Abstract
Aims
Individualization of cancer chemotherapy based on the patient’s genetic makeup holds promise for reducing side effects and improving efficacy. However, the relative contribution of genetics to drug response is unknown.
Materials & methods
In this study, we investigated the cytotoxic effect of 29 commonly prescribed chemotherapeutic agents from diverse drug classes on 125 lymphoblastoid cell lines derived from 14 extended families.
Results
The results of this systematic study highlight the variable role that genetics plays in response to cytotoxic drugs, ranging from a heritability of <0.15 for gemcitabine to >0.60 for epirubicin.
Conclusion
Putative quantitative trait loci for cytotoxic response were identified, as well as drug class-specific signatures, which could indicate possible shared genetic mechanisms. In addition to the identification of putative quantitative trait locis, the results of this study inform the prioritization of chemotherapeutic drugs with a sizable genetic response component for future investigation.
Keywords: cancer, cell line, chemotherapy, cytotoxicity, pharmacogenetics, pharmacogenomic, QTL
Important progress continues to be made in the treatment of most common cancers, but therapeutic benefit remains difficult to predict and severe or fatal adverse events occur frequently. The Human Genome Project has fueled the notion that genetic information can produce effective and cost-efficient selection of therapies for individual patients [1], but validated genetic signatures that predict response to most chemotherapy regimens remain to be identified. Numerous genes potentially influence drug response, but current candidate gene approaches aimed at discovering and characterizing pharmacogenetic effects are limited by the requirement of a priori knowledge about the genes involved [2]. While genome-wide association analyses represent unbiased approaches to trait mapping, the moderate size of most clinical trials often limits this avenue for cancer pharmacogenomics discovery [3]. Furthermore, many pharmacogenomic studies are performed with the unstated and untested assumption that the drug response is a heritable trait, potentially wasting scarce clinical and analytical resources if that assumption proves false.
In response to these limitations, we undertook a thorough, unbiased pharmacogenomic assessment of cytotoxic effects of the majority of US FDA approved cancer compounds. We used an in vitro model system to determine the heritability of drug-induced cell killing and performed genome-wide analysis for pharmacologic quantitative trait loci (pQTL). Promising results have been obtained previously using similar systems on a smaller scale, supporting the notion that genetics can influence cytotoxic activity of some drugs [4–7]. In addition to preliminary identification of putative response loci, the results of this study inform the prioritization of chemotherapeutic drugs with a sizable genetic response component for future investigation.
Materials & methods
Cell lines
The Centre d’Etude du Polymorphisme Humain (CEPH) cell lines are a set of immortalized lymphoblastoid cell lines (LCLs) collected from normal, healthy human volunteers, which are commercially available from the Coriell Cell Repositories (NJ, USA). This collection is unique because the cell lines were established from large multigenerational families and there is comprehensive, publically available genotype data available for these cells lines [7]. A total of 125 LCLs contained within the following CEPH family pedigrees were used in this study: 35, 45, 1334, 1340, 1341, 1345, 1350, 1362, 1408, 1420, 1447, 1451, 1454 and 1459. These cell lines were chosen to provide at least 80% power to detect a heritability (h2) of 0.3 or greater.
Fresh growing lymphoblastoid cells were received from Coriell in T25 flasks and were further cultured in Roswell Park Memorial Institute medium 1640 containing 2 mM l-glutamine (Gibco, CA, USA) and 15% fetal bovine serum (Sigma-Aldrich Co, MO, USA; lot 107K8408) at 37°C, 5% CO2 in a NUAIRE autoflow infrared direct heat CO2 incubator (Labrepco, PA, USA). No media antibiotics were used for these studies. Frozen aliquots of each cell line were stored in liquid nitrogen for use throughout the study. Individual cell lines were counted in a Z1 Coulter Particle Counter (Beckman Coulter, CA, USA) and plated at 4000 cells/well (45 µl per well) in a 384-well plate containing the drugs (as noted in the ‘Drugs’ section below). One cell line was assayed per 384-well plate and replicates of each cell line were performed using a new frozen aliquot grown at a different time. Individual cells lines were assayed in random order across assay plates (e.g., not assayed by family or family structure) to limit the influence of any unknown systematic assay bias on the results. Furthermore, prior to commencement of these studies, seeded cultures of cells were plated in 384-well plates containing vehicle controls and assayed as described below. Minimal edge effects were observed and the fluorescence intensity of the outer edge wells were consistently within one standard deviation of the mean of the entire plate.
Drugs
Gemcitabine, epirubicin hydrochloride, temozolomide, teniposide and topotecan were obtained from LKT laboratories (MN, USA). All other drugs were purchased from Sigma. Drugs were diluted in either H2O or dimethyl sulphoxide (DMSO) depending on solubility. Drugs with poor solubility in water, 0.9% NaCl, or DMSO and those requiring in vivo metabolic activation were not assessable. Final concentrations, after addition of 45 µl of cells and media, of DMSO were 1% or less in all wells, except for the negative control wells which contained 10% DMSO. Drug doses used were selected based on the results of testing a small number (n < 6) of unrelated cell lines with a broad range of doses. From that data, four doses spanning the observed viability range were selected for further study on the complete 125 cell line panel. A subset of drugs could not be dosed high enough to reach maximal cytotoxic effect due to drug solubility issues. Details about drugs and vehicles utilized are shown in Supplementary Figure 1 (see www.futuremedicine.com/doi/suppl/10.2217/pgs.11.92). Four different doses of each drug and four vehicle controls (0.1% DMSO, 1.0% DMSO, 10% DMSO and water) were plated in quadruplicate (5 µl/well) on a 384-well plate using a Biomek 3000 (Beckman Coulter) liquid handling robot. Drug-containing 384-well plates were sealed and stored at −80°C and thawed once before use in the cytotoxicity assay. Drug- and control-well treatment positions in the plate were held constant throughout the study.
Cytotoxicity measurements
The cytotoxic effects of the 29 drugs were determined using the nontoxic, colorimetric Alamar Blue assay (BIosource, CA, USA) [7]. Briefly, 45 µl of fresh cultures of cells were seeded in 384-well plates (Corning #3712) at a density of 4000 cell/well containing 5 µl of varying doses of 29 drugs. Plates were incubated for 72 h at 37°C, 5% CO2 before the addition of 5 µl of Alamar Blue (Biosource International, CA, USA). Plates were incubated an additional 24 h before fluorescence intensity was measured using an excitation filter of 535 nm and emission filter of 595 nm. Plates were read from above the plate for 0.1 s using Multimode Detection Software v2.1.0.17 on a DTX880 plate reader (Beckman Coulter). For each drug, percent viability was measured relative to the appropriate untreated control wells using the following formula: mean raw fluorescence units (RFU) of four drug-treated wells – mean RFU of four 10% DMSO wells/mean RFU of four vehicle only wells – mean RFU of four 10% DMSO wells
A 10% DMSO treatment condition was used as a negative control (i.e., 0% viability) in the calculation of viability for each cell line to control for background fluorescence due to serum, media or cellular debris. For each drug, viability was calculated relative to the vehicle only control well that was appropriate for that drug (i.e., 0.1% DMSO, 1.0% DMSO or water). For each of 125 LCLs, four doses of each drug were assayed in quadruplicate wells. Then, for each cell line, this entire process was repeated with cells from a separately seeded frozen culture.
Statistical methods
Cell cytotoxicity assay data conversion
Data were cleaned with a careful quality control pipeline prior to analysis. First, assay plates were checked for inappropriately mass cell death by comparing the wells RFUs with a threshold. Any plates whose 90th percentile of RFU values was below 2000 were removed, as the majority of the cells on these plates were dead for reasons unrelated to drug effect. Second, outliers on plates were identified and removed with a coefficient of variation screen, checking for values outside the range defined by the 99.5% coefficient of variation above and below the mean. Third, errant values in the 10% DMSO negative control were identified and removed, since an errant negative control reading greatly influences the viability calculation used (presented above) for further analysis. Again, a coefficient of variation threshold was used. Fourth, vehicle only wells that serve as positive controls in the assays were checked for deviant values. Just as with the DMSO negative controls, errant values in the vehicle only wells would greatly impact viability calculations and downstream analysis. There is high correlation between the mean vehicle viabilities for each cell line across all vehicles used (0.78 < r < 1.0), and these relationships are used to screen the data for errors. This provides a means to check if any one particular vehicle is far from the expectation. To test for outliers in vehicle well values, we performed a simple linear regression for each of the viabilities from one vehicle on the others. Therefore, each mean vehicle viability is involved as the response for four regressions. Any response that was more than one standard deviation away from its predicted value was considered deviant and was removed. If a particular response was deviant in three or four, out of the four regressions, it was removed. After careful quality control, average percentage viability was calculated from the two independent plates, with four technical replicates on each plate. In addition, for each vehicle used, growth rate was calculated as previously described [8].
Box plots for the viabilities observed for each drug across all doses used for 125 cell lines within the 14 families are shown for each drug, in Supplementary Figure 2. Box plots for the viabilities for each drug at all dose points across all individual cell lines are included in Supplementary Figure 3.
Heritability analysis
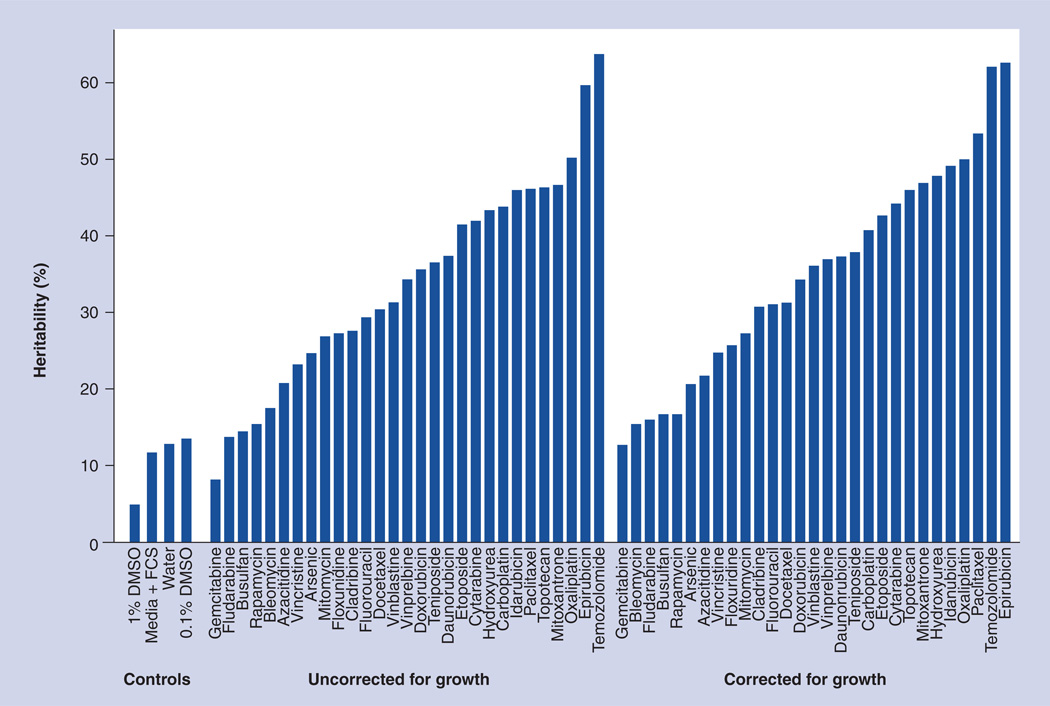
To estimate the proportion of variation in dose response due to inherited factors, variance components analysis as implemented in MERLIN® 1.1.2 [9,101] was used to calculate heritability. Heritability of the growth rate for each vehicle was calculated, and the heritability calculation of each drug and dose treatment combination was adjusted for the growth rate for the appropriate vehicle by using growth rate as a covariate in the variance components analysis. For each drug, the highest heritability for any dose is given in Figure 1. The doses that correspond to the highest heritability are listed in Supplementary Figure 4 for drugs with putatively significant linkage results.
Figure 1. Maximum heritability observed over all doses for commonly used anticancer drugs as well as vehicle only controls.
Heritability estimates are presented both uncorrected and corrected for growth rate.
DMSO: Dimethyl sulphoxide; FCS: Fetal calf serum.
Genotype data & error checking
Genotype data for each cell line was downloaded from version 10 of the CEPH database [102,10] using error checked markers. Genetic map information was downloaded from the Marshfield database [103,11]. Error checking for Mendelian incompatibility, misspecified relationships and unlikely recombinations was performed, as previously described [7]. A combined total of 8269 SNPs and microsatellite markers were used for linkage analysis.
Technical and biological reproducibility was high for our assay, with the average correlation for each cell line across replicates being 0.9067 (standard deviation = 0.0835). The average coefficient of variation for all drugs and vehicle and control replicates across all replicates was 6.57% (standard deviation = 6.28%).
Linkage analysis
While the current study was designed to estimate heritability, the publicly available data was also used for linkage analysis to identify putative regions of the genome linked to the drug response traits. Recognizing the limited power of the current study to detect pQTLs, we relied on a permutation and replication approach to prioritize pQTLs within and across drug response phenotypes. Given these limitations, it is crucial that these results be interpreted as ‘hypothesis generation’ rather than strict ‘hypothesis testing’ results.
Phenotypes of interest for linkage analysis were defined as cell viability at each drug and dose point, resulting in a total of 116 total phenotypes from the current study. For each phenotype, nonparametric linkage analysis was performed using MERLIN because it is robust to nonnormality in the phenotypic variables of interest. As described in detail in Abecasis et al. MERLIN constructs a likelihood ratio test for linkage based on inheritance vectors [9]. For quantitative traits, scores used to calculate the likelihood ratio test are defined as follows:
where S(ν) is the score for each inheritance vector, Sallele(ν) is the score for each founder allele, yi is the phenotype for each individual, μ is the mean phenotype for the population, and ν is the list of individuals who carry a specific founder allele such that the score for each inheritance vector is the summation of the squared score for each founder allele, and the score for each found allele is the sum of square deviation from all individuals that carry that allele.
For each phenotype of interest, QTL maps were constructed by displaying the logarithm of odds scores from the likelihood ratio tests across each chromosome, and QTL maps for different doses of the same drugs were overlaid. A complete set of QTL maps for each drug, by chromosome can be found in Supplementary Figure 5. This approach does not directly account for potential dependence between dose point phenotypes for individual cell lines, this expected dependence is incorporated into our prioritization pipeline described below.
Results prioritization & peak identification
To estimate the probability of obtaining false-positive evidence of linkage for each drug and dose combination under the null hypothesis of no linkage to observed phenotypes, we conducted gene-dropping permutations using MERLIN. Marker data were simulated under the null hypothesis of no linkage or association to observed phenotypes while retaining the same pedigree structures, maps, marker allele frequencies, and missing data patterns. We simulated 10,000 replicates for each phenotype of interest (discussed above), resulting in a total of 1.16 million simulated datasets, and conducted linkage analyses as described earlier for each replication. Based on these simulations, null distributions for each phenotype, for each chromosome were constructed.
Empirical results from the permutation testing were used to ascribe putative significance to the linkage results. The pQTL results from the dose of each drug that resulted in the highest heritability estimate were used to prioritize results for replication in additional doses.
Using the permutation distributions constructed across all the chromosomes for each phenotype evaluated, logarithm (base 10) of odds (LOD) score cut-offs corresponding to p-values less than or equal to 0.05 for each drug at the highest heritability dose were found, and used to define significant LOD score peaks. A complete list of LOD score significance cut-offs (for all drug and dose combinations) can be found in Supplementary Figure 6. The average LOD score cut-off indicating ‘significant linkage’ across all drugs, doses and chromosomes was 1.44. This significance cut-off corresponds to a family-wise error rate of 5% for each drug and dose tested (such that the false-positive rate is divided between chromosomes for each drug), and treats drug/dose combinations as independent tests. These LOD score cut-offs were used to define significant peaks for each drug, where a region was considered significant if the highest LOD score in the region was greater than or equal to the permutation testing derived cut-offs. A complete list of significant peaks is included in Supplementary Figure 4.
In addition to defining these putatively statistically significant peaks, suggestive pQTLs were identified using the permutation distributions. LOD score cut-offs that define suggestive peaks were determined as the minimum LOD score to achieve a p-value of 0.05 at each chromosome (not corrected for the entire genome). The average LOD score cut-off indicating ‘suggestive linkage’ across all drugs, doses and chromosomes was 0.601. Cut-offs that define suggestive linkage for each drug and dose combination are also listed in Supplementary Figure 4.
The region encompassing each significant pQTL was defined as the contiguous chromosomal segment with LOD scores above a minimum value that was either: the peak LOD score minus one or the LOD score cut-off that defines suggestive linkage, whichever is higher. The LOD score cut-offs used for defining the bounds of pQTLs are listed in Supplementary Figure 6.
Results
We investigated the cytotoxic effect of 29 commonly prescribed chemotherapeutic agents from diverse drug classes (Table 1) on 125 LCLs derived from 14 extended families from the CEPH collection. A high-throughput assay, employing a 384-well plate format and the fluorescent viability indicator Alamar Blue, was used to determine the cytotoxicity of the drugs on the LCLs. Viability after drug treatment was calculated relative to cells treated with vehicle only. For each drug, four doses were chosen in an effort to capture the exponential portion of the cytotoxicity curve.
Table 1.
Drugs used in this study
| Drug name | Mechanism of action |
|---|---|
| Cladribine | Ribonucleotide reductase inhibitor |
| Fludarabine | Ribonucleotide reductase inhibitor |
| Gemcitabine | Ribonucleotide reductase inhibitor |
| Azacitidine | Ribonucleotide reductase inhibitor |
| Cytarabine | Ribonucleotide reductase inhibitor |
| 5-fluorouracil | Thymidylate synthase inhibitor |
| Floxuridine | Thymidylate synthase inhibitor |
| Epirubicin | DNA intercalators and topoisomerase 2 inhibitors |
| Doxorubicin | DNA intercalators and topoisomerase 2 inhibitors |
| Daunorubicin | DNA intercalators and topoisomerase 2 inhibitors |
| Idarubicin | DNA intercalators and topoisomerase 2 inhibitors |
| Mitoxantrone | DNA intercalators and topoisomerase 2 inhibitors |
| Vincristine | Microtubule destabilizers |
| Vinorelbine | Microtubule destabilizers |
| Vinblastine | Microtubule destabilizers |
| Docetaxel | Microtubule stabilizers |
| Paclitaxel | Microtubule stabilizers |
| Oxaliplatin | DNA crosslinkers |
| Carboplatin | DNA crosslinkers |
| Etoposide | Topoisomerase 2 inhibitors |
| Teniposide | Topoisomerase 2 inhibitors |
| Arsenic trioxide | Other |
| Bleomycin | Other |
| Busulfan | Other |
| Hydroxyurea | Other |
| Mitomycin | Other |
| Rapamycin | Other |
| Temozolomide | Other |
| Topotecan | Other |
Heritability was calculated for each drug and dose outcome, as well as growth rate for each control vehicle in the current study using variance-components methods as implemented in MERLIN [9]. The heritability of cellular growth rate was <0.14 across the drug vehicles, and heritability was calculated with and without using cell growth as a covariate for the cytotoxic response traits (Figure 1).
Results of drug cytotoxicity assays showed variation across families and individual cell lines (Supplementary Figures 2 & 3). The maximum heritability of the drugs studied varied from 0.06 to 0.64. In total, 19 of the 29 drugs yielded a maximum heritability of >0.3. Overall, only two of the 29 drugs had a maximum heritability at or below that observed for drug vehicle.
Nonparametric linkage analysis was performed to identify pQTLs for each drug at each dose. A permutation approach was employed to define LOD score cutoffs indicating significant and suggestive evidence for linkage (see “Results prioritization & peak identification”). To prioritize results, primary emphasis was placed on the dose of each drug that resulted in the highest heritability estimate for cytotoxicity, with supporting evidence provided by results in other doses.
A total of 31 regions with peak LOD scores above the putative significance cut-offs were observed among the pQTL results for 26 of the drugs at the dose demonstrating the highest heritability (Supplementary Figure 4). Of the three drugs for which no significant LOD scores were found, two drugs (gemcitabine and fludarabine) had the lowest heritabilities of all the drugs tested, while the third (cytarabine) showed moderate heritability.
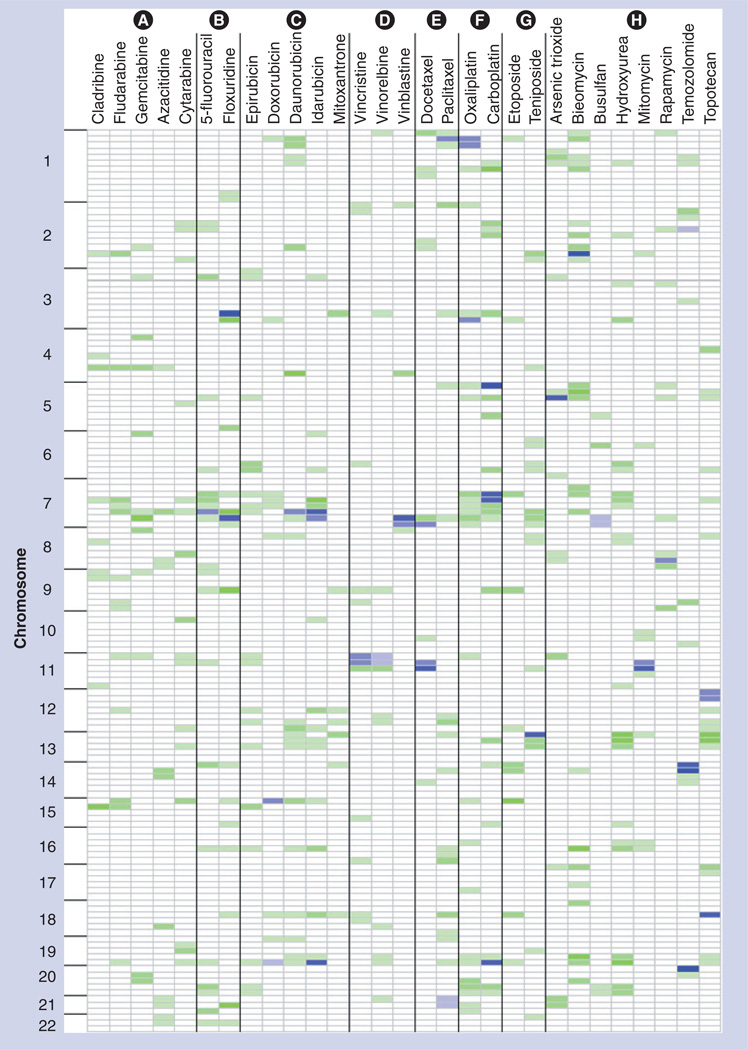
For all significant peaks identified in the dose point with the highest heritability, these regions were evaluated for replication at a suggestive or significant level in additional dose points for the drug. A region was considered replicated if any portion of the region that defined the pQTL showed evidence for linkage at an additional dose. Of the 31 significant pQTLs across all drugs, nine replicated at least a suggestive level at one additional dose; 13 replicated at two additional doses; and six replicated at three additional doses (i.e., all four dose points evaluated). In addition, for regions with suggestive linkage in the highest heritability dose, the pQTL mapping results from additional doses were evaluated for replication at a suggestive or significant level. There were a total of 31 regions that reached significant linkage. In addition, a total of 589 peaks reached ‘suggestive’ cut-offs in the highest heritability dose. Of these suggestive regions, 333 showed evidence of at least suggestive linkage in at least one additional dose. Figure 2 displays the results of this prioritization for significant peaks, as well as suggestive peaks that replicate in a minimum of one additional dose. Details of the peaks identified are found in the supplemental materials (Supplementary Figures 4 & 7).
Figure 2. Genome-wide patterns of quantitative trait loci for all drugs, arranged by mechanistic classes.
Each chromosome was partitioned into 25-cm regions. For each drug at the dose that resulted in the highest heritability estimate, any putative significant pharmacologic quantitative trait loci regions (log10 of odds > empirical significance cut-off) is indicated in blue. Intensity of the shading is dependent on the total number of additional doses that indicated a replicating signal at either a suggestive or significant level, with higher intensity indicating higher numbers of doses. Regions that also had at least a suggestive pharmacologic quantitative trait loci, as well as at least one additional replication in another dose, are shown in green, where again intensity of the color represent a higher number of dose replications. Drugs are grouped according to their mechanism of actions, where the groups are labeled as follows: Group (A): ribonucleotide reductase inhibitors; Group (B): thymidylate synthase inhibitors; Group (C): DNA intercalators and topoisomerase 2 inhibitors; Group (D): microtubule destabilizers; Group (E): microtubule stabilizers; Group (F): DNA crosslinkers; Group (G): topoisomerase 2 inhibitors; Group (H): other.
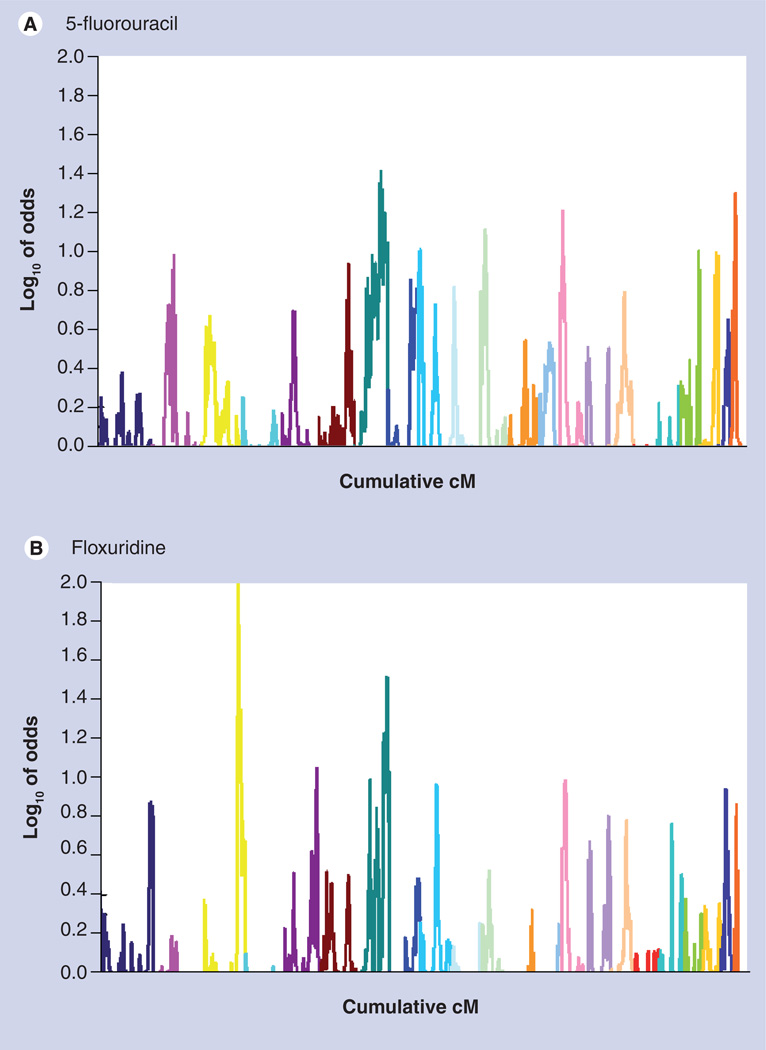
Pharmacologic QTLs were observed to occur in similar locations across doses for a given drug, and also across drugs families with common chemical structures or presumed mechanisms of action. For example, overlapping pQTLs were found in the same region of chr7 for the fluoropyrimidines 5-fluorouracil and floxuridine, two drugs thought to share a similar mechanism of action (Figure 3). Peak co-localization was observed for other drug classes as well, specifically the DNA crosslinkers, vinca alkaloids and anthracyclines (Figure 2). Regions with significant peak LOD scores for growth in the three different drug vehicle controls did not overlap peaks for any of the significant drug cyotoxicity results (Data Not Shown), indicating that genetic effects on growth rates of the different cell lines are not likely to be responsible for the loci identified by the cytotoxicity analysis. These pQTL results were formally evaluated for over-representation in drugs of similar mechanistic classes. The hypergeometric distribution was used to test whether significant pQTLs were overrepresented in mechanistic drug classes, and indicated there is a nonrandom distribution of peaks amongst classes (p < 0.05).
Figure 3. Genome-wide quantitative trait loci map.
(A) 5-fluorouracil and (B) floxuridine, two structurally and functionally related drugs. Each chromosome is represented by a different color.
Discussion
The results of this study have several important implications for the field of cancer pharmacogenomics. Heritability estimates varied widely across the drugs studied, which implies quantitative differences in the role that genetic polymorphism plays in response to chemotherapeutic agents. The majority of drugs (66%) had a maximum heritability of >0.3, a threshold value for many genetic studies of complex human traits. This provides an objective basis for the prioritization of whole genome discovery studies in valuable clinical trial DNA samples. Indeed, if the results of the study presented here using this model system can be extended to future in vivo studies, the application of genome wide association studies to a drug with low heritability is unlikely to produce significant results.
By using multiple drugs within and across classes, evaluating common and unique pQTLs across drug classes may reveal important aspects of the genetic etiology of drug response. In addition, as replication has become the gold standard in human genetics applications, evaluation of different dose points for a single drug, other members of drug classes or structurally similar drugs could provide a measure of internal replication for the pQTLs identified. This replication approach can be used to help prioritize regions for follow-up in future studies. Similarly, further study of pQTLs that are common within and across drug classes may help elucidate potential unifying downstream mechanisms of action of the drugs. However, given our limited power to determine genome-wide significant linkage peaks, it should be noted that the results of the current study should be evaluated in future, well powered studies for finer genetic mapping.
As with any model system, the limitations of the system must be considered when interpreting the results. This in vitro model of drug-induced cytotoxicity simplifies the effect of these drugs on cancer patients in clinical settings, where pharmacokinetic and other factors will also play a role in patient response. For the results of this study to have clinical relevance, the genetic pathways of chemotherapeutic response in host or tumor response in vivo would need to be similar to what was seen in this study. Unfortunately, performing a study with similar aims to the one presented here using intact, extended human families would not be feasible due to ethical, logistical and cost hurdles.
While LCLs have been frequently used as a model system for uncovering the genetics of gene expression [12], radiation response [13] and drug induced cytotoxicity [4–7], a recent study [8] highlighted the need for proper study design and interpretation of the data. In order to limit the effect of technical and biological noise on the results of this study, each drug and dose measurement was performed in quadruplicate and was then repeated from a separately seeded culture of LCLs. Technical and biological reproducibility for the assay was extremely high, with a strong correlation among replicate measures (r = 0.9067). In addition, the average coefficient of variation for all drugs and vehicle controls across all replicates was 6.57% (standard deviation = 6.28%), indicating low variation across replicates. We also included baseline growth rate, calculated for each vehicle control as a factor in our variance components analysis, which was itself minimally heritable (h2 < 0.14). Furthermore, testing for over-representation of pQTLs within drug classes suggests that drugs with similar chemical structures had similar cytotoxic response, indicating that LCL cytotoxicity was specific to each drug and not an artifact of the experimental design. Unfortunately, we did not test for Ebstein–Barr virus copy number levels in our assay, and thus this remains a limitation of the study.
The results of this study lay the groundwork for future studies to uncover the underlying genes and variants influencing chemotherapeutic response. Whole-genome association studies, using LCLs or DNA derived from clinical trials should be undertaken for drugs found to have the highest heritability. This phenotype selection approach provides a clear justification for where investment in extensive genomic analysis should be prioritized to reach the goal of effective selection of therapy for individual patients.
Conclusion
The promise of pharmacogenomics is the individualization of cancer chemotherapy based on the patient’s genetic makeup. However, the relative contribution of genetics to drug response for specific chemotherapeutic drugs is largely unknown. By systematically investigating 29 common prescribed chemotherapeutic agents using an in vitro model system, we have been able to estimate the proportion of cytotoxic response that is attributable to genetic factors as well as identify putative QTLs. The results of this our study highlight the variable role that genetics plays in response to cytotoxic drugs, ranging from a heritability of <0.15 for gemcitabine to >0.60 for epirubicin. In this study we were also able to identify several putative significant QTLs, many of which replicated across drug doses. These results are useful for the prioritization of chemotherapeutic drugs and genomic regions for further studies using complementary methods such as further in vitro studies, use of animal models or clinical research.
Executive summary.
-
▪
Pharmacogenomics holds promise for individualizing cancer treatment by reducing the risk of adverse events and nonresponse.
-
▪
No systematic study has been performed to determine the relative contribution of genetics on cytotoxic response across this broad of a set of chemotherapeutic drugs.
Materials & methods
-
▪
The study described here used a systematic, in vitro approach to determine the influence of genetic factors on cytotoxic response of 29 chemotherapeutic drugs at several drug doses.
Results
-
▪
The relative contribution of genetic factors (heritability) to drug response varied widely across drugs, from <0.15 (gemcitabine) to >0.60 (epirubicin).
-
▪
The results of our analysis identified several quantitative trait loci including 31 putative significant linkage peaks, nine of which replicated in additional doses.
-
▪
Significant correlation was observed between linkage peaks and drug mechanistic class, indicating possible shared mechanism.
Discussion
-
▪
These results are valuable for the prioritization of chemotherapeutic drugs and genomic regions for future studies using complementary methods such as further in vitro studies, use of animal models or clinical research.
Supplementary Material
Acknowledgments
This work was supported by the NIH Pharmacogenetics Research Network (U01 GM63340), NIEHS 2 T32 ES007329, the University North Carolina University Cancer Research Fund, and the Triad Golfers Against Cancer.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
- 1.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auman JT, McLeod HL. Cancer pharmacogenomics: DNA genotyping and gene expression profiling to identify molecular determinants of chemosensitivity. Drug Metab. Rev. 2008;40:303–315. doi: 10.1080/03602530801952427. [DOI] [PubMed] [Google Scholar]
- 3.Ratain MJ, Miller AA, McLeod HL, Venook AP, Egorin MJ, Schilsky RL. The cancer and leukemia group B pharmacology and experimental therapeutics committee: a historical perspective. Clin. Cancer Res. 2006;12:3612S–3616S. doi: 10.1158/1078-0432.CCR-06-9008. [DOI] [PubMed] [Google Scholar]
- 4.Duan S, Bleibel WK, Huang RS, et al. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang RS, Duan S, Bleibel WK, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc. Natl Acad. Sci. USA. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am. J. Hum. Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc. Natl Acad. Sci. USA. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. ▪ Used similar in vitro methodology to investigate the influence of genetic variation on two chemotherapeutic drugs.
- 8.Choy E, Yelensky R, Bonakdar S, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 10.Cohen D, Chumakov I, Weissenbach J. A first-generation physical map of the human genome. Nature. 1993;366:698–701. doi: 10.1038/366698a0. [DOI] [PubMed] [Google Scholar]
- 11.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheung VG, Conlin LK, Weber TM, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. ▪ Innovative study that helped to show the utility of in vitro methods for interrogating the influence of genetic variation on quantitative traits.
- 13.Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Websites
- 101.MERLIN Software Homepage. www.sph.umich.edu/csg/abecasis/Merlin/index.html.
- 102.CEPH Genptype Database. ftp://ftp.cephb.fr/ceph_genotype_db/ceph_db/Ver_10/mkr.
- 103.Marshfield Genetic Map Mammalian Genotyping Service. http://research.marshfieldclinic.org/genetics.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.