The dynamic genome is a concept associated with the discovery of transposable elements by Barbara McClintock. Her Nobel lecture concluded with a challenge to biologists considering this issue (1). She wrote,
We know about the components of genomes that could be made available for such restructuring. We know nothing, however, about how the cell senses danger and instigates responses to it that are often truly remarkable. (ref. 1, pp. 800–801)
Fifteen years later we still know nothing about the mechanisms underlying genome-restructuring events in response to environmental cues. This is despite numerous studies, especially in plants, suggesting a connection between growth conditions and one form of genome restructuring, a change in genome size. In many plants there is impressive variation in total genomic DNA content among individuals and populations (2) including Helianthus annuus [sunflower, 50% within-plant reduction (3, 4)], Pisum sativum [pea, 1.29-fold variation (5)], Linum usitatissimum [flax, 1.16-fold (6)], and Glycine max [soybean, 1.15-fold variation (7)]. Although this variation is correlated with environmental gradients or growth conditions in a number of species (2–4), in no cases have the specific genomic components of DNA content change been identified.
In this issue of PNAS, Kalendar et al. (8) document an example of genome size variation in natural populations of the wild barley Hordeum spontaneum. This paper shows that an abundant and active component of the barley genome, namely the BARE-1 long terminal repeat (LTR)-retrotransposon, displays nearly a three-fold intraspecific copy number variation. Furthermore, correlations between BARE-1 copy number, genome size, and local environmental conditions suggest, for the first time, a testable molecular mechanism linking habitat with retrotransposon induction in natural populations.
LTR retrotransposons are members of the retroelement or Class 1 family, which also includes retroviruses, long interspersed nuclear elements (LINEs, also known as non-LTR retrotransposons), and short interspersed nuclear elements (SINEs). LTR retrotransposons are flanked by long terminal repeats and usually encode all of the proteins required for their transposition, including a capsid (Gag), protease, integrase, reverse transcriptase, and RNase H. For all Class 1 elements, it is the element-encoded transcript (mRNA), and not the element itself, that forms the transposition intermediate. Transcription of most of the active plant elements characterized to date is largely quiescent during normal development but can be induced by biotic and/or abiotic stresses, including cell culture, wounding, and pathogen attack (9, 10). For two elements, the tobacco Tnt1 and the rice Tos17, increased transcription is correlated with retrotransposition (11, 12).
LTR retrotransposons are the most abundant transposable element class in grass genomes, of which barley is a member (reviewed in ref. 13). In fact, differential amplification of LTR retrotransposons largely accounts for the C-value paradox in this group of organisms. The C-value paradox is the observed lack of correlation between DNA content and organismal complexity (14). It has been documented for both animal and plant species but, to date, only appears to be “solved” for the members of the grass tribe. That is, the fraction of the genome contributed by LTR retrotransposons increases with genome size from rice, the smallest characterized grass genome [430 Mbp, ≈14% LTR retrotransposons (15)], through maize [≈3,200 Mbp, 50–80% retrotransposons (16)] to barley [≈4,800 Mbp, >70% retrotransposons (17)]. For maize, SanMiguel et al. (18) made the remarkable discovery that the majority of the retrotransposon insertion events occurred very recently, within the last two to six million years.
As discussed above, we are beginning to understand the relative contribution and time scale of retrotransposition among different grass species. However, little is known about the dynamics of transposable element copy-number evolution within and among natural populations, or its significance with respect to natural selection. In a similar vein, it is well known that transposition events may lead to modified patterns of gene expression, but this process has rarely been demonstrated to be selectively relevant within natural populations. Thus, the possible connections between genome size variation and adaptive genic evolution (as illustrated in Fig. 1) have remained elusive.
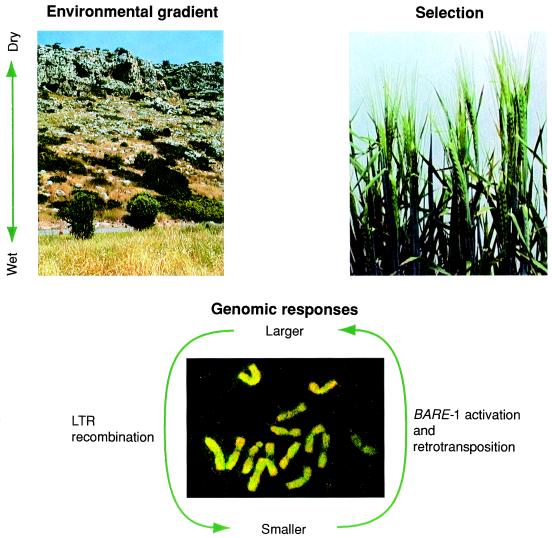
Figure 1.
Genome evolution on a local ecological scale. Wild barley plants are distributed along ecological gradients both regionally and locally, and vary nearly three-fold in copy number for the retrotransposon BARE-1 (in situ hybridization to barley chromosomes, bottom). Illustrated here are the intriguing interconnections between local adaptation to a moisture gradient (upper left) in a single canyon in Israel and the correlated distribution of BARE-1 copy-number (8). Local adaptation conceivably may be facilitated by direct selection on genome size (genome-level selection) or from functionally relevant physiological effects of individual BARE-1 insertions (gene-level selection). Credits: M Kemppinen (barley, upper right); K. Anamthawat-Jónsson (BARE-1 in situ hybridization); A. Schulman (canyon); A. Gardner (assistance with illustration).
Kalendar et al. (8) may have taken a first step toward intertwining these once disparate threads. In a study of natural populations of wild barley (Hordeum spontaneum) from a single canyon in Israel, they describe patterns of retrotransposon accumulation on a local spatial scale. Their data demonstrate a striking degree of population-level genome dynamics and suggest what well may be an example of retrotransposon-mediated adaptive evolution.
The barley plants studied derived from six natural populations distributed across a 300-m transect of a single canyon. Ten individuals were sampled from each population, which were selected to span the spectrum of local edaphic and microclimatic conditions present in the canyon, including potentially important ecological variables such as level of solar irradiation and aridity. Each individual was genetically fingerprinted, and copy number was estimated for the barley retrotransposon BARE-1, a relatively high copy-number (average of 14,000 copies/Hordeum species) family of elements that earlier was shown to be transcriptionally (19) and translationally active and assembled into virus-like particles (20). In the present study, full-length BARE-1 retroelements were shown to comprise an average of nearly 3% of the approximately 4.5-pg haploid wild barley genome, accumulating to a mean of 14,000 copies per genome. Although this observation is not in itself especially noteworthy, the variance in copy number among closely spaced natural plants is unprecedented and remarkable: a nearly three-fold range in copy number (8,300–22,100) was observed among individuals of the six populations, corresponding to 1.77–4.70% of the nuclear DNA.
Such extraordinary variation in retroelement copy number among spatially adjacent plant populations implicates a history of recent transposition, a suggestion supported by “REMAP” DNA fingerprinting. This technique, which couples PCR priming sites in BARE-1 LTRs with those designed from simple sequence repeats, was used by Kalendar et al. to show that wild barley plants from this single erosion gorge have high levels of interindividual polymorphism for REMAP fragments. An important implication of these observations, when considered in light of the striking local variation in BARE-1 copy-number, is that retroelement proliferation may contribute to genome size evolution within and among local populations (Fig. 1). Extrapolated to a more global level, this study may provide a snapshot of the dynamics that underlie patterns of C-value evolution.
A more provocative implication of the Kalendar et al. study emerges from consideration of the spatial distribution of BARE-1 copy-number among wild barley plants. When the REMAP genetic fingerprinting data were subjected to multivariate analysis, populations from the north- and south-facing slopes of the gorge clearly were distinguished. On both slopes, but particularly on the drier, south-facing slope, there was a significant positive correlation between height in the canyon and BARE-1 copy-number. These data parallel regional trends observed in a broader sampling of H. spontaneum populations collected from across Israel (17) and suggest a relationship between retroelement accumulation and one or more ecological variable related to the sampled populations. The most obvious variable is moisture availability; higher sites and those from the south-facing slope are the driest and thus potentially the most water-stressed. By far the highest BARE-1 accumulation is in the highest site from the south-facing slope. Kalendar et al. note a remarkable connection between the presence, within the BARE-1 promoter, of ABA (abscissic acid)-response elements, found in water stress-induced genes (19), and BARE-1 copy number variation, suggesting that BARE-1 proliferation in wild barley populations may be stress-induced. With the important caveat that the data are correlative rather than causal, it is tempting to speculate that other examples of interpopulational DNA content variation (2) will similarly be found to result from stress-induced retrotranspositional activity.
An intriguing aspect of the BARE-1 data of Kalendar et al. concerns the relative abundance of full-length elements and solo LTRs. The latter, which are relatively rare in the maize genome (21) but are common in yeast and Hordeum species (17, 22, 23), are thought to arise from intraelement or perhaps intrachromosomal recombination between transiently paired LTRs. Kalendar et al. used dot-blot reconstruction to estimate copy number for both LTRs and BARE-1 integrase genes and found an average of 5.4-fold more LTRs than internal domains. These data show that recombinational loss of BARE-1 elements is an important factor limiting element accumulation in wild barley populations. Significantly, the geographical sites with the highest BARE-1 copy number, i.e., those from the most stressed sites, have the highest ratio of full-length to solo LTRs, suggesting once again a connection between environmental sensing and either rates of recombinational loss (favored explanation of Kalendar et al.) or recent bursts of retrotranspositional activity.
A central question that emerges from this study concerns the role, if any, of BARE-1 element proliferation in the stress response. One might postulate, for example, that water stress-induced epigenetic modifications have led to release from suppression of BARE-1 retrotransposition in the higher, drier sites, but that this burst of element activity has been independent of the actual adaptively significant physiological responses. Under this scenario, local adaptation may be taking place in wild barley populations, but this adaptation is postulated to arise from genetic and/or epigenetic changes unrelated to BARE-1 activity. Alternatively, perhaps the relationships between BARE-1 activity, water-stress, and adaptation are not only correlative but causal. To the extent that this is true, the mode of action of natural selection in the process remains mysterious, as does the organizational level on which selection might be manifested. It may be, for example, that selection is operating on one or more aspects of genome size that we presently do not perceive of as adaptively relevant. In contrast to whole-genome selection, perhaps retroelement activation has led to adaptively relevant insertions that affect drought-tolerant pathways or other ecologically relevant physiologies. Given that thousands of insertions appear to distinguish wild barley populations from adjacent sites, it may be a daunting challenge to ferret out the adaptively significant insertions.
Notwithstanding the number of remaining issues, the study of Kalendar et al. provides perhaps the best example yet of the dynamic nature of plant genome evolution on a local ecological scale, and hints at retrotransposon-mediated adaptive evolution. In this regard, the authors have taken the first significant step toward addressing McClintock's challenge to figure out how cells restructure their genomes in response to perceived danger.
Footnotes
See companion article on page 6603.
References
- 1.McClintock B. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 2.Price H J. Evol Trends Plants. 1988;2:53–60. [Google Scholar]
- 3.Price H J, Johnston J S. Proc Natl Acad Sci USA. 1996;93:11264–11267. doi: 10.1073/pnas.93.20.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston J S, Jensen A, Czechin D G, Price H J. Am J Bot. 1996;83:1113–1120. [Google Scholar]
- 5.Cavallini A, Natali L, Cionini G, Gennai D. Heredity. 1993;70:561–565. [Google Scholar]
- 6.Evans G M, Durant A, Rees H. Nature (London) 1966;212:697–699. [Google Scholar]
- 7.Graham M J, Nickell C D, Rayburn A L. Theor Appl Genet. 1994;88:429–432. doi: 10.1007/BF00223656. [DOI] [PubMed] [Google Scholar]
- 8.Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman A H. Proc Natl Acad Sci USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wessler S R. Curr Biol. 1996;6:959–961. doi: 10.1016/s0960-9822(02)00638-3. [DOI] [PubMed] [Google Scholar]
- 10.Grandbastien M-A. Trends Plant Sci. 1998;3:181–189. [Google Scholar]
- 11.Hirochika H. EMBO J. 1993;12:2521–2528. doi: 10.1002/j.1460-2075.1993.tb05907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirochika H, Sugimoto K, Otsuki Y, Kanda M. Proc Natl Acad Sci USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar A, Bennetzen J L. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 14.Thomas C A. Annu Rev Genet. 1971;5:237–256. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- 15.Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A. Plant Cell. 2000;12:381–391. doi: 10.1105/tpc.12.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SanMiguel P, Bennetzen J L. Ann Bot (London) 1998;81:37–44. [Google Scholar]
- 17.Vicient C M, Suoniemi A, Anamthawat-Jónsson K, Tanskanen J, Behavav A, Nevo E, Schulman A H. Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.SanMiguel P, Gaut B S, Tikhonov A, Nakajima Y, Bennetzen J L. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 19.Suoniemi A, Narvanto A, Schulman A H. Plant Mol Biol. 1996;31:295–306. doi: 10.1007/BF00021791. [DOI] [PubMed] [Google Scholar]
- 20.Jääskelainen M, Mykkanen A-H, Arma T, Vicient C, Suoniemi A, Kalendar R, Savilahti H, Schulman A H. Plant J. 1999;20:413–422. doi: 10.1046/j.1365-313x.1999.00616.x. [DOI] [PubMed] [Google Scholar]
- 21.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 22.Jordan I K, McDonald J F. Genetics. 1999;151:1341–1351. doi: 10.1093/genetics/151.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirasu, K., Schulman, A. H., Lahaye, T. & Schulze-Lefert, P. (2000) Genome Res., in press. [DOI] [PMC free article] [PubMed]