Abstract
High-throughput and high-content databases are increasingly important resources in molecular medicine, systems biology, and pharmacology. However, the information usually resides in unwieldy databases, limiting ready data analysis and integration. One resource that offers substantial potential for improvement in this regard is the NCI-60 cell line database compiled by the US National Cancer Institute, which has been extensively characterized across numerous genomic and pharmacological response platforms. In this report we introduce a CellMiner1 web application designed to improve use of this extensive database. CellMiner tools allowed rapid data retrieval of transcripts for 22,217 genes and 360 microRNAs along with activity reports for 18,549 chemical compounds including 91 drugs approved by the US Food and Drug Administration. Converting these differential levels into quantitative patterns across the NCI-60 clarified data organization and cross comparisons using a novel pattern-match tool. Data queries for potential relationships among parameters can be conducted in an iterative manner specific to user interests and expertise. Examples of the in silico discovery process afforded by CellMiner were provided for multidrug resistance analyses and doxorubicin activity; identification of colon-specific genes, microRNAs and drugs; microRNAs related to the miR-17-92 cluster; and drug identification patterns matched to erlotinib, gefitinib, afatinib, and lapatinib. CellMiner greatly broadens applications of the extensive NCI-60 database for discovery by creating web-based processes that are rapid, flexible, and readily applied by users without bioinformatics expertise.
Keywords: Systems Pharmacology, systems biology, omics, pharmacogenomics, biomarkers
Introduction
Access to bioinformatics frequently acts as a choke-point in the flow of information between large-scale technologies and the researchers who have the expertise to assess the data. Difficulty in fluid data access leads to restricted ability to integrate diverse data types, reducing understanding of complex biological and pharmacological systems. One such large-scale information set with multiple genomic and drug response platforms is the NCI-60 cancer cell line data base. These cell lines, due to the extensive pharmacology and genomic data available are prime candidates for data integration and broad public access.
The NCI-60 cell line panel was initially developed as an anti-cancer drug efficacy screen by the Developmental Therapeutics Program2 (DTP) of the US National Cancer Institute (NCI). Many thousands of compounds have been and continue to be applied to the NCI-60 (1, 2). In parallel, multiple platforms have been used to characterize the cells including, i) array comparative genomic hybridization (aCGH) (3), karyotypic analysis (4), ii) DNA mutational analysis (5), iii) DNA fingerprinting (6), iv) microarrays for transcript expression (7–9), v) microarrays for microRNA expression (9, 10), and vi) protein reverse-phase lysate microarrays (11).
An emphasis within our group is integration and open-access dissemination of molecular biology and molecular pharmacology information. One form of data integration we have developed over the last several years is the combination of multiple transcript microarray platforms. This integration saves time by preventing researchers from having to review data from each platform individually, and improves the accuracy and reliability of the results. Starting with a three-platform integration (12), we next tested the use of z score averages for probes to facilitate integration of results of different platforms done at different times (13). A z score is a mathematical transformation that for each probe measurement, for example ABCB1 gene expression across the NCI-60, subtracts the mean (to center the data), and then divides by the standard deviation (to normalize the range). This approach has recently been expanded to integrate five platforms (14), and has proven to be both reliable and informative (15–18). Here we show this approach can be adopted for the DTP drug activity as well.
In the current study, we introduce for non-informaticists a set of web-based tools accessible through our CellMiner web-application (19) that allow rapid access to and comparison of transcript expression levels of 22,217 genes, 360 microRNAs, and 18,549 compounds including 91 Food and Drug Administration (FDA)-approved drugs. The tools allow easy identification of drugs with similar activity profiles across the NCI60. The gene and drug assessments, having been derived from widely varying numbers of probes or experiments, include all probe or experimental results that pass quality control, allowing the assessment of data reliability. In addition, we introduce our “Pattern Comparison” tool, which rapidly searches for robust connections between these parameters, as well as any independent pattern of interest, and allows the user to mine data not only for specific genes or drugs, but also for systems biology and systems pharmacology investigations.
Materials and Methods
Quantitation of gene transcript expression levels in the NCI-60 using five microarray platforms
Transcript expression for each gene was determined through the integration of all pertinent probes from five platforms. From Affymetrix (Affymetrix Inc., Sunnyvale, CA) the Human Genome U95 Set (HG-U95, GEO accession GSE5949) (8); the Human Genome U133 (HG-U133, GEO accession GSE5720) (8); the Human Genome U133 Plus 2.0 Arrays (HG-U133 Plus 2.0, GEO accession GSE32474) (13); and the GeneChip Human Exon 1.0 ST array (GH Exon 1.0 ST, GEO accession GSE29682) (14). From Agilent (Agilent Technologies, Inc., Santa Clara, CA) we used the Whole Human Genome Oligo Microarray (WHG, GEO accession GSE29288) (9). Affymetrix microarrays were normalized by GCRMA (20). All WHG mRNA probes detected in at least 10% of cell lines were normalized using GeneSpring GX by i) setting gProcessedSignal values less than 5 to 5, ii) transforming gProcessedSignal or gTotalGeneSignal to Logbase 2, and iii) normalizing per array to the 75th percentile (9). Data for these microarrays are accessible at our web-based data retrieval and integration tool, CellMiner1 (19). Affymetrix probe-sets are referred to as “probes” for ease of description within this manuscript.
Quality control for genes is done as follows. For every probe for that gene, the intensity range across the NCI-60 is determined, and all probes with a range of ≤ 1.2 log2 are dropped. The number of probes that pass this criterion is determined, and 25% of that number calculated (keeping a minimum of 2 and a maximum of 122). For the remaining probes, Pearson’s correlations are determined for all probe/probe combinations. The average correlation for each probe is determined compared to all other (remaining) probes. Probes whose average correlations are less than 0.30 (p< 0.02 for 60 cell lines in the absence of multiple comparisons correction) and not correlated to any other individual probes at ≥ 0.30 are dropped. The average correlation for remaining probes is then recalculated. Next, for probes with average correlations less than 0.60 (p<0.0000004 for 60 cell lines), the lowest of these are dropped, and average correlations recalculated for all remaining probe combinations. Probes are dropped until either all are ≥ to 0.60, or the 25% (of probes that passed the 1.2 log2 range criteria) level is reached. This ensures significant pattern match across probes.
Probe intensity values that pass these quality controls are transformed to z-scores (21). Average z-scores were determined for each gene for each cell line. Probes with only one experiment that passes the ≤ 1.2 log2 range test are included as they are potentially informative, but must be considered less reliable.
Quantitation of drug and compound activity levels
Drug activity levels expressed as 50% growth inhibitory levels (GI50s) are determined by the Developmental Therapeutics Program2 at 48 hours using the sulphorhodamine B assay (22). Repeat experiments must pass quality control criteria, similar to those for gene transcript levels. Experiments with range less than 1.2 log10 or with information on less than 35 cell lines are dropped. This serves to eliminate non-responsive and out of proper range data. The number of experiments that pass these criteria is determined, and 25% of that number calculated (keeping a minimum of 2 and a maximum of 122). Pearson’s correlations are determined for all remaining possible experiment/experiment combinations. Experiments whose average correlations are less than 0.334 (p<0.05 for the 35 cell line minimum in the absence of multiple comparisons correction) and were not correlated to individual probes at ≥ 0.334 are dropped. For the remaining experiments with average correlations less than 0.60 (p<0.00014 for the 35 cell line minimum), the lowest is dropped, and the correlations recalculated for all remaining possible experiment/experiment combinations. Experiments are dropped in this fashion until either all are ≥ to 0.60, or the 25% (of experiments that passed the 1.2 log2 range criteria) level is reached. This ensures significant pattern match across experiments. Drugs with only one experiment, but pass that the ≤1.2 log10 range test are included as output as they are potentially informative, but must be considered less reliable. For the purpose of this manuscript, compounds that have not yet undergone clinical trials will sometimes be referred to as drugs, as well as those that have.
MicroRNA expression levels
MicroRNA expression levels were determined as described previously (9) for the Agilent Technologies 15k feature Human miRNA Microarray (V2) following manufactures recommendations, and are available at CellMiner1 as well as at GEO (accession GSE22821).
Pattern comparisons
The comparisons of gene transcript expression, microRNA expression, and drug activity across the NCI-60 use the data prepared as described in the previous three sections. Pearson’s correlations between these parameters were calculated using Java 5.0.
Results
Web-based bioinformatics tool access for the NCI-60
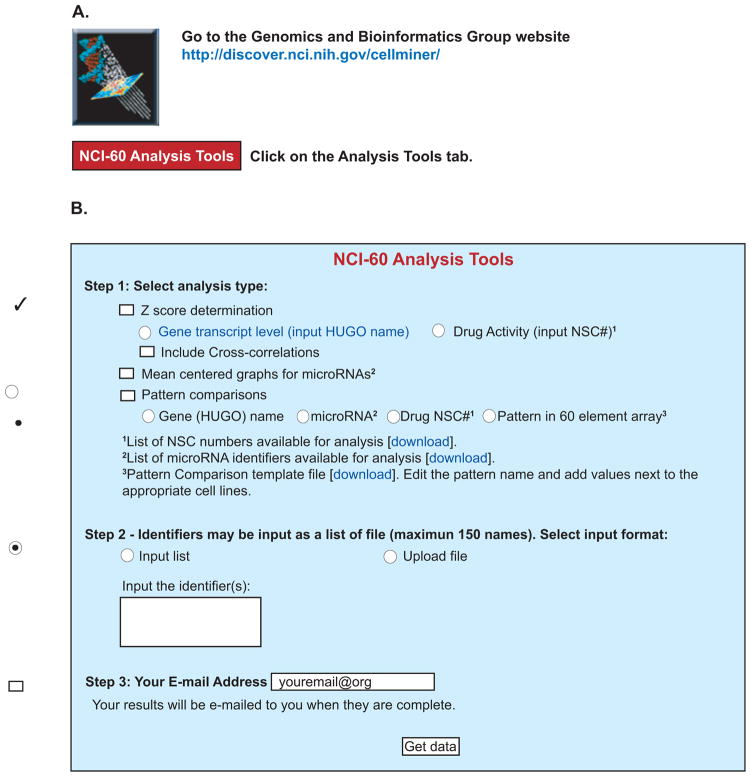
Several important forms of bioinformatics analyses, made to be easily accessible for the non-informaticist, are now available at the Genomics and Bioinformatics Group’s CellMiner1 site (Figure 1A). To access those, click the “NCI-60 Analysis Tools” tab. These tools (Figure 1B) allow the user to rapidly obtain information for the NCI-60 that would otherwise require lengthy data retrieval, compilation, and assessment. Included are data for relative transcript and microRNA levels, as well as drug activity. The tools include pattern comparison functionality that enables the identification of relationships between these and other parameters, as driven by the user’s interest.
Figure 1.
Snapshot of the NCI-60 Analysis Website, a suite of web-based tools designed to facilitate rapid pharmacologic and genomic bioinformatics for the NCI-60 cell lines. A. These tools are accessible at the CellMiner website (http://discover.nci.nih.gov/cellminer/) by clicking on the NCI-60 Analysis Tools tab. B. The analysis of interest (Z score determination, Mean centered graphs for microRNAs, or Pattern comparisons) is selected in Step1 using the check boxes. The specific identifier or pattern of interest is selected in Step 2, either by typing in an identifier using the “Input list” function, or by uploading a file using the “Upload file” function. A maximum of 150 identifiers (genes, microRNAs or drugs) can be input at once. The results are e-mailed to the address entered in Step 3. Multiple check boxes may be selected for a single input. Radio buttons (circles) for an analysis type are mutually exclusive.
In each case (Figure 1B), users select the tool in “Step1”. Identifiers or patterns are typed in if one chooses “Input list”, or uploaded as a .txt or .xls file if “Upload file” is chosen in “Step 2”. Next, users enter their e-mail address for receipt of the data in “Step 3”, and click “Get data” to receive data (as an Excel file). Data are generally available within a few minutes. Error files are returned if no results are provided, stating the reason. These tools are described with examples in the subsequent figures.
Relative transcript levels tool
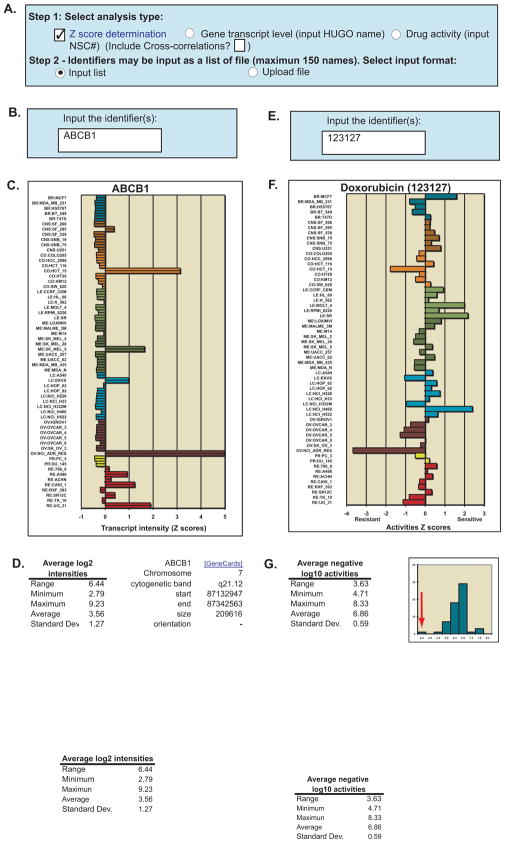
To obtain high-definition, easily interpretable transcript level data select the “Z score determination” tool and “Gene transcript level” shown in Step 1 of Figure 2A. Check “Input list” and input your gene of interest in “Step 2” (Figure 2A and B). Official gene names are required (see the Human Genome Organization, HUGO3). In the example, ABCB1 is the HUGO name of the gene encoding the P-glycoprotein MDR (P-gp) drug efflux transporter. The tool compiles all probe information available for the selected gene from 5 microarray platforms, the Affymetrix HG-U95, HG-133, HG-U133 Plus 2.0, and the GeneChip Human Exon 1.0 ST array. Quality control is included based on single probe variation, as well as probe-to-probe correlation to eliminate poor quality, dead, or background level probes.
Figure 2.
Relative transcript expression and drug activity levels in the NCI-60. A. Check the “Z score determination” check-box, and then choose either the “Gene transcript level” or the “Drug activity” radio button in step 1. In those instances in which multiple genes or drugs are entered, a cross correlation of all genes or drugs entered may be included by checking the “Include cross-correlations” check box. In Step 2, the user chooses whether to type in the input, or upload it as a file by selecting “Input list” or “Upload file” (as .txt or .xls), respectively. B. For relative transcript expression levels, input the gene name(s) using the “official” (HUGO) name. C. A graphical z score composite of all transcript probes that pass quality control is generated, along with the numerical values (data not shown). D. Mean log2 intensity values for range, minimum, maximum, average, and standard deviation for all Affymetrix probes are included to assist in data interpretation, as well as chromosomal location. E. For relative drug activity levels, input the drug “official” NSC number(s) (see “Download NSCs” file from Figure 1B, Step 1, for a listing of these). F. A graphical z score composite of all drug experiments that pass quality control is generated, along with the numerical values (data not shown). G. Mean log10 intensity values for range, minimum, maximum, average, and standard deviation for all experiments are included to assist in data interpretation, as well as a histogram of the cell lines average activities. For the histogram, the x-axis is the average experiment activity for the cell lines, and the y-axis the frequency at which they occur. The red arrow indicates the most resistant cell line to doxorubicin, NCI-ADR-RES.
The tool output includes relative transcript intensity presented as z scores and visualized as a bar graph (Figure 2C). The bars for each cell line are color coded by tissue of origin. Parameters of the Affymetrix probe intensities (range, minimum, maximum, average, and standard deviation) are included (Figure 2D) to provide an untransformed reflection of transcript level variation. Chromosomal location is also included (Figure 2D). In the example shown in Figure 2C, NCI-ADR-RES and HCT-15 are the top two expressers of ABCB1. Notably, most cell lines appear to the left of the mean with uniformly low values. Those low values all approach 2.79 log2 intensity units (Figure 2D), which is within background and indicates that all those cell lines have undetectable or low ABCB1 transcript levels.
In each data-file for individual genes, the user also receives; i) the total number of probes for the gene (including those that failed), ii) all intensity values (for those probes that passed quality control) to a maximum of 122, iii) the z score transforms for each probe, iv) the platform that each probe originated from, v) the exon to which the probe hybridizes, vi) the average z score for each cell line, and vii) the distribution of the individual cell line average intensities presented as a histogram. Currently information on 22,217 genes is available.
The “Z score determination” tool also allows multiple gene entries in the input box (Figure 2B; with each gene on a separate line). Each gene is then returned as separate Excel file with its own plots and data. In the case of multiple genes being entered, a cross correlation table of the resultant z scores can be generated by clicking the “Include cross-correlations” box in Figure 2A, Step 1.
Relative drug activities tool
To obtain curated GI50 data (50% growth inhibition), select the “Z score determination” tool shown in Figure 1A (top right), and select “Drug activity”. Specify the compound using the National Service Center number (NSC) as the input (Figure 2E, doxorubicin’s NSC is 123127). This tool will compile all experimental information available from the DTP2 for the selected compound(s). Quality control is included based on single experiment variation, as well as experiment-to-experiment correlation to eliminate out of appropriate concentration, weak or invariant response, and irreproducible experiments. The compound must have a minimum of 35 cell lines with activity information to be included. The tool output includes relative compound activity presented as z scores and visualized as a bar graph (Figure 2F). Parameters of the compound activities (range, minimum, maximum, average, and standard deviation) are included (Figure 2G) to provide the user with an untransformed reflection of compound activity. The distribution of the individual cell line average responses is presented as a histogram. In the example shown, NCI-ADR-RES and HCT-15 are the two cell lines most resistant to doxorubicin in the NCI-60, which is consistent with the fact they over-express ABCB1, whose gene product P-gp pumps the drug from the cell (23, 24). In the case of multiple drugs being entered, a cross correlation table of the resultant z scores for each drug may also be generated by clicking the “Include cross-correlations” box in Figures 2A and 1B, Step 1.
The user also receives: i) the total number of experiments done for the compound (including those that failed), ii) whether the compound has been FDA approved, iii) all activity values (for those experiments that passed quality control, to a maximum of 122), iv) the z score transforms for each experiment, v) and the average z score for each cell line. Currently, 47,540 compounds have passed through these filters, resulting in data for 18,549 compounds. NSC numbers are the required input (download the list of currently available NSCs at “List of NSC numbers available for analysis”, Figure 1B). NSC numbers can be cross-referenced to other identifying parameters, including their chemical structures4.
Relative microRNA levels tool
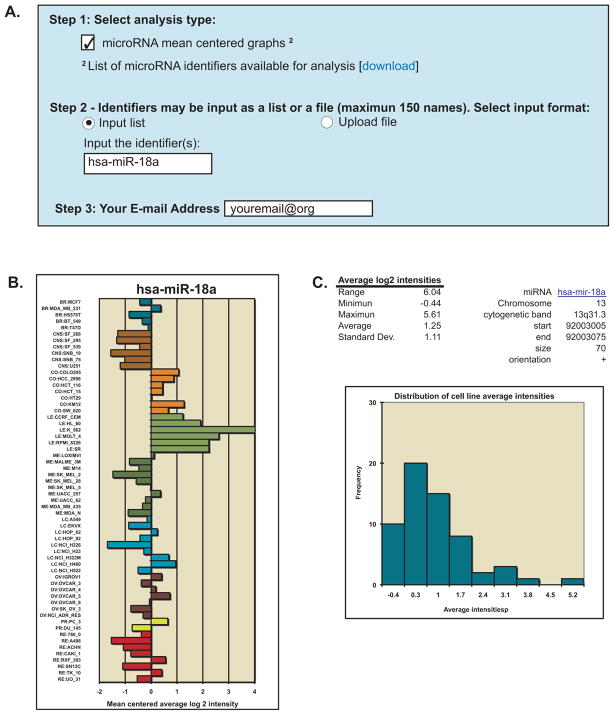
To rapidly obtain the log2 intensity values for microRNAs, the “microRNA mean centered graphs” tool is selected as shown in Step 1 of Figure 3A. Input your microRNA(s) of interest in “Step 2” using the name from the provided “List of microRNA identifiers available for analysis” accessed by clicking [download] in “Step 1”. The microRNA tool provides access to the average log2 intensity values for the Agilent Technologies Human miRNA Microarray (V2) (9). Z scores are not used, as this is a single platform analysis done at a single time. The tool output includes relative transcript intensities visualized as a bar graph. In the example shown in Figure 3B, we queried the data for a microRNA from the miR-17-92 oncogenic cluster (25). Other microRNAs from the same cluster showed the same pattern of expression, with high levels in leukemia and colon cancer lines (data not shown; see Discussion). Parameters of probe intensities (range, minimum, maximum, average, and standard deviation), as well as the chromosomal location are included (Figure 3C). The distribution of the individual cell line average intensities is presented as a histogram. Currently information on 360 microRNAs is available.
Figure 3.
Relative microRNA transcript expression levels in the NCI-60. A. Input of data. Check the “microRNA mean centered graphs” check-box in Step 1. To access the list of microRNAs names to use, go to footnote 2, “List of microRNA identifiers” and click “download”. In Step 2, the user chooses whether to type in the input, or upload it as a file (.txt or .xls) by selecting “Input list” or “Upload file”, respectively. If typing in the input, input the microRNA name(s) in the “Input the identifiers” box. In Step 3, enter the e-mail address to send the results to. B. The output includes a plot of the mean-centered average log2 intensities along with their numerical values (data not shown). C. Mean log2 intensity values for range, minimum, maximum, average, and standard deviation for all cell lines are included to assist in data interpretation, as well as a histogram of the cell lines average intensities. For the histogram, the x-axis is the average intensity for the cell lines, and the y-axis the frequency at which they occur.
Pattern comparison tools
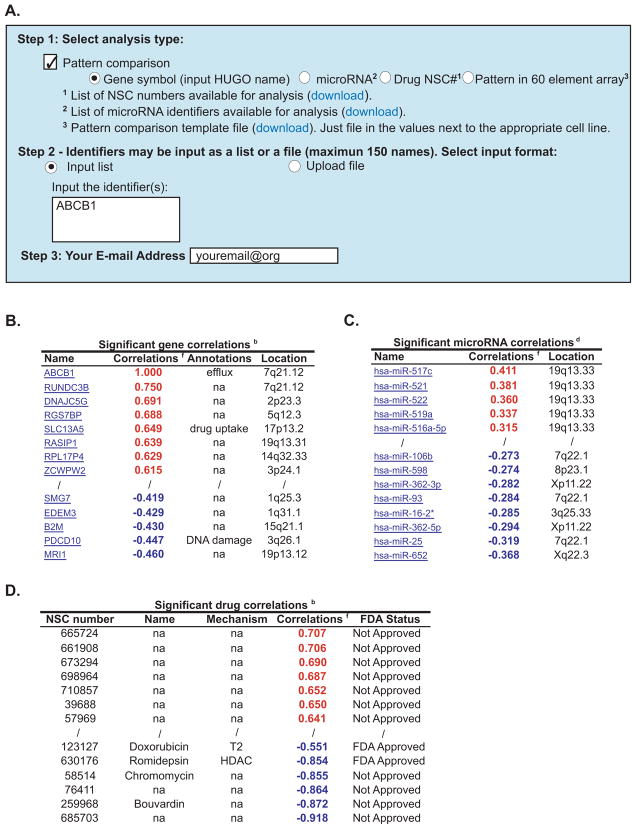
To make comparisons between an input pattern of interest (including expression of a gene or microRNA, activity of a drug, or any pattern of interest) and i) gene expression, ii) microRNA expression, iii) and drug activity levels, start by selecting the “Pattern comparison” tool shown in Step 1 of Figure 4A, and one of the four possible types of input, “Gene symbol”, “microRNA”, “Drug NSC#”, or “Pattern in 60 element array”. Input your identifier(s) in “Step 2”, using a gene, microRNA, drug or pattern. The first three of these are the same 22,217 gene expression levels, 18,549 drugs activity levels, and 360 microRNA expression levels, described above (see Figures 1–3). The forth option is to input any pattern across the NCI-60 that is of interest to the user. A “Pattern comparison template file” for this option is accessible for download in Step 1, footnote 3 (Figure 4A).
Figure 4.
Pattern comparison to transcript expression, microRNA expression and drug activity levels in the NCI-60. A. Choose your input type by checking the “Pattern comparison” check-box, and then choose either the “Gene symbol”, “microRNA”, “Drug NSC#”, or “Pattern in 60 element array” radio buttons in Step 1. In Step 2, the user chooses whether to type in the input, or upload it as a file by selecting “Input list” or “Upload file”, respectively (use the same identifiers as in Figure 2 and 3). To input your own template, use the “Pattern comparison template file” download from footnote 3, with your numerical values and “na” for missing or to be ignored values. B. Significant gene correlation output, given for all genes that match your input pattern at a significance level of p<0.05. C. Significant microRNA correlations output, given for all microRNAs that match your input pattern at a significance level of p<0.05. D. Significant drug correlations output, given for all compounds that match your input pattern at a significance level of p<0.05. Only the top and bottom of the lists are shown in each case.
The output is returned as a single Excel spreadsheet file that includes six main sections. Those are: “All” and “Significant gene correlations” (Figure 4B), “All” and “Significant microRNA correlations” (Figure 4C), and “All” and “Significant drug correlations” (Figure 4D) (the three “All” columns are not shown in panels B,C,D). The top and bottom portions of these sets from the input of ABCB1 are shown in Figure 4. The “all” versions of these outputs contain the same columns as do the “significant”, with the exception of the “FDA Status” column in Figure 4D, but include all possible correlations and are in either alphabetical (for genes and microRNAs) or numerical (for drugs by NSC number) order. The “Significant correlations” outputs, are based on statistical significance (p <0.05) in the absence of multiple comparisons correction, and are ordered by descending correlations with color-coding (red- and blue-bold fonts for positive and negative significant correlations, respectively). The data used for the correlation calculations for the genes, drugs, and microRNAs are those generated in the gene transcript level, drug activity, and microRNA tools in Figures 2 and 3, respectively. Information included for the genes sections are annotations of several pharmacology, cancer, or gene regulation categories (see the “Annotations” column, Figure 4B), as well as the chromosomal locations for the “significant correlations” section (see the “Location” column, Figure 4B). Gene names for the “Significant correlations” section are hyperlinked to GeneCards4. Information included for the “Significant correlations” microRNA section is the chromosomal locations of the microRNAs (see the “Location” column, Figure 4C). MicroRNA names for the “significant correlations” section are hyperlinked to miRBase5. Information included for the drug sections are name (where available, see the “Name” column, Figure 4D), mechanism of action (for 243 drugs currently, see the “Mechanism” column, Figure 4D), and for the “Significantly correlated” section, the Food and Drug Administration approval status (see the “FDA Status” column, Figure 4D). A footnotes page is included with the output to provide details on the definitions, approaches, abbreviations, and background.
Additional uses of the pattern tools (starting with a predefined pattern or a given drug) are presented as examples in the discussion (see Figures 5 and 6).
Figure 5.
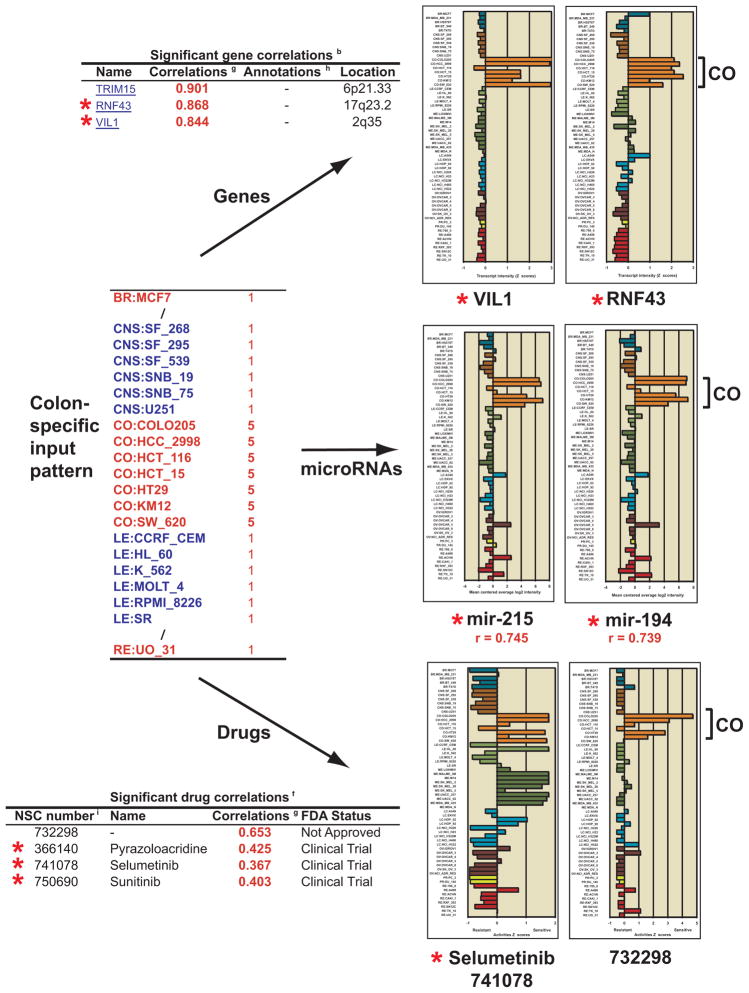
Pattern comparison example for a colon-specific input pattern. The input for this analysis consisted of ones for all non-colon cell lines, and fives for all colon. The three forms of output described in Figure 4, genes, microRNAs, and drugs, are shown. The top three genes by correlation are shown in tabular fashion. Bar graphs for two of the genes, generated as described in Figure 2, display the data visually. The top two microRNAs by correlation are shown next in graphical fashion, generated as described in Figure 3. The top three drugs with either the FDA-approved or clinical trials by correlation, and one clinically untested compound are shown in tabular fashion. Bar graphs for two of these, generated as described in Figure 2, are displayed. The red star in all cases indicates prior literature association with colon cancer.
Figure 6.
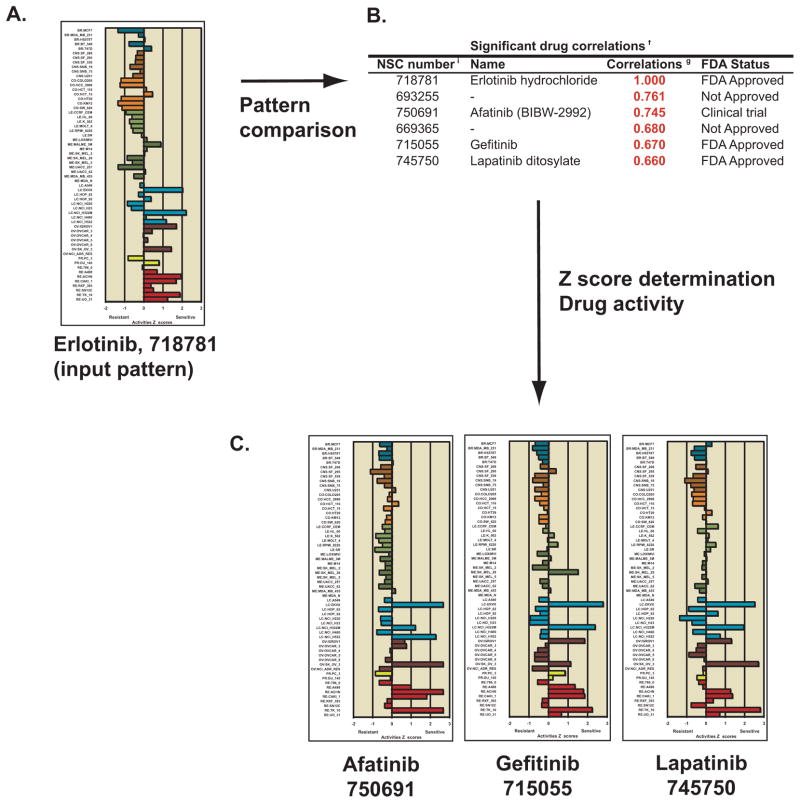
Recognition of drugs with similar mechanism of action, using erlotinib as an input for pattern comparison. A. Use the “Pattern comparison” tool (Figure 4) selected for “Drug NSC” and input 718781 (the NSC for erlotinib). The bar graph shown is that from the “Z score determination” tool from Figure 2, selected for “Gene transcript level”. B. The top six rows of the “Significant drug correlations” output from the “Pattern comparison” tool, identifying two other FDA-approved drugs and one in advanced clinical trials. C. The bar graphs shown are from the “Z score determination” tool from Figure 2, selected for “Gene transcript level” for the two FDA-approved, and one in clinical trials drugs identified in B.
Discussion
The sheer quantity of data generated by current high-throughput platforms have encumbered their use and access. However, due to the emergence of new “omic” technologies and the questions to be asked in dealing with issues of human disease, it is extremely important to open such data to clinicians, molecular biologists, and others with insights into aspects of disease. This is clearly the case in the difficult and complex case of cancer.
Our web-based tools provide assistance in this area for the NCI-60. They allow rapid determination of i) a composite “best” gene transcript expression level pattern from five microarray platforms, ii) drug activity from all experimental repeats done by the DTP, and iii) transcript levels of microRNAs from duplicate microarrays (9) (Figures 2 and 3). The “Pattern comparison” tools (Figures 4–6) allow rapid, global exploration of relationships between these parameters and any input pattern of interest.
This accession of relative levels of transcript expression (Figure 2C) provides several advantages. The tool i) compiles all probes for a single gene, ii) incorporates built-in quality control, and iii) includes probe-remapping based on HG-19, eliminating the need to spend time or have expertise in those areas. Allowing the user to see the input probes for a single gene allows assessment of reliability and accuracy. For example, an expression pattern for a gene with 46 highly correlated probes (as is the case for ABCB1 in Figure 2C) demonstrates both high reliability and accuracy. Conversely, a gene with two probes that barely pass quality control should be considered both less reliable and accurate. Availability of reliable patterns of relative transcript expression in turn facilitates comparison to other data, such as drug activity and microRNA expression. For ABCB1 and doxorubicin (Figure 2), there is obvious correlation between high ABCB1 expression and doxorubicin resistance, consistent with prior results (23, 24).
During development of the tools derived for gene transcript z score patterns, we have validated and exploited them to elucidate novel gene regulation mechanisms for MYC, TOP1 and CHEK2 (15–17). The cross correlation function illustrated in Figure 2A, Step1, identified transcriptional co-regulation among kinetochore genes (17) and across genes driving cell migration and adhesion (26).
That regulatory elements may be identified using these tools is shown by the example of the cell-cell adhesion factor CDH1 (E-cadherin). Using the pattern comparison tool with CDH1 as input, one finds the transcriptional repressors ZEB1 (TCF8), SNAI2 (SLUG), ZEB2 (SIP1), and TWIST1 each has significant negative correlation to CDH1 transcript levels, at −0.63, −0.47, −0.37, and −0.47, respectively. These repressors have been reported previously to negatively regulate CDH1 (27–31). In addition, they show robust positive correlations to hsa-miR-200c, 200a, 200b, 200b* and 200a*, with values of 0.73, 0.59, 0.55, 0.48, and 0.41 respectively. The microRNA-200 family has been previously associated with E-cadherin regulation through ZEB1 and 2 targeting (32, 33).
The microRNA mean-centered graph tool (Figure 3A and B) simply provides raw data and mean-centered graphical representation of the data determined previously (9). In the example shown, miR-18a exhibits notable specificities for colon and especially leukemia cell lines. MiR-18a has previously been shown to be part of a polycistronic oncogenic microRNA cluster, which includes 6 consecutive mature microRNAs, miR-17, miR-18a, miR-19a, miR-19b, miR-20a and miR-92a co-expressed as a single primary transcript (25, 34). Notably, the pattern comparison tool for miR-18a retrieves the 5 other microRNAs at the top of the “Significantly correlated microRNA” list (with Pearson correlation coefficients between 0.96 and 0.77) and MYC ranked 37th in the gene list with a correlation coefficient of 0.61. The MYC correlation is consistent with the fact that the miR-17-92 cluster is a transcriptional MYC target (35).
The ability to rapidly compare patterns of transcript expression, microRNA expression, and drug activity (Figures 2 and 3) both to themselves, and other patterns of interest provides a powerful and flexible tool to the user. The tool automatically i) allows the input of multiple types of data, ii) calculates the correlation to three types of data, and iii) identifies those correlations that are statistically significant. Allowing the user to see the correlation level and order in which genes, microRNAs, or compounds are ranked for any single input, provides criteria by which to identify potential relationships between disparate parameters and to access the robustness of these relationships. Of the named drugs in Figure 4D other than doxorubicin, romidepsin (FK228) has previously been shown to undergo efflux by ABCB1 (36), and bouvardin to have cross-resistance with doxorubicin (37). To the best of our knowledge, chromomycin had not been previously associated with ABCB1. Significant microRNA expression versus gene expression, gene expression versus drug activity, and microRNA expression versus drug activity correlations have previously been identified in this fashion (14, 17).
A novel example is provided in Figure 5, in which a colon specific pattern is entered into the pattern comparison tool. The input was 1’s for all non-colon cell lines, and 5’s for all colon cell lines. The top three genes from the “Significant gene correlations” output are shown. The top gene, TRIM15 is a little studied tripartite motif family gene, but its high correlation (0.901) to the colon specific input, makes it a candidate for being a colon cancer specific marker. RNF43, is an ubiquitin ligase known to be up-regulated in colon cancer (38). VIL1, is an actin-binding protein previously identified as a diagnostic marker for colon cancer (39). The bar graphs for VIL1 and RNF43, derived from the “Z score determination” tool (Figure 2) illustrate their colon specificity. The bar plots for the top two microRNAs from the “Significant microRNA correlations” are shown next. Both miR-215 and 194 (with correlations of 0.745 and 0.739, respectively) have previously been described as prognostic indicators for colorectal cancer (40, 41). The bar graphs, from the “microRNA mean centered graphs” tool from Figure 3, illustrate their colon specificity. Four of the top seven drugs from the “Significant drug correlations” list are also shown. Of these, three have some clinical trial history. Pyrazoloacridine, has been tested in colon cancer with modest success (42). Selumetinib (AZD6244) has been proposed as a potential therapy for colorectal cancer (43). From our bar graph, it appears to have specificity for melanoma in addition to colon cancers. Sunitinib is in clinical trial for colorectal cancer (44). The additional compound presented as a bar graph, NSC732298, has better specificity for colon that any of these (r = 0.653), but has not gone through clinical trials. The bar graphs for selumetinib and 732298, from the “Z score determination” tool from Figure 2 illustrate their level of colon specificity. Together, these genes, microRNAs, and drugs illustrate how one can, from a single pattern input, rapidly obtain multiple types of both known and novel information relevant to a users area of interest.
The ability to access relative drug activity levels and patterns (Figure 2F) provides advantages similar to those for relative transcript levels. The tool automatically i) compiles all experiments for a single drug from all experiments done, and ii) incorporates quality control into the approach. It allows the user to see the input experiments, allowing assessment of reliability and accuracy. This is especially important for the activity data, as there are many compounds with small numbers of experiments, and some with reduced numbers of cell lines tested. An example of a highly reliable activity pattern is that for doxorubicin, which has 122 highly correlated experiments (Figure 2F). The identification of a reliable pattern of relative compound activity facilitates its comparison to other types of data. Drug patterns derived in this fashion have been used previously for comparison to mRNA and microRNA expression (14).
A final example of the flexibility of the new tools is depicted in Figure 6. As illustrated, the pattern comparison tools can be readily adapted to perform COMPARE-like (1, 45) drug analyses, in which the user can query the 18,549 drugs and chemicals from the DTP database to identify similar drugs. In Figure 6, the input is erlotinib (NSC 718781), an inhibitor of the epidermal growth factor receptor (EGFR) tyrosine kinase. Pleasingly, the two other FDA-approved EGFR-targeted drugs gefitinib and lapatinib ranked 5th and 6th, and afatinib (BIBW2992), which is in advanced clinical trials and also targets EGFR-ERB kinase ranked 3rd among the 18,549 drugs (Figure 6B). Compound 693255, which ranked 2nd is a tyrphostin derivative, a class of drugs shown to inhibit tyrosine kinase including EGFR (46). The plots shown in Figure 6C were obtained using the z score tool (see Figure 2F). They demonstrate pattern similarity among the four highly correlated drugs. Conversely, the piperidinium compounds (618757, 636676, 638634, and 630602 with r = −0.587, −0.536, −0.532, and −0.504, respectively) have consistently high inverse correlation. That is, they work well when the EGFR-inhibitors work poorly. This example illustrates the tools usefulness in the comparison of drugs with similar mechanisms of action, including those that are in clinical development (such as afatinib), as well as identifying novel compounds that either might work in a similar fashion, or are inverse to it.
The ease, rapidity and flexibility of the new tools provide users with an important new data integration capacity. One of our goals is to update and enhance these tools forwarding an ongoing fashion. Our next two additional tools will be the Comparative Genomic Hybridization tool and database (aCGH), which will assist in interpretation of DNA copy number using our Roche NimbleGen 385k CGH array, and the Whole Exome Sequencing tool and database (WES), which will provide access to the whole genome sequences for all exons across the NCI-601. Together, these databases and tools provide publicly available and unique opportunities for systems biology and systems pharmacology investigations.
Acknowledgments
This work is supported by the Center for Cancer Research, the intramural program of NCI, and the Developmental Therapeutics Program (DTP), Division of Cancer Treatment and Diagnosis (DCTD), NCI.
Footnotes
There are no conflicts of interest to report.
References
- 1.Holbeck SL, Collins JM, Doroshow JH. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol Cancer Ther. 2010;9:1451–60. doi: 10.1158/1535-7163.MCT-10-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoemaker R. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6(10):813–23. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 3.Bussey K, Chin K, Lababidi S, Reimers M, Reinhold W, Kuo W, et al. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 cell line panel. Mol Cancer Ther. 2006;5:853–67. doi: 10.1158/1535-7163.MCT-05-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roschke A, Tonon G, Gehlhaus K, McTyre N, Bussey K, Lababidi S, et al. Karyotypic Complexity of the NCI-60 Drug-Screening Panel. Cancer Research. 2003;63:8634–47. [PubMed] [Google Scholar]
- 5.Ikediobi O, Davies H, Bignell G, Edkins S, Stevens C, O’Meara S, et al. Mutation analysis of twenty-four known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–12. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorenzi P, Reinhold W, Varma S, Hutchinson A, Pommier Y, Chanock S, et al. DNA fingerprinting of the NCI-60 cell line panel. Mol Cancer Ther. 2009;8:713–24. doi: 10.1158/1535-7163.MCT-08-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scherf U, Ross D, Waltham M, Smith L, Lee J, Tanabe L, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–44. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 8.Shankavaram U, Reinhold W, Nishizuka S, Major S, Morita D, Reimers M, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6:820–32. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- 9.Liu H, D’Andrade Petula, Fulmer-Smentek Stephanie, Lorenzi Philip, Kohn Kurt W, Weinstein John N, et al. mRNA and microRNA expression profiles integrated with drug sensitivities of the NCI-60 human cancer cell lines. MCT. 2010;9(5):1080–1091. doi: 10.1158/1535-7163.MCT-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blower PE, Verducci JS, Lin S, Zhou J, Chung J, Dai Z, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6:1483–91. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 11.Nishizuka S, Charboneau L, Young L, Major S, Reinhold W, Waltham M, et al. Proteomic profiling of the NCI60 cancer cell lines using new high-density ‘reverse-phase’ lysate microarrays. Proc Natl Acad Sci U S A. 2003;100:14229–34. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfister TD, Reinhold WC, Agama K, Gupta S, Khin SA, Kinders RJ, et al. Topoisomerase I levels in the NCI-60 cancer cell line panel determined by validated ELISA and microarray analysis and correlation with indenoisoquinoline sensitivity. Mol Cancer Ther. 2009;8:1878–84. doi: 10.1158/1535-7163.MCT-09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhold WC, Mergny JL, Liu H, Ryan M, Pfister TD, Kinders R, et al. Exon array analyses across the NCI-60 reveal potential regulation of TOP1 by transcription pausing at guanosine quartets in the first intron. Cancer Res. 2010;70:2191–203. doi: 10.1158/0008-5472.CAN-09-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gmeiner WH, Reinhold WC, Pommier Y. Genome-wide mRNA and microRNA profiling of the NCI 60 cell-line screen and comparison of FdUMP[10] with fluorouracil, floxuridine, and topoisomerase 1 poisons. Mol Cancer Ther. 2010;9:3105–14. doi: 10.1158/1535-7163.MCT-10-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoppoli G, Douarre C, Dalla Rosa I, Liu H, Reinhold W, Pommier Y. Coordinated regulation of mitochondrial topoisomerase IB with mitochondrial nuclear encoded genes and MYC. Nucleic Acids Res. 2011;39:6620–32. doi: 10.1093/nar/gkr208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zoppoli G, Solier S, Reinhold WC, Liu H, Connelly JW, Jr, Monks A, et al. CHEK2 genomic and proteomic analyses reveal genetic inactivation or endogenous activation across the 60 cell lines of the US National Cancer Institute. Oncogene. 2011 doi: 10.1038/onc.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinhold WC, Erliandri I, Liu H, Zoppoli G, Pommier Y, Larionov V. Identification of a predominant co-regulation among kinetochore genes, prospective regulatory elements, and association with genomic instability. PLoS One. 2011;6:e25991. doi: 10.1371/journal.pone.0025991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeeberg BR, Reinhold W, Snajder R, Thallinger GG, Weinstein JN, Kohn KW, et al. Functional Categories Associated with Clusters of Genes That Are Co-Expressed across the NCI-60 Cancer Cell Lines. PLoS One. 2012;7:e30317. doi: 10.1371/journal.pone.0030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, Reinhold WC, et al. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics. 2009;10:277. doi: 10.1186/1471-2164-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol. 2005;12:882–93. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- 21.Larsen RJ, Marx ML. An Introduction to Mathematical Statistics and Its Applications. 3. 2000. [Google Scholar]
- 22.Rubinstein LV, Shoemaker RH, Paull KD, Simon RM, Tosini S, Skehan P, et al. Comparison of in vitro anticancer-drug-screening data generated with a tetrazolium assay versus a protein assay against a diverse panel of human tumor cell lines. J Natl Cancer Inst. 1990;82:1113–8. doi: 10.1093/jnci/82.13.1113. [DOI] [PubMed] [Google Scholar]
- 23.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15665–70. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szakacs G, Annereau JP, Lababidi S, Shankavaram U, Arciello A, Bussey KJ, et al. Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell. 2004;6:129–37. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 25.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohn KW, Zeeberg BR, Reinhold WC, Sunshine M, Luna A, Pommier Y. Gene expression profiles of the NCI-60 human tumor cell lines define molecular interaction networks governing cell-matrix attachments in migrating cells. PLoS ONE. 2012 doi: 10.1371/journal.pone.0035716. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinhold WC, Reimers MA, Lorenzi P, Ho J, Shankavaram UT, Ziegler MS, et al. Multifactorial regulation of E-cadherin expression: an integrative study. Mol Cancer Ther. 2010;9:1–16. doi: 10.1158/1535-7163.MCT-09-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Tillo E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 29.Montserrat N, Gallardo A, Escuin D, Catasus L, Prat J, Gutierrez-Avigno FJ, et al. Repression of E-cadherin by SNAIL, ZEB1, and TWIST in invasive ductal carcinomas of the breast: a cooperative effort? Hum Pathol. 2011;42:103–10. doi: 10.1016/j.humpath.2010.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7:1267–78. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 31.Bolos V, Peinado H, Perez-Moreno M, Fraga M, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 32.Xiong M, Jiang L, Zhou Y, Qiu W, Fang L, Tan R, et al. MiR-200 family regulates TGF-{beta}1-induced renal tubular epithelial to mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J Physiol Renal Physiol. 2011 doi: 10.1152/ajprenal.00268.2011. [DOI] [PubMed] [Google Scholar]
- 33.Tryndyak VP, Beland FA, Pogribny IP. E-cadherin transcriptional down-regulation by epigenetic and microRNA-200 family alterations is related to mesenchymal and drug-resistant phenotypes in human breast cancer cells. Int J Cancer. 2010;126:2575–83. doi: 10.1002/ijc.24972. [DOI] [PubMed] [Google Scholar]
- 34.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Haaften G, Agami R. Tumorigenicity of the miR-17-92 cluster distilled. Genes Dev. 2010;24:1–4. doi: 10.1101/gad.1887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao JJ, Foraker AB, Swaan PW, Liu S, Huang Y, Dai Z, et al. Efflux of depsipeptide FK228 (FR901228, NSC-630176) is mediated by P-glycoprotein and multidrug resistance-associated protein 1. J Pharmacol Exp Ther. 2005;313:268–76. doi: 10.1124/jpet.104.072033. [DOI] [PubMed] [Google Scholar]
- 37.Chitnis MP, Joshi SS, Gude RP, Menon RS. Induced resistance in leukaemia L1210 to adriamycin and its cross-resistance to vincristine and bouvardin. Chemotherapy. 1982;28:209–12. doi: 10.1159/000238078. [DOI] [PubMed] [Google Scholar]
- 38.Yagyu R, Furukawa Y, Lin YM, Shimokawa T, Yamamura T, Nakamura Y. A novel oncoprotein RNF43 functions in an autocrine manner in colorectal cancer. Int J Oncol. 2004;25:1343–8. [PubMed] [Google Scholar]
- 39.Nishizuka S, Chen S, Gwadry F, Alexander J, Major S, Scherf U, et al. Diagnostic markers that distinguish colon and ovarian adenocarcinomas: identification by genomic, proteomic, and tissue array profiling. Cancer Res. 2003;63(17):5243–50. [PubMed] [Google Scholar]
- 40.Kahlert C, Klupp F, Brand K, Lasitschka F, Diederichs S, Kirchberg J, et al. Invasion front-specific expression and prognostic significance of microRNA in colorectal liver metastases. Cancer Sci. 2011 doi: 10.1111/j.1349-7006.2011.02023.x. [DOI] [PubMed] [Google Scholar]
- 41.Karaayvaz M, Pal T, Song B, Zhang C, Georgakopoulos P, Mehmood S, et al. Prognostic Significance of miR-215 in Colon Cancer. Clin Colorectal Cancer. 2011 doi: 10.1016/j.clcc.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dees EC, Rowinsky EK, Noe DA, O’Reilly S, Adjei AA, Elza-Brown K, et al. A phase I and pharmacologic study of pyrazoloacridine and cisplatin in patients with advanced cancer. Invest New Drugs. 2003;21:75–84. doi: 10.1023/a:1022924511602. [DOI] [PubMed] [Google Scholar]
- 43.Yoon J, Koo KH, Choi KY. MEK1/2 inhibitors AS703026 and AZD6244 may be potential therapies for KRAS mutated colorectal cancer that is resistant to EGFR monoclonal antibody therapy. Cancer Res. 2011;71:445–53. doi: 10.1158/0008-5472.CAN-10-3058. [DOI] [PubMed] [Google Scholar]
- 44.Yoshino T, Yamazaki K, Hamaguchi T, Shimada Y, Kato K, Yasui H, et al. Phase I Study of Sunitinib plus Modified FOLFOX6 in Japanese Patients with Treatment-naive Colorectal Cancer. Anticancer Res. 2012;32:973–9. [PubMed] [Google Scholar]
- 45.Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, et al. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of a mean graph and COMPARE algorithm. J Natl Cancer Inst. 1989;81:1088–92. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- 46.Levitzki A, Mishani E. Tyrphostins and other tyrosine kinase inhibitors. Annu Rev Biochem. 2006;75:93–109. doi: 10.1146/annurev.biochem.75.103004.142657. [DOI] [PubMed] [Google Scholar]