Retroviruses. 2000;16:1047–1053. doi: 10.1089/08892220050075309. [DOI] [PubMed] [Google Scholar]
- 5.The Los Alamos HIV Sequence Database. www.hiv.lanl.gov www.hiv.lanl.gov
- 6.Leitner T. Escanilla D. Marquina S, et al. Biological and molecular characterization of subtype D, G and A/D recombinant HIV-1 transmissions in Sweden. Virology. 1995;209:136–146. doi: 10.1006/viro.1995.1237. [DOI] [PubMed] [Google Scholar]
- 7.Leitner T. Korovina G. Marquina S. Smolskaya T. Albert J. Molecular and biological characterization of Russian HIV-1 strains. AIDS Res Hum Retroviruses. 1996;12:1595–1603. doi: 10.1089/aid.1996.12.1595. [DOI] [PubMed] [Google Scholar]
- 8.Skar H. Axelsson M. Berggren I, et al. Dynamics of two separate but linked HIV-1 CRF01_AE outbreaks among injection drug users in Stockholm, Sweden, and Helsinki, Finland. J Virol. 2011;85:510–518. doi: 10.1128/JVI.01413-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 10.Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 11.Guindon S. Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 12.Drummond AJ. Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skar H. Borrego P. Wallstrom TC, et al. HIV-2 genetic evolution in patients with advanced disease is faster than in matched HIV-1 patients. J Virol. 2010;84:7412–7415. doi: 10.1128/JVI.02548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leitner T. Albert J. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc Natl Acad Sci USA. 1999;96:10752–10757. doi: 10.1073/pnas.96.19.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]