Abstract
Mutations were introduced to the domain-swapped homodimer of the antiviral lectin griffithsin (GRFT). Whereas several single and double mutants remained dimeric, insertion of either two or four amino acids at the dimerization interface resulted in a monomeric form of the protein (mGRFT). Monomeric character of the modified proteins was confirmed by sedimentation equilibrium ultracentrifugation and by their high resolution X-ray crystal structures, whereas their binding to carbohydrates was assessed by isothermal titration calorimetry. Cell-based antiviral activity assays utilizing different variants of mGRFT indicated that the monomeric form of the lectin had greatly reduced activity against HIV-1, suggesting that the antiviral activity of GRFT stems from crosslinking and aggregation of viral particles via multivalent interactions between GRFT and oligosaccharides present on HIV envelope glycoproteins. Atomic resolution crystal structure of a complex between mGRFT and nonamannoside revealed that a single mGRFT molecule binds to two different nonamannoside molecules through all three carbohydrate-binding sites present on the monomer.
Highlights
► Native griffithsin is a domain-swapped dimer with potent antiviral properties ► Several monomeric mutants of griffithsin were obtained by structure-guided design ► A HIV gp120-associated oligosaccharide was shown to crosslink molecules of griffithsin ► Dimeric structure and crosslinking are necessary for maintaining antiviral activity
Introduction
A variety of infections caused by enveloped viruses are currently the subject of extensive research aimed at discovery and development of preventive and therapeutic antiviral agents. In particular, infections caused by viruses such as HIV, SARS-associated corona virus, ebola hemorrhagic fever virus, and hepatitis virus are in need of novel ways of prevention and therapy. One of the promising, although not well-developed strategies for combating these viral infections is inhibiting viral entry with lectins such as cyanovirin-N (CV-N), scytovirin (SVN), or griffithsin (GRFT) (Balzarini, 2007). GRFT, originally isolated from extracts of red alga Griffithsia sp., has demonstrated antiviral activity against both HIV (Mori et al., 2005) and SARS (O'Keefe et al., 2010, Zeitlin et al., 2009). It has the most potent activity of all known lectins against various HIV-1 isolates, with EC50 in the midpicomolar range. Furthermore, GRFT has been shown to be amenable to large-scale, agricultural manufacturing (O'Keefe et al., 2009). The stability and low immunogenicity of GRFT make it a candidate for formulation as a component of vaginally or rectally applicable microbicides (Zeitlin et al., 2009).
Earlier work in our laboratories has resulted in elucidation of the structural, biophysical, and biochemical properties of GRFT, along with its mode of binding to several mono- and disaccharides (Giomarelli et al., 2006, Mori et al., 2005, Ziólkowska et al., 2006, Ziólkowska et al., 2007b). However, we were not able to prepare soluble complexes of GRFT with branched carbohydrates corresponding to those present on viral envelopes, such as a synthetic nonamannoside or a slightly larger Man9 (Figure 1 ), since such complexes precipitated from solution even at very low concentration. For that reason, previously we used molecular modeling to derive the putative structure of the GRFT-Man9 complex (Ziólkowska et al., 2007a). GRFT is a domain-swapped homodimer with the first 16 amino acids of one monomer completing the β-prism-I fold of the other monomer (Ziólkowska et al., 2006). Each monomer comprises one binding surface at opposite ends of a double-prism homodimer, and each binding surface is composed of three carbohydrate-binding pockets, for a total of six binding pockets per GRFT dimer. Still to be answered are questions about the exact mechanism for the antiviral activity of GRFT and its mode of binding to high-mannose, branched carbohydrates. Logical models for the mechanism of GRFT's activity include (i) coating of viral particles and blocking interaction with cell surface receptors, (ii) inhibiting viral entry postattachment, (iii) aggregation/agglutination of viral envelope glycoproteins or particles, and (iv) enhancement of the cellular immune response. One approach to finding answers to these questions appeared to be through engineering, expression, and purification of a monomeric form of GRFT (here called mGRFT). The availability of such a protein was expected to allow us to directly address the role of multivalency and crosslinking in GRFT binding to carbohydrates and to elucidate how these binding interactions relate to antiviral activity. It was also anticipated that an antiviral protein of reduced size might exhibit potentially more advantageous physiological attributes.
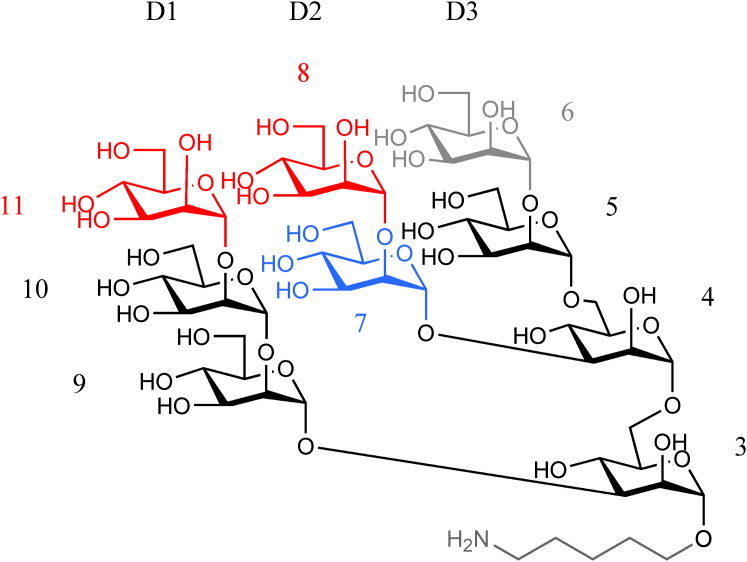
Figure 1.
Chemical Structure of the Nonamannoside Used in This Study
The three branches are labeled D1–D3, and the individual mannose moieties are numbered in a manner consistent with the nomenclature used for Man9 (Man9GlcNAc2) in the earlier studies of GRFT binding to branched carbohydrates (Ziólkowska et al., 2007a). Man9 differs from the nonamannoside by replacement of the pentyl chain by two N-acetyl glucosamines connected to mannose 3 by a β1-4 linkage. Carbohydrate residues that bind to one molecule of mGRFT are red, the residue bound to its symmetry mate is blue, and the atoms not visible in the electron density map of the complex of the nonamannoside with 1GS-S are gray.
We have engineered several variants of mGRFT and studied their physical properties using ultracentrifugation, isothermal titration calorimetry, and X-ray crystallography, as well as investigated their biological activity using anti-HIV infectivity assays. We found that the monomers were fully capable of binding carbohydrates, but were less stable thermodynamically than GRFT and had greatly diminished antiviral activity. The latter result provides a clue to the mechanism responsible for the antiviral activity of GRFT.
Results
Attempts to Create mGRFT by Mutation of Amino Acids
Analysis of the previously solved X-ray crystal structures of GRFT (Ziólkowska et al., 2006, Ziólkowska et al., 2007b) pointed to several possible approaches to engineering of mGRFT. Our initial efforts at creating monomeric GRFT were aimed at stabilizing its hinge region (residues 15–19) so as to allow the monomeric form to predominate, and on removing the hydrogen bonded interactions with the N-terminal peptide. We have thus created a series of mutants, starting from the replacement of Ser65 (which makes a hydrogen bond with the amide group of Gly8) by an aromatic residue, such as tyrosine or tryptophan. Additional mutations were made at positions 2 (L2V), 6 (K6V), 106 (S106E), and 119 (E119I), both singly and in combination with the mutation S65Y. None of these initial attempts resulted in the formation of monomeric GRFT and the resulting proteins retained their dimeric character.
Creation of mGRFT by Insertion of Residues into the Hinge Region (1GS and 2GS)
Another approach to creation of monomeric GRFT relied on the detailed analysis of the region responsible for domain swapping. The dimerization interface of GRFT consists of a “hinge” formed by the swapped strands (containing Ser16 and Gly17), as well as a “lock” created by hydrophobic interactions of Leu2, Ile101, and Tyr117 (Figure 2A). Furthermore, structures of GRFT in complex with maltose and N-acetylglucosamine (Ziólkowska et al., 2007b) contain an ethylene glycol molecule spanning the 7.4 Å gap between the swapped strands at the dimerization interface. We hypothesized that the gap between the swapped strands can be bridged by insertion of two or more residues between Ser16 and Gly17, at least one being a serine (to provide polar interactions similar to those of ethylene glycol). Additionally, sequences containing GS are commonly found in natural linkers (Lubkowski et al., 1999); thus, we selected either one or two of these dipeptides in designing mGRFT.
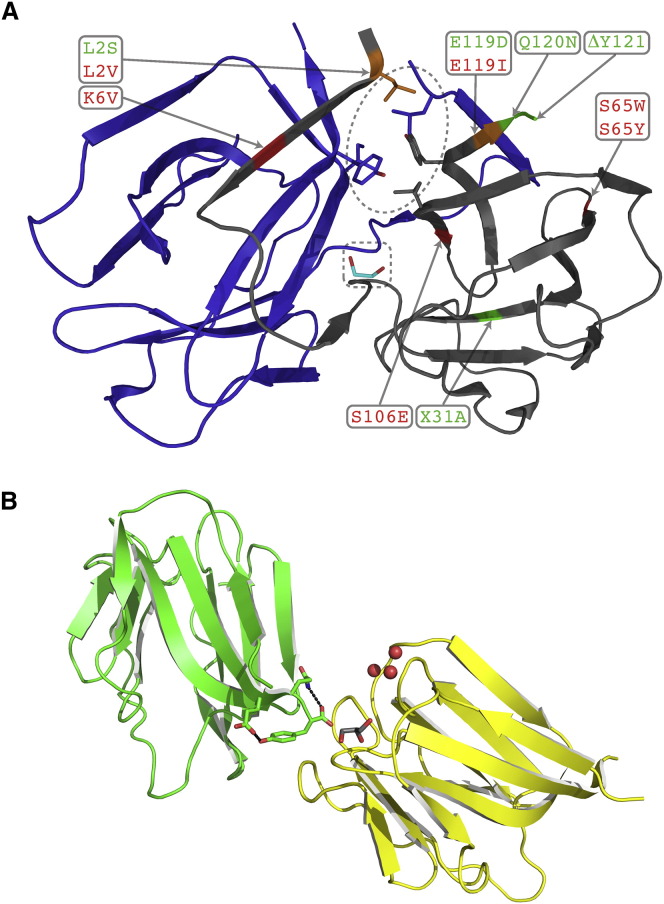
Figure 2.
Structural Features of griffithsin
(A) Dimerization interface of GRFT. Individual monomers are shown in blue and gray. The hinge region of the swapped strands is shown in the dashed rectangle with the glycerol molecule seen in two structures colored in cyan. The lock region is shown in the dashed oval with the relevant hydrophobic residues (L2, I101, and Y117) from both chains in stick configuration. All mutations other than the insertions at the hinge region relevant to this study are mapped onto the gray monomer. Successful and unsuccessful mutants are colored green and red, respectively, on the chain and labels. Positions that yielded mixed results are colored orange on the chain.
(B) Carbohydrate binding surface of mGRFT (1GS). The relative orientation and interaction of two symmetry related molecules (green and yellow) are shown with the C-terminal residues of one monomer in stick configuration. Hydrogen bonds that stabilize Y121 are shown in black dashed lines. Water (red sphere) and glycerol (gray) molecules in adjacent carbohydrate binding pockets are shown for the yellow molecule.
Both constructs, here called 1GS and 2GS, were successfully expressed, purified, and crystallized. Their predominantly monomeric nature at lower concentrations was initially confirmed with analytical ultracentrifugation. The dissociation constants of 1GS and 2GS calculated by sedimentation equilibrium ultracentrifugation (SEU) were 0.12 and 1.17 mM, respectively. The weight-averaged molecular mass obtained using a single-species model was 15.96 ± 0.05 kDa and 15.11 ± 0.08 kDa for the 1GS and 2GS mutants, respectively. These data can be interpreted as showing limited self-association of monomers that does not involve domain swapping.
The 1GS protein crystallized in the space group P61 with a monomer in the asymmetric unit and its structure was solved at the resolution of 1.1 Å (Table 1 ). Each of the three carbohydrate binding pockets of this form of mGRFT was occupied by molecules present in the crystallization and cryoprotection solutions. Pocket 1 (Ziólkowska et al., 2006), adjacent to Asp112, contained a glycerol molecule. Pocket 2, near Asp30, was filled by the terminal carboxylate group belonging to a symmetry-related molecule (Figure 2B), whereas pocket 3, near Asp70, contained two water molecules. The 2GS mGRFT crystallized in the space group P6522 and the structure was solved at 1.7 Å (Table 1). Each of the potential carbohydrate-binding pockets contained a glycerol molecule. Electron density for the inserted sequences on both of these mGRFT molecules were clearly seen in the |Fo|-|Fc| maps (Figure 3 ). As predicted, these insertions led to formation of loops, allowing the first 16 amino acids to complete the β-prism fold of the same molecule.
Table 1.
Data Collection and Refinement Statistics
| 1GS | 2GS | 1GS-SDNΔY | 1GS-Sm9 | |
|---|---|---|---|---|
| Data Collection | ||||
| Space group | P61 | P6522 | P3121 | I41 |
| Cell dimensions | ||||
| a, b, c (Å) | 48.6, 48.6, 94.2 | 55.1, 55.1, 156.7 | 55.4, 55.4, 61.8 | 90.5, 90.5, 27.8 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 30–1.10 (1.12–1.10) | 30–1.70 (1.76–1.70) | 30–0.97 (1.00–0.97) | 30–0.97 (1.00–0.97) |
| Rmerge | 6.3 (58.3) | 9.0 (69.0) | 8.2 (78.0) | 5.5 (50.7) |
| I / σI | 28.8 (2.0) | 24.4 (2.1) | 24.2 (2.0) | 27.6 (2.0) |
| Completeness (%) | 97.9 (99.7) | 97.9 (93.5) | 99.9 (99.4) | 99.8 (98.4) |
| Redundancy | 6.2 (3.5) | 11.4 (5.7) | 7.1 (5.2) | 4.8 (3.4) |
| Refinement | ||||
| Resolution (Å) | 30-1.11 | 30-1.70 | 30-0.97 | 30-0.97 |
| No. reflections | 47848 | 15073 | 64277 | 65888 |
| Rwork/Rfree | 15.87/15.90 | 18.39/21.02 | 15.16/16.50 | 14.34/15.46 |
| No. atoms | ||||
| Protein | 942 | 934 | 1020 | 933 |
| Ligand/ion | 41 | 29 | 1 | 122 |
| Water | 117 | 135 | 146 | 165 |
| B-factors | ||||
| Protein | 14.73 | 18.42 | 12.57 | 12.40 |
| Ligand/ion | 23.12 | 25.90 | 10.23 | 19.68 |
| Water | 31.11 | 31.10 | 25.47 | 27.99 |
| Rmsd | ||||
| Bond lengths (Å) | 0.020 | 0.020 | 0.018 | 0.020 |
| Bond angles (°) | 1.967 | 1.875 | 2.304 | 2.579 |
| 3LKY | 3LL0 | 3LL1 | 3LL2 | |
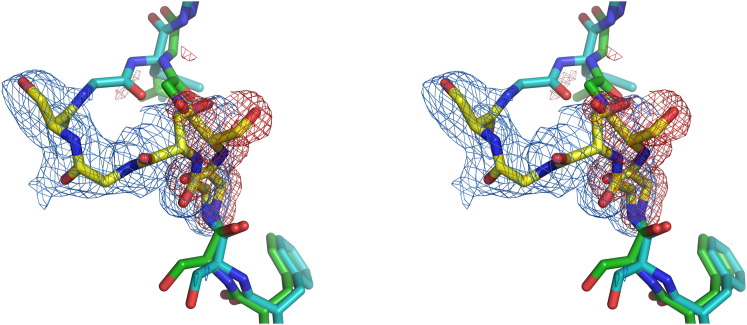
Figure 3.
Superposition of the Monomeric 1GS and 2GS Constructs of mGRFT, Showing the Area of the Dimerization/Insertion Region
Bonds involving carbon atoms of 1GS are rendered in green and those of 2GS in cyan, with both inserts in yellow. The omit maps, contoured at 2.5 σ, are red and blue for 1GS and 2GS, respectively.
Increasing Monomerization and Sensitivity to TEV Protease (1GS-S)
To facilitate monomerization we also reduced the interactions at the hydrophobic patch opposite to the strand swap region (the lock) by creating the L2S variant of the 1GS mGRFT (hereon called 1GS-S). An unexpected result of the L2S mutation was that the N terminus of mGRFT was dramatically more susceptible to cleavage by TEV protease (Phan et al., 2002) for the removal of the N-terminal hexahistadine affinity tag (Parks et al., 1994) (not shown). Because of the ease of its purification, this mutant was used for crystallization of the Man9 complex and for titration isothermal calorimetry studies. This mutant was uniformly monomeric with a weight-averaged molecular mass obtained for this mutant by SEU analysis of 11.58 ± 0.11 kDa.
Elimination of Residual Monomer-Monomer Interactions (1GS-SDNΔY)
Crystal packing of the 1GS variant of mGRFT was such that the C terminus of one molecule of mGRFT was bound into the carbohydrate binding pocket 2 of an adjacent mGRFT molecule (Figure 2B). In order to eliminate this interaction we mutated the C terminus of 1GS-S, changing its sequence from EQY to DN (hereon referred to as 1GS-SDNΔY). The stability of this mutant was reduced (see Table 2 available online) and its weight-averaged molecular mass appeared to be the lowest of all mGRFT mutants (11.03 ± 0.99 kDa). 1GS-SDNΔY crystallized in the space group P3121 and, although the crystals still diffracted to atomic resolution, showed a high degree of disorder in the carbohydrate-binding surface, with two binding pockets containing only water molecules and the third one incorporating the guanidinium moiety from Arg80 of a symmetry-related molecule, along with a chloride ion. The chloride ion mediates most of the interactions between the symmetry-related Arg80 and the binding pocket.
Table 2.
Comparison of the Thermodynamic Binding Parameters of GRFT and 1GS-S
| Affinity (μM) | ΔH (kcal/mol) | ΔG (kcal/mol) | TΔS (kcal/mol) | ||
|---|---|---|---|---|---|
| Mannosea | GRFT | 102.0 ± 13.0 | −0.081 ± 0.002 | −5.53 ± 0.08 | 5.54 ± 0.08 |
| 1→2α- mannobiose |
GRFT | 53.9 ± 6.6 | −0.173 ± 0.003 | −5.91 ± 0.07 | 5.74 ± 0.07 |
| 1GS-S | 55.9 ± 14.3 | −0.292 ± 0.012 | −5.80 ± 0.21 | 5.50 ± 0.21 | |
| 1→3α- mannobiose |
GRFT | 50.5 ± 8.3 | −0.169 ± 0.005 | −5.95 ± 0.10 | 5.76 ± 0.10 |
| 1GS-S | 46.1 ± 8.4 | −0.310 ± 0.008 | −5.91 ± 0.13 | 5.60 ± 0.13 | |
| 1→6α- mannobiosea |
GRFT | 83.3 ± 17.5 | −0.072 ± 0.002 | −5.65 ± 0.12 | 5.58 ± 0.12 |
| 1GS-S | 84.7 ± 13.7 | −0.202 ± 0.008 | −5.55 ± 0.09 | 5.35 ± 0.09 | |
| Mannopentaose | GRFT | 18.9 ± 2.8 | −1.20 ± 0.14 | −6.54 ± 0.88 | 5.35 ± 0.14 |
| 1GS-S | 15.1 ± 1.3 | −1.83 ± 0.05 | −6.57 ± 0.05 | 4.74 ± 0.05 | |
| Nonamannoside | GRFT | 0.50 ± 0.16 | −13.50 ± 0.11 | −8.73 ± 0.31 | −4.75 ± 0.31 |
| 1GS-S | 0.51 ± 0.22 | −11.70 ± 1.73 | −8.58 ± 0.56 | −3.08 ± 1.10 | |
| gp120a | GRFT | 0.008 ± 0.004 | −30.40 ± 0.26 | −11.20 ± 0.28 | −19.16 ± 0.28 |
| 1GS-S | 0.112 ± 0.050 | −35.50 ± 0.56 | −9.47 ± 0.68 | −25.99 ± 0.67 |
Previously published (O'Keefe et al., 2010, Ziólkowska et al., 2007b, Ziólkowska et al., 2007a).
Structure of 1GS-S in Complex with Nanomannoside (1GS-Sm9)
Crystallization trials of 1GS-S in the presence of nonamannoside resulted in the growth of crystals under a wide range of conditions. However, regardless of the crystallization conditions, all tested crystals were isomorphous in the tetragonal space group I41. Diffraction data were obtained at a resolution of 0.97 Å (Table 1). The electron density maps show very clearly that the complex was formed, delineating unambiguously the position of the D1 and D2 branches of the nonamannoside (Figure 4A) and of the two connecting β-mannoses. The density was partial and weak in the region where mannose 5 of the D3 branch would be expected. The binding mode of nonamannoside to 1GS-S is different in some important respects from our previous computational model (Ziólkowska et al., 2007a), or from the recently proposed mode of binding of Man9 to actinohivin, a lectin isolated from Longispora albida which is also presumed to contain three carbohydrate-binding pockets creating an equilateral triangle (Tanaka et al., 2009). The terminal mannose 11 of the D1 branch and mannose 8 of the D2 branch of the nonamannoside bind into two pockets of 1GS-S (pockets 3 and 1 according to the previously introduced nomenclature; Ziólkowska et al., 2006), with the rest of the nonamannoside molecule spanning an arch between these two binding pockets. In addition, binding pocket 2 is occupied by mannose 7 of the D2 branch of a symmetry-related nonamannoside molecule, and hence the overall ratio between polysaccharide and lectin is still one to one. The weight-averaged molecular mass of the nonamannoside-saturated complex was 16.73 ± 0.13 kDa, suggesting a two-to-one ratio of nonamannoside to 1GS-S in solution. This apparent stoichiometric ambiguity can be explained by the difference in the primary structure of the three binding pockets. Tyrosines 27, 68, and 110 form the internal walls of the mannose-binding pockets and create an almost perfectly symmetric triangle at the center of the binding surface. The preceding residues are Asp67 and Asp109 in pockets 3 and 1, respectively, whereas Ser26 forms part of pocket 2. This difference in primary structure leads to significant alteration of the electrostatic landscape of the carbohydrate-binding surface (Figure 4B). Whereas the surface potential is highly negative in pockets 1 and 3, it is much less negative in pocket 2. In addition, Asp67 and Asp109 are both within hydrogen bond distance from the nonamannoside, while Ser26 in pocket 2 is too far to contribute to binding. It is thus likely that pockets 1 and 3 offer better binding to the terminal mannoses of two branches of nonamannoside, while pocket 2 is occupied more transiently. It can be postulated that when this site becomes temporarily occupied by β-mannose 7 of a nonamannoside bound to another protein molecule, the resulting symmetric interactions lead to creation of a stable crystal lattice, which ultimately changes the apparent stoichiometry of the complexes. These results point out that the originally proposed model in which only the terminal mannoses of each branch bound to a single molecule of GRFT (Ziólkowska et al., 2007a) was most likely oversimplified, with the observed mode of crosslinking being only one of many that lead to prevention of the viral entry by this class of lectins.
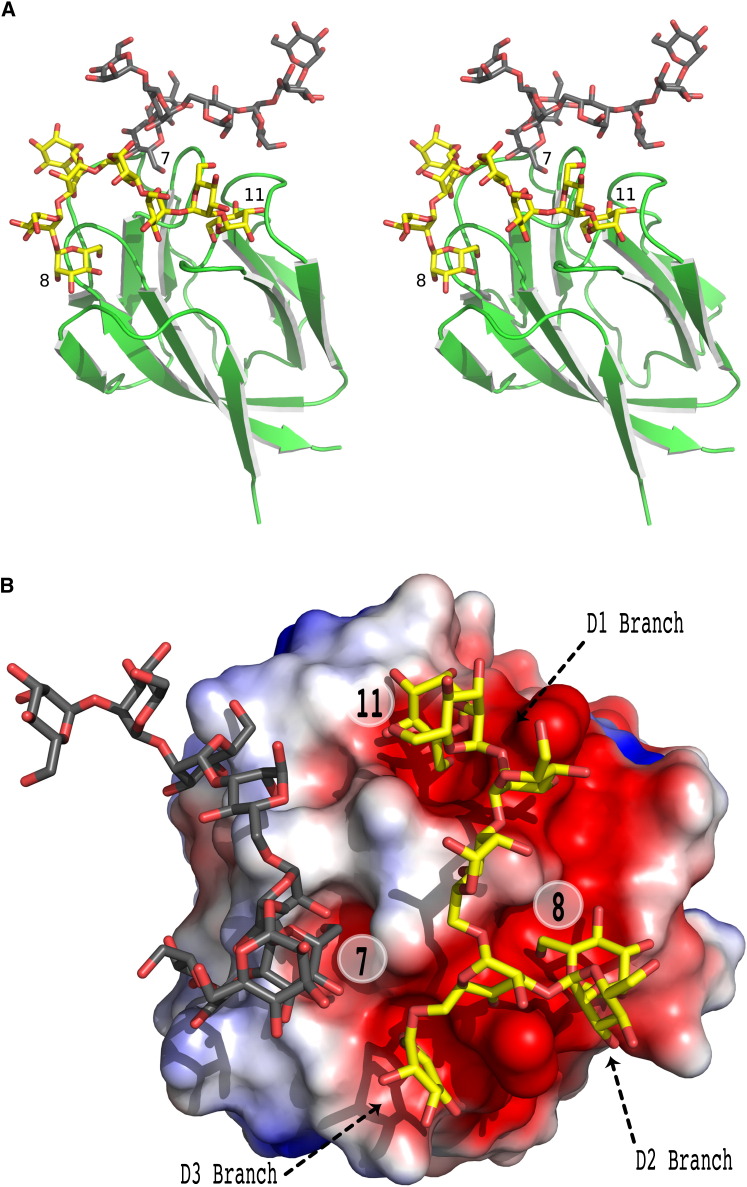
Figure 4.
Binding of Nonamannoside to the 1GS-S Construct of GRFT
(A) Stereo view of the complex of nonamannoside and 1GS-S. One molecule of nonamannoside (yellow) bridges carbohydrate-binding pockets 1 and 3, whereas a symmetry-related molecule of nonamannoside (gray) binds into pocket 2. The β-mannose monomers bound in the binding pockets are numbered as in Figure 1.
(B) Computed surface electrostatic potential of 1GS-S monomer (space filling representation) colored by charge from negative (red) to positive (blue), with bound nonamannosides shown as sticks. One nonamannoside molecule (yellow) binds to two carbohydrate-binding pockets, with negative electrostatic potential due principally to Asp67 and Asp109. The second nonamannoside molecule (gray) binds to the third binding pocket formed in part by Ser26 instead of Asp in the equivalent position and carrying a more positive electrostatic potential.
Differential Scanning Calorimetry
To further quantitate the GRFT monomerization process, the thermal stabilities of the GRFT monomer variants were studied using differential scanning calorimetry (DSC). Here, the heat-capacity change upon unfolding was monitored with respect to temperature, and the extrapolated midpoint of the transition, the melting temperature (Tm), and integrated area under the curve, the enthalpy of unfolding (ΔHunf), were obtained (Table 3 ). Dimeric GRFT melted at a Tm of 78.82°C, with an enthalpy of unfolding (ΔHunf) corresponding to 248 kcal/mol (Table 3). The 1GS mGRFT construct, where a GS sequence was introduced between Ser16 and Gly17, melted at a dramatically decreased Tm of 67.33°C and ΔHunf of 91 kcal/mol. Insertion of two GS sequences in the 2GS GRFT construct resulted in a similar unfolding as observed in the 1GS construct. The 2GS GRFT appeared to have greater conformational stability (ΔHunf = 125 kcal/mol), although it melted at a temperature about a degree less than its 1GS counterpart. The 1GS-S GRFT melted at an even further reduced Tm of 63.48°C and appeared to have an almost 2-fold greater structural stability (ΔHunf = 163 kcal/mol) compared with 1GS, suggesting that the L2S mutation increased the monomerization potential while stabilizing the overall domain structure of the protein (Table 3). Last, and in complete agreement with the analytical ultracentrifugation experiments, the elimination of residual monomer-monomer interactions in the 1GS-SDNΔY was corroborated in the DSC experiments, this protein melting at the lowest temperature (Tm = 51.34°C) of all the tested variants of mGRFT. The enthalpy of unfolding of 1GS-SDNΔY GRFT was integrated as 117 kcal/mol, and the ΔΔHunf = 46 kcal/mol, between this monomer variant and the 1GS-S mGRFT, pointed to possible intermolecular monomer-monomer contacts that stabilized the 1GS-S but were missing in the 1GS-SDNΔY construct.
Table 3.
Thermal Stability of GRFT and mGRFT Constructs as Determined by DSC
| Tm (°C) | ΔH (kcal/mol) | |
|---|---|---|
| GRFT | 78.82 ± 0.03 | 248 ± 2 |
| 1GS | 67.33 ± 0.06 | 91 ± 2 |
| 2GS | 66.42 ± 0.05 | 125 ± 2 |
| 1GS-S | 63.48 ± 0.06 | 163 ± 3 |
| 1GS-SDNΔY | 51.34 ± 0.04 | 117 ± 1 |
Isothermal Titration Calorimetry
We previously used isothermal titration calorimetry (ITC) to characterize the thermodynamics of binding of GRFT to mono- and disaccharides and showed that the protein preferentially binds to mannose and mannose-containing sugars (Ziólkowska et al., 2006, Ziólkowska et al., 2007b). We have extended this analysis here to study larger branched oligomannose polysaccharides to derive a more biologically relevant model for the antiviral activity of griffithsin. A panel of linear and branched oligomannose sugars was assembled and their binding to dimeric GRFT and to the 1GS-S mutant was examined and compared using isothermal titration calorimetry (ITC). The derived thermodynamic binding parameters are shown in Table 2.
In terms of energetics, all interactions observed between the sugars and the proteins were exothermic, indicating favorable binding contacts. In addition, entropically favored bindings (positive TΔS values) were mediated by all carbohydrates except for the nonamannoside, suggesting that the conformational entropy of the sugars (mannose, mannobioses, and mannopentaoses) and/or of the protein regions at the binding interface were not compromised during complex formation. Among the dimannoses, 1→6α mannobiose was the weakest GRFT binding partner with a Kd value almost twice that of the other disaccharides and with very slight enthalpy contributions. Unlike the 1→6α dimannose (Ziólkowska et al., 2007a), the 1→2α dimannose and 1→3α dimannose bound GRFT with a 2-fold better binding affinity as compared with mannose alone, pointing to the importance of proper presentation of the terminal mannoses in the tested sugars. The enthalpy values among the mannobioses also pointed to possible differences in complex formation between monomeric and dimeric forms of GRFT, the sugars appearing to mediate more contacts with the mGRFT protein (negative ΔH), although crystal structures of dGRFT (Ziólkowska et al., 2006) and mGRFT showed nearly identical integration of the sugar hydroxyl and carbon backbones within the protein's binding sites. Since in ITC experiments the binding interactions were studied in solution, naturally the results would report all global and dynamic effects of complex formation, the proteins and sugars freely tumbling and accessing varying conformational states of their binding partner. Thus, according to the calorimetric data with mGRFT, the sugars appeared to access more surface area contacts that were probably precluded in the dimer structure due to tumbling or steric effects. However, these slight differences in binding enthalpy did not amount to a corresponding difference in binding affinity, suggesting that the increased surface area contacts with mGRFT were nonspecific in nature, and indeed the monomeric and dimeric forms of the protein exhibited marked similarities in binding to any particular sugar (Table 2).
As seen in Table 2, a significant shift in affinity (and enthalpy) occurred when the GRFT proteins were titrated with branched oligomannoses (mannopentaose and nonamannoside). Compared with the dimannose titration experiments, titrations of GRFT and mGRFT with mannopentaose resulted in a 3-fold increase in affinity, presumably due to the three terminal nonreducing mannoses of the mannopentaose. However, the 6-fold increase in enthalpy could not be accounted for by a simple additive enthalpic contribution of the exposed terminal mannoses in the mannopentaose (ΔHmannopentaose > > 2ΔHα1,3 dimannose + ΔHα1,6 dimannose), suggesting that chemically equivalent nonreducing mannoses in the context of a branched sugar structure (mannopentaose) yielded better binding contacts and better binding affinity with GRFT as compared with the linear dimannoses. In the absence of a crystal structure of this branched sugar with GRFT, it is unclear how these terminal mannoses occupy the sugar binding sites. However, from examining the entropy of binding it was apparent that crosslinking of proteins by the mannopentaose did not occur, as this thermodynamic parameter remained favorable (Table 2, positive TΔS). Similar to the dimannoses, the mannopentaose appeared to mediate more contacts with the mGRFT protein (negative ΔH) than with dGRFT.
In the nonamannoside titrations of GRFT and mGRFT, more than a 100-fold increase in affinity over the dimannoses, and more than a 30-fold increase in affinity over the mannopentaose was measured (Table 2). In terms of energetics, the bindings were largely enthalpically driven (negative ΔH; ΔG = ΔH – TΔS), with 50-fold increase in enthalpy over the dimannoses, and an approximate 10-fold increase in enthalpy over the mannopentaose. Such an increase in enthalpy indicated the presence of numerous favorable contacts between the sugar and the protein, in addition to those mediated by the sugar-binding pockets. This was suggested by the observed crystallographic results (Figure 4) that showed how a single nonamannoside molecule spanned an arch of ∼15 Å on 1GS-S mGRFT. The expanse of protein surface area bridged by the sugar could have allowed the two binding partners to come close enough together to mediate multiple, short-distance contacts to account for the large increase in enthalpy observed in the calorimetry experiments. Moreover, the observed calorimetric stoichiometry of two nonamannosides bound per mGRFT (data not shown) indicated multisite binding and additionally supported analytical ultracentrifugation experiments which showed a weighted average of 16.73 kDa for the nonamannoside-mGRFT complex and which suggested that, on average and in solution, two molecules of nonamannoside were bound to one molecule of 1GS-S mGRFT. Where the binding entropy of the smaller sugars was favorable, the unfavorable binding entropy (negative TΔS) of nonamannoside-GRFT complex strongly suggested that nonamannoside acted as a multivalent ligand, binding separate protein molecules and greatly limiting the conformational freedoms of both binding partners as well as potentially trapping more water molecules and further reducing the entropy of the system. The calorimetry data, taken together with the crystallographic structures and the analytical ultracentrifugation data, indicated that the multisite binding by GRFT and the multivalent binding mediated by the nonamannoside contributed to the tight affinity/avidity of the binding system. The avidity of the multisite, multivalent interaction between GRFT and nonamannoside was also seen in dynamic light scattering experiments (data not shown) where the percent polydispersity indicated the presence of multiple species ranging from monomer to higher combinations of the protein and oligosaccharide.
The ability of the mGRFT and dGRFT molecules to mediate multisite, multivalent interactions with oligomannoses was next tested in experiments with HIV-1 glycoprotein gp120, which contains multiple high-mannose oligosaccharides believed to be the recognition elements of antiviral proteins such as griffithsin and cyanovirin-N. Previously, the antiviral lectin cyanovirin-N (CV-N), which has two carbohydrate-binding domains, has been shown to be able to simultaneously bind to both HIV gp120 and gp41 (O'Keefe et al., 2000). This interaction was later shown to be due to specific multivalent interactions between CV-N and high mannose oligosaccharides on the surface of HIV envelope glycoproteins (Shenoy et al., 2001). Thus, it was reasonable to investigate the effect that multivalency might play in the carbohydrate-binding and biological activity of both mGRFT (with three unidirectional binding sites) and dGRFT (with a bidirectional composition of six binding sites) with gp120. As can be seen from the results of the ITC experiments (Table 2), dGRFT shows approximately 15-fold greater affinity for gp120 than mGRFT. This difference in measured affinity is likely more related to a change in avidity resulting from the decreased off-rate of dGRFT (which caused significant precipitation of gp120 in solution) as compared with mGRFT (which did not cause similar precipitates). This result was additionally supported by dynamic light scattering experiments (data not shown) which showed the dGRFT-gp120 complex to be fully precipitated, whereas the mGRFT-gp120 complex to be in a fluid, transitory, and fully soluble system composed of a number of differently sized species. Hence, the reduction in crosslinking of HIV envelope glycoproteins by the more limited carbohydrate-binding interface of mGRFT, as evidenced by the lack of precipitates, resulted in a significant reduction in the measured affinity/avidity of this molecule for gp120. The functional relevance of this lack of crosslinking on antiviral activity was further investigated via assessment of the anti-HIV activity of the various GRFT mutants.
Cell-Based Anti-HIV Activity Assay
To assess the antiviral activity of the various mutant forms of GRFT, we utilized a previously described assay that measures the cytopathic effect of HIV infection on T-lymphoblastic cells (Gulakowski et al., 1991). Nine different mutant forms of griffithsin were tested for their activity in protecting CEM-SS cells from HIV. As can be seen in Table 4 , dGRFT was potently active against HIV with and EC50 < 0.2 nM. Several additional mutants made in the effort to design an obligate monomeric from of GRFT were also tested. The mutant forms of GRFT which did not form monomers (S65W, S65W/S106E, E119I/S65Y, K6V/S65Y, L2V/S65Y) retained potent anti-HIV activity with EC50 values below 1 nM. However, all monomeric forms of GRFT exhibited severely diminished anti-HIV activity. Interestingly, such activity seems to be directly correlated to the thermal stability of the mutants and inversely correlated to their propensity to self-aggregate (Table 3, Table 4). HIV-1 gp120 has been estimated to bear 24 N-linked oligosaccharides of which 11 are high mannose (Zhang et al., 2004). As our ITC results have shown that the binding affinity of GRFT and mGRFT for branched high-mannose oligosaccharides are essentially identical, the antiviral results argue for the conclusion that it is the crosslinking of these oligosaccharides on individual or close neighbor gp120 molecules that results in the striking antiviral activity displayed by GRFT.
Table 4.
Anti-HIV Activity of GRFT Mutants
| Sample | EC50 (nM) | Form |
|---|---|---|
| GRFT | <0.2 | Dimer |
| S65W | <0.3 | Dimer |
| S65Y/S106E | <0.2 | Dimer |
| S65Y/E119I | 0.5 | Dimer |
| S65Y/K6V | 0.4 | Dimer |
| S65Y/L2V | 0.5 | Dimer |
| 1GS | 7 | Monomer |
| 2GS | 17 | Monomer |
| 1GS-S | 323 | Monomer |
| 1GS-SDNΔY | 305 | Monomer |
Discussion
Analysis of the previously published crystal structures of GRFT (Ziólkowska et al., 2006, Ziólkowska et al., 2007a, Ziólkowska et al., 2007b) has led us to a conclusion that this protein should be considered as “a candidate for domain swapping,” as defined by Schlunegger et al. (1997). Unlike in the case of true domain-swapped proteins, there was no previous indication of the existence of a monomeric form of this lectin, although the general fold of GRFT is the same as of the members of jacalin family (Bourne et al., 2002). Whereas jacalin and related proteins have been shown to form oligomers, none of them make domain-swapped dimers (Ziólkowska and Wlodawer, 2006). However, the reported dimeric nature of banana lectin (Meagher et al., 2005, Singh et al., 2005) may be related to its comparatively high antiviral activity (Swanson et al., 2010). It has been also pointed out that other known antiviral lectins, such as CV-N (Bewley et al., 1998) and SVN (Moulaei et al., 2007), form at least quasidimers (Botos and Wlodawer, 2005). Domain-swapped dimers and a variety of multimeric forms have been reported for the members of the structurally unrelated β-prism-II family (Chandra et al., 1999) and served as a basis of evaluating multivalency of binding to carbohydrates, using molecular modeling (Ramachandraiah et al., 2003). Creation of a monomeric form of GRFT was considered by us as a step in the direction of correlating the quaternary structure of the antiviral lectins with their biological activity, but the successful engineering of mGRFT also yielded an unexpected benefit of allowing a crystallographic study of a complex of this protein with the branched carbohydrate nonamannoside. The latter results form the basis of a proposed model that explains the role of multivalency of carbohydrate binding on the antiviral activity of β-prism-I family.
Initial attempts at forming stable monomers of GRFT centered on stabilizing the hinge region in GRFT that provides the flexibility necessary for domain swapping, as well as on preventing hydrogen bonding between the N-terminal peptide and the body of the protein. These mutants, involving aromatic substitutions of Ser65, and sometimes additional 1–2 amino acids, did not have the desired effect and thus an alternative approach was necessary. A clue to the best way of designing mGRFT was provided by the presence of a glycol molecule in the vicinity of residues 16 and 17′ (Ziólkowska et al., 2007b), suggesting a path for their connection. Similarly to the case of an engineered Cro monomer (Albright et al., 1996), it appeared that insertion of two or more residues into the hinge region might prevent domain swapping and result in a monomeric form of GRFT. Whereas utilization of a string consisting of five different residues (DGEVK) led to engineered monomeric Cro, it was sufficient for GRFT to insert only one or two repeats of the sequence GS, frequently found in linking regions of proteins due to its flexibility (Lubkowski et al., 1999).
Both the 1GS and 2GS constructs yielded stable lectins, with almost identical thermal stability as judged by their melting temperatures of ∼67°C (Table 3). Interestingly, both constructs were less stable than the dimeric GRFT, and several additional mutations destabilized them even further. The most telling of these additional mutations were the 1GS-S and 1GS-SDNΔY forms that resulted in melting temperature of ∼63 and ∼51°C, respectively, a loss of up to 27° of thermal stability compared with GRFT. The results of the DSC studies suggest that the mutations necessary to form true monomeric GRFT result, in this instance, in a less stable protein. These studies were followed up with experiments designed to assess the effect of nonamannoside binding on the stability of both GRFT and various mGRFT mutants. As shown in Table 3, although GRFT shows no change in stability coincident with carbohydrate binding, mGRFT showed a definite increase in thermal stability upon binding to nonamannoside. With the overall reduced thermal stability of mGRFT compared with GRFT, the increase in melting temperature upon binding nonamannoside is suggestive of conformational changes leading to such enhanced stability.
Not surprisingly, in isothermal titration calorimetry experiments both the monomeric and dimeric forms of GRFT exhibited similar affinity to disaccharides, pentamannoside, and virtually identical avidity of binding to the branched oligosaccharide nonamannoside. The progression of both binding enthalpies and dissociation constants indicates that both GRFT and mGRFT share similar binding interfaces to carbohydrates. What is telling, however, is the significant increase in the strength of the binding interaction when one moves from likely single-site binding interactions (i.e., mannobioses) to those that potentially involve multiple binding sites on individual GRFT monomers (e.g., pentamannoside and, to a greater degree, nonamannoside). Despite the similarity of their interactions with isolated oligosaccharides, the binding of GRFT and mGRFT to the viral envelope protein gp120 differed significantly, with the avidity of mGRFT being approximately 15-fold lower. Another major difference was observed in their antiviral properties, with GRFT exhibiting EC50 of <0.2 nM, whereas the activity of mGRFT was reduced by three orders of magnitude under similar conditions (EC50 of 323 nM). This observation is in agreement with the results reported for the monomeric and dimeric forms of CV-N, where the presence of at least two binding sites, whether located on a single molecule or on two molecules, was shown to be necessary to for their antiviral properties (Liu et al., 2009). Furthermore, the activity of CV-N mutants that were inactive as monomers was restored by their dimerization (Matei et al., 2010). However, another quasidimeric lectin, SVN, behaves in a different way, since even just its N-terminal domain (SD1) exhibits significant antiviral properties (Xiong et al., 2006). It should be noted that several other mutants of GRFT created for this study did not show either decreased antiviral activity or true monomeric character. The antiviral assay and thermodynamic results described here detail the close association of multivalent interactions between dGRFT and gp120 with its antiviral activity. The significant reduction in the affinity of mGRFT for gp120 is mirrored by its drastic reduction in antiviral activity. The results showing that both dGRFT and mGRFT bind with similar enthalpies and dissociation constants to the Man9 analog nonamannoside indicate that the interactions between individual high mannose oligosaccharide moieties and individual monomeric units of GRFT are not sufficient to engender significant antiviral activity. It is only in the crosslinking of multiple high-mannose oligosaccharides present on gp120 when the carbohydrate-binding capabilities of lectins such as GRFT confer an antiviral effect. These results further explain our previous results which detailed the large difference in anti-HIV EC50 values for a number of monovalent and multivalent monosaccharide-specific lectins (Ziólkowska et al., 2007b).
The ability to grow crystals of the complex of mGRFT with nonamannoside provides an indirect evidence of the most likely mode of the antiviral activity of GRFT. All previous attempts to grow crystals of the complexes of GRFT with Man9 or nonamannoside failed due to precipitation of the material from solution (T.M. and A.W., unpublished data). This was the case even when both components were mixed at very low concentration and then concentrated. These results indicate that the activity of GRFT might be compared with that of an antibody, which requires the presence of dimers in order to crosslink the antigens. Whereas the absence of such crosslinking would not influence carbohydrate binding, it would abolish the ability to interfere with viral infection. A similar explanation of the basis of antiviral activity was also recently proposed for CV-N (Matei et al., 2010).
The mode of binding of nonamannoside to mGRFT seen in the crystals was quite unexpected and did not correspond fully to the previously published model of their interactions (Ziólkowska et al., 2007a). The model structure assumed that the three terminal mannoses of branches D1-D3 of the nonamannoside would each occupy sites 1–3 on a GRFT monomer (as defined by Ziólkowska et al., 2006), whereas the remaining mannoses would participate in only limited or no interactions. In agreement with that model, we found that in the crystals of mGRFT one nonamannoside molecule inserts its mannose 11 from the D1 branch into site 3 on the protein, whereas mannose 8, terminal in branch D2, is located in site 1. However, it is mannose 7, the second unit on the D2 branch from a symmetry-related nonamannoside molecule, that is located in site 2 of mGRFT. It is very likely that this mode of binding is a crystallization artifact, which, incidentally, must have helped in the growth of that particular crystal form of the complex. However, this fortuitous mode of binding may actually be used to explain why GRFT, at least in its dimeric form, possesses strong antiviral activity, whereas jacalin (with one binding site per monomer), or β-prism II lectins such as garlic lectin, despite their three sites, do not. A major difference between GRFT and garlic lectin is the distance between carbohydrate-binding sites on a monomer, ∼15 Å for the former and a minimum of 22 Å for the latter. Thus, a single molecule of nonamannoside cannot reach more than one site on the garlic lectin (Ramachandraiah et al., 2003) but can easily reach two or three on GRFT (Ziólkowska et al., 2007a). If all three GRFT sites were occupied by the terminal residues of a single nonamannoside molecule, there would not be any more branches left to provide crosslinking. However, with two sites that are bound to terminal mannoses in the D1 and D2 branch offering different electrostatic properties than the third one (Figure 4B), the latter (and presumably weaker) site may provide an anchoring point for other nonamannosides and thus initiate crosslinking. The presence of two, almost identical, sites that can be reached by a single molecule vastly increases the avidity of binding, whereas the third site enhances crosslinking. Such organization is unique to GRFT (other lectins mentioned above either have fewer sites, or the existing sites cannot be spanned by a single complex carbohydrate molecule). With dimers allowing much more elaborate crosslinking patterns than monomers, their antiviral activity is enhanced. While highly likely, this interpretation will still require further verification through mutagenesis studies that would involve modification of the individual binding sites.
Experimental Procedures
Mutagenesis and Protein Expression
All restriction enzymes where purchased from New England Biolabs. All chemicals were obtained from American Bioanalytical, unless otherwise stated. An expression vector containing the gene for GRFT downstream of a hexahistidine tag and thrombin cleavage site (Giomarelli et al., 2006) was used as a PCR template. The amplified cassette was cloned into pET-15b expression vector in between the NcoI and XhoI sites. Standard site-directed mutagenesis technique was used to mutate the thrombin cleavage sequence to TEV cleavage sequence and to create all mutant forms of GRFT using mutagenic primers (IDT), PfuUltra DNA polymerase (Stratagene), and DpnI (NEB). All mutants were expressed in BL21(DE3) pLysS cells (Novagen) at 37°C and 226 rpm. Induction was initiated at optical density of 0.7 measured at 600 nm by addition of IPTG to a final concentration of 0.5 μM and continued for 10 hr.
Protein Purification
Cell pellets were lysed in 50 mM Tris (pH 8.0), 500 mM NaCl, and 5% v/v BugBuster 10× Protein Extraction Reagent (Merck), and the lysate was centrifuged at 17,000 rpm in a SS-34 rotor for 30 min. The supernatant was loaded onto a Ni-NTA Superflow column (QIAGEN), equilibrated with five column volumes of buffer A (20 mM Tris [pH 8.0], 100 mM NaCl). The column was washed with buffer A and eluted with buffer A containing 250 mM imidazole. TEV protease was added in 1:100 molar ratio of protease to eluted protein and the sample was incubated at room temperature for 1 hr. At this stage, two different strategies were used to complete the purification. For 1GS and 2GS proteins, the sample was loaded onto an amylose column equilibrated with buffer A. The column was washed with buffer A and eluted with buffer A containing 250 mM maltose. For 1GS-S and 1GS-SDNΔY proteins, the sample was heated at 50°C for 10 min and then centrifuged at 12,000 rpm in a SS-34 rotor for 10 min. The supernatant was loaded onto a Ni-NTA column (QIAGEN) equilibrated with buffer A and the flowthrough was collected. For all versions of mGRFT, the purified protein was dialyzed against deionized, double-distilled water overnight at 4°C, and then concentrated.
Crystallization, Data Collection, and Refinement
All proteins were concentrated to 25 mg/ml. To create the complex with nonamanoside, the carbohydrate was dissolved in water to a concentration of 10 μM and then titrated into a 1 mg/ml sample of 1GS-S, 2 μl at a time, followed by gentle mixing. The final molar ratio of nonamanoside to 1GS-S was 1.5:1. The complex was subsequently concentrated to 25 mg/ml. Initial screens for crystallization conditions were performed using a Phoenix liquid handling robot (Art Robbins Instruments) in 96-well Intelli-Plates (Hampton Research), with protein to well solution volumetric ratios of 1:2, 1:1, and, 2:1.
Initial crystallization conditions for the 1GS mutant were obtained from EasyXtal Ammonium Sulfate crystallization suite (QIAGEN). These conditions were optimized and diffraction quality crystals were grown in 0.1 M sodium acetate (pH 4.0), 0.15 M ammonium sulfate, and 25% w/v PEG 3350 at 20°C by hanging drop vapor diffusion method. The protein and well solution were mixed in 1:1 volumetric ratio. All other crystals of mGRFT mutants were obtained directly from crystallization suites, with 1:1 volumetric ratio of protein to well solution.
Crystals of the 2GS construct were grown in 0.1 M sodium acetate (pH 4.6), 0.1 M cadmium chloride, and 30% w/v PEG 400 (QIAGEN EasyXtal Classics Suite, #75). The 1GS-SDNΔY crystals were grown in 0.1 M citric acid (pH 3.5) and 3 M sodium chloride (QIAGEN EasyXtal Classics II, #7).
The 1GS-Sm9 crystallized under a variety of conditions from a number of crystallization suites. Nevertheless, all crystals were isomorphous in the space group I41. The highest resolution diffracting crystal was grown in 0.1 M mixture of imidazole, sodium cacodylate, MES, bis-tris (pH 6.5), 0.1 M mixture of L-Na-glutamate, alanine, glycine, lysine HCl and serine, and 30% w/v mixture of PEG MME 550 and PEG 20000 (Molecular Dimensions Morpheus, #85).
All crystals were soaked for 3 min in a cryoprotectant solution consisting of 4:1 volumetric ration of mother liquor to 80% v/v glycerol and rapidly cooled in a nitrogen stream (100 K). Data for the 2GS protein were collected with a MAR345 dtb image plate detector mounted on a Rigaku MicroMax-007HF generator operated at 40 kV and 30 mA. All other data were measured at the SER-CAT beamline 22-ID, at the Advanced Photon Source, on a MAR 300CCD detector. The atomic resolution data for 1GS-Sm9 were measured in two passes, with a low-resolution (30–3.0 Å) pass and a high-resolution (5.0–1.0 Å) pass. The measured intensities for both passes were scaled and merged together. All data were processed and scaled using the HKL3000 package (Minor et al., 2006). Initial phases were calculated by molecular replacement method using the program Phaser (McCoy et al., 2007) and a model derived from the structure of GRFT (PDB ID 2HYR), using residues 17–121 of chain A and 1–16 of chain B. The structures were refined with the program Refmac5 (Murshudov et al., 1997) and rebuilt with Coot (Emsley and Cowtan, 2004). Statistics of data processing and refinement are given in Table 1.
Isothermal Titration Calorimetry
ITC experiments were performed using a MicroCal VP-ITC microcalorimeter (MicroCal, Northampton, MA). The oligomannose sugars were diluted in the same exact buffer (20 mM Tris, 150 mM NaCl [pH 7.5]) used to dialyze the GRFT protein variants as well as gp120. The concentrations of the oligomannose sugars were determined by oligosaccharide composition analysis by HPAEC at the Complex Carbohydrate Research Center (CCRC, University of Georgia, Athens, GA). The concentrations of all proteins were assessed by amino acid analysis. In titrations with the oligomannose sugars, the protein solutions were in the range of 0.1–0.3 mM, and the sugars in the concentration range of 1–15 mM. In a typical titration experiment, the oligomannose sugar was placed in the syringe injector and the GRFT protein solution was placed in the calorimeter cell. In the experiments with HIV-1 glycoproteins, a protein solution of gp120 was placed in the calorimeter cell at a concentration of 1.0 μM, and the GRFT proteins were loaded into the syringe injector at a concentration of 0.175 mM. In all experiments, a total of 55 injections (5 μl per injection) were made into a rapidly mixing solution (300 rpm), with 600 s spacing between injections. The temperature was set at 25°C and the reference power to 7 μcal/s.
The heats of dilution of the oligomannoses were accounted for by blank titrations, where the oligomannoses were titrated into a buffer solution. After subtracting these baseline thermograms from experimental thermograms, the data were fit using Origin 5.0 nonlinear least-squares program supplied by the manufacturer, and values for the binding enthalpy (ΔH) and the dissociation constant (Kd) were derived.
Differential Scanning Calorimetry
DSC experiments were performed using a MicroCal VP-DSC microcalorimeter (MicroCal, Northampton, MA). The oligomannose sugars were diluted in the same exact buffer (20 mM Tris, 150 mM NaCl [pH 7.5]) used to dialyze the GRFT monomer variants. The concentrations of all proteins were assessed by amino acid analysis. In a typical experimental format, buffer (20 mM Tris, 150 mM NaCl [pH 7.5]) was placed in both the reference and sample cells and the calorimeter was allowed to cycle at a heating rate of 60°C/hr, scanning from 10°C to 90°C, with data recordings every 15 s. The calorimeter cells were self-pressurized to approximately 30 psi of positive pressure to prevent evaporation of solutions at the higher temperatures. During the down-scan at 25°C, the buffer solution in the sample cell was quickly and efficiently replaced by a GRFT protein solution (60 μM), positive pressure was reapplied, and the calorimetric cells were allowed to cycle between a total of six alternating up- and down-scans from 10°C to 90°C to measure reversibility of the unfolding process. Using Origin DSC Analysis software provided by the manufacturer, the melting profiles of all samples were corrected for buffer effects by subtracting the first up-scan of each experiment. To take into account the heat-capacity changes between the native and denatured states of the protein, a cubic baseline was extrapolated from pretransition and posttransition baselines and was subtracted from the melting profile. The resulting thermogram was fit to a two-state melting model and the calorimetric transition enthalpy (ΔHunf) was obtained from the numerical integration of the area under the excess heat-capacity peak, and the midpoint of the transition was extrapolated as the melting temperature.
Sedimentation Equilibrium Ultracentrifugation
Sedimentation equilibrium ultracentrifugation experiments were performed in a Beckman Optima XL-A ultracentrifuge at 20°C with absorbance optics at 280 nm. All protein samples were in a buffer containing 20 mM Tris (pH 8.0) and 100 mM NaCl. Samples were spun in a Beckman An-60 Ti rotor using 1.2 cm two-sector charcoal-fillled Epon centerpieces with quartz windows at speeds of 21,000, 25,000, and 30,000 rpm. Equilibrium was reached after 24 hr for each speed and verified by comparison with a second scan taken 2 hr later. All data were analyzed using the programs Sedfit and Sedphat (Schuck, 2000) with a buffer density of 1.00293 g/cm3 and a buffer viscosity of 0.01023 Poise as calculated using the program Sedenterp. Partial specific volumes were also calculated with Sednterp to be 0.7164 (1GS), 0.7155 (2GS), 0.7142 (1GS-S), and 0.7134 (1GS-SDNΔY) in units of g/cm3. All data were fit using the simplex algorithm.
Whole-Cell anti-HIV Assays
An XTT-tetrazolium based assay was used to determine the anti-HIV activity of GRFT against a T-tropic laboratory strain (HIV-1RF) in CEM-SS cells as previously described (Gulakowski et al., 1991). In brief, CEM-SS cells were maintained in RPMI 1640 media without phenol red and supplemented with 10% fetal bovine serum (BioWhittaker), 2 mM L-glutamine (BioWhittaker), and 50 μg/mL gentamicin (BioWhittaker). Exponentially growing cells were washed and resuspended in medium, and a 50 μl aliquot containing 5 × 103 cells was added to individual wells of a 96-well round-bottom microtiter plate containing serial dilutions of GRFT, GRFT mutants, or AZT in a volume of 100 μl medium. Stock supernatants of HIV-1RF were diluted in medium to yield sufficient cytopathicity (80%–90% cell kill in 6 days), and a 50 μl aliquot was added to appropriate wells. Plates were incubated for 6 days at 37°C, then stained for cellular viability using 2,3-bis-[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide inner salt (XTT).
Acknowledgments
We thank Chi-Huey Wong and Sheng-Kai Wang (Scripps) for the gift of the nonamannoside, Nikolay V. Dokholyan (University of North Carolina) for suggesting several mutations of GRFT, Nicole LaRonde-LeBlanc (University of Maryland) for her assistance with ultracentrifugation experiments and helpful discussions, and David Eisenberg (UCLA) for discussion of the phenomenon of domain swapping. We would also like to thank Jennifer Wilson (MTL, CCR, NCI) for anti-HIV bioassay support, and Marzena Dyba (SBL, SAIC-Frederick, CCR, NCI) and Sergei Tarasov (SBL, CCR, NCI) for their assistance with dynamic light scattering experiments. We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT), located at the Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and by the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health grant to A.W. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Published: September 7, 2010
Accession Numbers
Atomic coordinates and structure factors for 1GS, 2GS, 1GS-SDNΔY, and 1GS-Sm9 versions of GRFT have been deposited in the Protein Data Bank under accession codes 3LKY, 3LL0, 3LL1, and 3LL2, respectively.
References
- Albright R.A., Mossing M.C., Matthews B.W. High-resolution structure of an engineered Cro monomer shows changes in conformation relative to the native dimer. Biochemistry. 1996;35:735–742. doi: 10.1021/bi951958n. [DOI] [PubMed] [Google Scholar]
- Balzarini J. Carbohydrate-binding agents: a potential future cornerstone for the chemotherapy of enveloped viruses? Antivir. Chem. Chemother. 2007;18:1–11. doi: 10.1177/095632020701800101. [DOI] [PubMed] [Google Scholar]
- Bewley C.A., Gustafson K.R., Boyd M.R., Covell D.G., Bax A., Clore G.M., Gronenborn A.M. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat. Struct. Biol. 1998;5:571–578. doi: 10.1038/828. [DOI] [PubMed] [Google Scholar]
- Botos I., Wlodawer A. Proteins that bind high-mannose sugars of the HIV envelope. Prog. Biophys. Mol. Biol. 2005;88:233–282. doi: 10.1016/j.pbiomolbio.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Bourne Y., Astoul C.H., Zamboni V., Peumans W.J., Menu-Bouaouiche L., Van Damme E.J., Barre A., Rouge P. Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose. Biochem. J. 2002;364:173–180. doi: 10.1042/bj3640173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra N.R., Ramachandraiah G., Bachhawat K., Dam T.K., Surolia A., Vijayan M. Crystal structure of a dimeric mannose-specific agglutinin from garlic: quaternary association and carbohydrate specificity. J. Mol. Biol. 1999;285:1157–1168. doi: 10.1006/jmbi.1998.2353. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Giomarelli B., Schumacher K.M., Taylor T.E., Sowder R.C., Hartley J.L., McMahon J.B., Mori T. Recombinant production of anti-HIV protein, griffithsin, by auto-induction in a fermentor culture. Protein Expr. Purif. 2006;47:194–202. doi: 10.1016/j.pep.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Gulakowski R.J., McMahon J.B., Staley P.G., Moran R.A., Boyd M.R. A semiautomated multiparameter approach for anti-HIV drug screening. J. Virol. Methods. 1991;33:87–100. doi: 10.1016/0166-0934(91)90010-w. [DOI] [PubMed] [Google Scholar]
- Liu Y., Carroll J.R., Holt L.A., McMahon J., Giomarelli B., Ghirlanda G. Multivalent interactions with gp120 are required for the anti-HIV activity of Cyanovirin. Biopolymers. 2009;92:194–200. doi: 10.1002/bip.21173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubkowski J., Hennecke F., Plückthun A., Wlodawer A. Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA. Structure. 1999;7:711–722. doi: 10.1016/s0969-2126(99)80092-6. [DOI] [PubMed] [Google Scholar]
- Matei E., Zheng A., Furey W., Rose J., Aiken C., Gronenborn A.M. Anti-HIV activity of defective cyanovirin-N mutants is restored by dimerization. J. Biol. Chem. 2010;285:13057–13065. doi: 10.1074/jbc.M109.094938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallograhic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher J.L., Winter H.C., Ezell P., Goldstein I.J., Stuckey J.A. Crystal structure of banana lectin reveals a novel second sugar binding site. Glycobiology. 2005;15:1033–1042. doi: 10.1093/glycob/cwi088. [DOI] [PubMed] [Google Scholar]
- Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. HKL-3000: The integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Mori T., O'Keefe B.R., Sowder R.C., Bringans S., Gardella R., Berg S., Cochran P., Turpin J.A., Buckheit R.W., Jr., McMahon J.B., Boyd M.R. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 2005;280:9345–9353. doi: 10.1074/jbc.M411122200. [DOI] [PubMed] [Google Scholar]
- Moulaei T., Botos I., Ziolkowska N.E., Bokesch H.R., Krumpe L.R., McKee T.C., O'Keefe B.R., Dauter Z., Wlodawer A. Atomic-resolution crystal structure of the antiviral lectin scytovirin. Protein Sci. 2007;16:2756–2760. doi: 10.1110/ps.073157507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- O'Keefe B.R., Shenoy S.R., Xie D., Zhang W., Muschik J.M., Currens M.J., Chaiken I., Boyd M.R. Analysis of the interaction between the HIV-inactivating protein cyanovirin-N and soluble forms of the envelope glycoproteins gp120 and gp41. Mol. Pharmacol. 2000;58:982–992. doi: 10.1124/mol.58.5.982. [DOI] [PubMed] [Google Scholar]
- O'Keefe B.R., Vojdani F., Buffa V., Shattock R.J., Montefiori D.C., Bakke J., Mirsalis J., d'Andrea A.L., Hume S.D., Bratcher B., et al. Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical microbicide component. Proc. Natl. Acad. Sci. USA. 2009;106:6099–6104. doi: 10.1073/pnas.0901506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe B.R., Giomarelli B., Barnard D.L., Shenoy S.R., Chan P.K., McMahon J.B., Palmer K.E., Barnett B.W., Meyerholz D.K., Wohlford-Lenane C.L., McCray P.B., Jr. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J. Virol. 2010;84:2511–2521. doi: 10.1128/JVI.02322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks T.D., Leuther K.K., Howard E.D., Johnston S.A., Dougherty W.G. Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal. Biochem. 1994;216:413–417. doi: 10.1006/abio.1994.1060. [DOI] [PubMed] [Google Scholar]
- Phan J., Zdanov A., Evdokimov A.G., Tropea J.E., Peters H.K., III, Kapust R.B., Li M., Wlodawer A., Waugh D.S. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 2002;277:50564–50572. doi: 10.1074/jbc.M207224200. [DOI] [PubMed] [Google Scholar]
- Ramachandraiah G., Chandra N.R., Surolia A., Vijayan M. Computational analysis of multivalency in lectins: structures of garlic lectin-oligosaccharide complexes and their aggregates. Glycobiology. 2003;13:765–775. doi: 10.1093/glycob/cwg095. [DOI] [PubMed] [Google Scholar]
- Schlunegger M.P., Bennett M.J., Eisenberg D. Oligomer formation by 3D domain swapping: a model for protein assembly and misassembly. Adv. Protein Chem. 1997;50:61–122. doi: 10.1016/s0065-3233(08)60319-8. [DOI] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S.R., O'Keefe B.R., Bolmstedt A.J., Cartner L.K., Boyd M.R. Selective interactions of the human immunodeficiency virus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J. Pharmacol. Exp. Ther. 2001;297:704–710. [PubMed] [Google Scholar]
- Singh D.D., Saikrishnan K., Kumar P., Surolia A., Sekar K., Vijayan M. Unusual sugar specificity of banana lectin from Musa paradisiaca and its probable evolutionary origin. Crystallographic and modelling studies. Glycobiology. 2005;15:1025–1032. doi: 10.1093/glycob/cwi087. [DOI] [PubMed] [Google Scholar]
- Swanson M.D., Winter H.C., Goldstein I.J., Markovitz D.M. A lectin isolated from bananas is a potent inhibitor of HIV replication. J. Biol. Chem. 2010;285:8646–8655. doi: 10.1074/jbc.M109.034926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Chiba H., Inokoshi J., Kuno A., Sugai T., Takahashi A., Ito Y., Tsunoda M., Suzuki K., Takenaka A., et al. Mechanism by which the lectin actinohivin blocks HIV infection of target cells. Proc. Natl. Acad. Sci. USA. 2009;106:15633–15638. doi: 10.1073/pnas.0907572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong C., O'Keefe B.R., Byrd R.A., McMahon J.B. Potent anti-HIV activity of scytovirin domain 1 peptide. Peptides. 2006;27:1668–1675. doi: 10.1016/j.peptides.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Zeitlin L., Pauly M., Whaley K.J. Second-generation HIV microbicides: continued development of griffithsin. Proc. Natl. Acad. Sci. USA. 2009;106:6029–6030. doi: 10.1073/pnas.0902239106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Gaschen B., Blay W., Foley B., Haigwood N., Kuiken C., Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- Ziólkowska N.E., Wlodawer A. Structural studies of algal lectins with anti-HIV activity. Acta Biochim. Pol. 2006;53:617–626. [PubMed] [Google Scholar]
- Ziólkowska N.E., O'Keefe B.R., Mori T., Zhu C., Giomarelli B., Vojdani F., Palmer K.E., McMahon J.B., Wlodawer A. Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure. 2006;7:1127–1135. doi: 10.1016/j.str.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziólkowska N.E., Shenoy S.R., O'Keefe B.R., McMahon J.B., Palmer K.E., Dwek R.A., Wormald M.R., Wlodawer A. Crystallographic, thermodynamic, and molecular modeling studies of the mode of binding of oligosaccharides to the potent antiviral protein griffithsin. Proteins. 2007;67:661–670. doi: 10.1002/prot.21336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziólkowska N.E., Shenoy S.R., O'Keefe B.R., Wlodawer A. Crystallographic studies of the complexes of antiviral protein griffithsin with glucose and N-acetylglucosamine. Protein Sci. 2007;16:1485–1489. doi: 10.1110/ps.072889407. [DOI] [PMC free article] [PubMed] [Google Scholar]