Abstract
Background
Excision repair cross complementing group 5 (ERCC5 or XPG) plays an important role in regulating DNA excision repair, removal of bulky lesions caused by environmental chemicals or UV light. Mutations in this gene cause a rare autosomal recessive syndrome, and its functional single nucleotide polymorphisms (SNPs) may alter DNA repair capacity phenotype and cancer risk. However, a series of epidemiological studies on the association between the ERCC5 Asp1104His polymorphism (rs17655, G>C) and cancer susceptibility generated conflicting results.
Methodology/Principal Findings
To derive a more precise estimation of the association between the ERCC5 Asp1104His polymorphism and overall cancer risk, we performed a meta-analysis of 44 published case-control studies, in which a total of 23,490 cases and 27,168 controls were included. To provide additional biological plausibility, we also assessed the genotype-gene expression correlation from the HapMap phase II release 23 data with 270 individuals from 4 ethnic populations. When all studies were pooled, we found no statistical evidence for a significantly increased cancer risk in the recessive genetic models (His/His vs. Asp/Asp: OR = 0.99, 95% CI: 0.92–1.06, P = 0.242 for heterogeneity or His/His vs. Asp/His + Asp/Asp: OR = 0.98, 95% CI: 0.93–1.03, P = 0.260 for heterogeneity), nor in further stratified analyses by cancer type, ethnicity, source of controls and sample size. In the genotype-phenotype correlation analysis from 270 individuals, we consistently found no significant correlation of the Asp1104His polymorphism with ERCC5 mRNA expression.
Conclusions/Significance
This meta-analysis suggests that it is unlikely that the ERCC5 Asp1104His polymorphism may contribute to individual susceptibility to cancer risk.
Introduction
Exposure to environmental carcinogens can cause different types of DNA damage that subsequently lead to carcinogenesis of different tissues, if left unrepaired. During the evolution, humans have developed a versatile DNA repair machinery to ensure genome integrity in response to the insults of cancer-causing agents. DNA repair is a complex biological process consisting of several distinct pathways. To date, more than 150 human DNA repair genes have been identified in five major pathways: nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR), double-strand break repair (DSBR), and transcription coupled repair (TCR). Of those pathways, NER is the most studied DNA repair mechanism responsible for various types of DNA damage, including thymidine dimers, oxidative DNA damage, bulky adducts cross-links, and alkylating damage [1]. At least eight core genes (i.e., ERCC1, XPA, XPB/ERCC3, XPC, XPD/ERCC2, XPE/DDB1, XPF/ERCC4, and XPG/ERCC5) in the NER pathway play vital roles in repairing DNA damage and maintain genome integrity [2], [3].
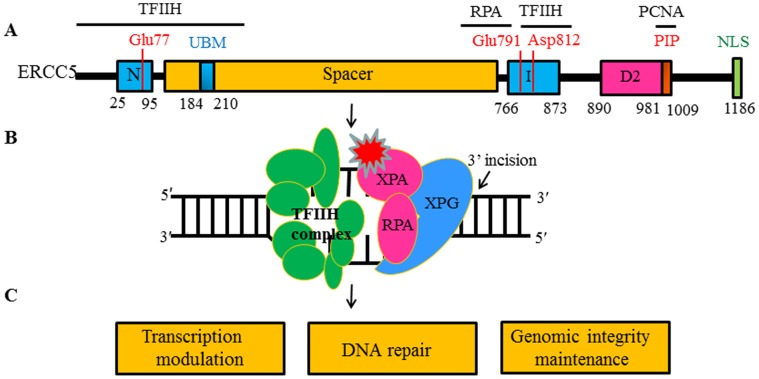
The excision repair cross complementing group 5 (ERCC5) gene, also known as the xeroderma pigmentosum group G (XPG) gene, is a member of the flap structure-specific endonuclease 1 (FEN1) family and encodes a protein of 1186 amino acids. The primary structure of human ERCC5 protein harbors the N- and I-nuclease domains that are highly conserved, which together form the nuclease core [4]. Mutations of several conserved residues in the active site, including Glu77, Glu791 and Asp812, abolish the catalytic activity of the protein [5], [6]. N- and I-nuclease domains are separated by the 600 amino acid spacer region that is highly acidic for protein-protein interactions including with TFIIH [7], [8], [9] and RPA [10] and therefore recruits ERCC5 to the sites of NER [11]. Additionally, ERCC5 contains ubiquitin-binding motif (UBM) as well as a PIP domain that mediates interactions with PCNA [12], [13]. Such an interaction between ERCC5 and PCNA could be involved in triggering the 3′ incision in NER. ERCC5 cleaves the 5′ flap, splayed arm and a variety of bubble substrates at ss/dsDNA junctions with the 5′overhang and makes the 3′ incision in NER [14] ( Figure 1 ).
Figure 1. Structural characteristics and function of ERCC5 protein modified from the picture in the reference [14].
(A) Human ERCC5 protein harbors the N- and I-nuclease domains (blue) and 600 amino acid spacer region (Orange). Conserved residues (Glu77, Glu791 and Asp812) located in the active site (red). Interaction regions with TFIIH, RPA, and PCNA (PIP) and the ubiquitin-binding domain (UBM) are indicated. (B) ERCC5 cleaves 5′ flap, splayed-arm and bubble substrates at ss/dsDNA junctions and makes the 3′ incision in NER. (C) ERCC5 protein plays versatile roles in cellular processes including DNA repair, genomic integrity maintenance and modulation of gene transcription.
As a structure-specific endonuclease and also a 5′-3′ exonuclease, the ERCC5 protein is required for two sub-pathways in NER. One is TCR, which preferentially removes DNA damage in the transcribed DNA strand of active genes; the other is global genomic repair (GGR), which removes lesions throughout the genome [15], [16]. Additionally, ERCC5 also possesses some secondary functions independent of its cleavage activity in supporting a role of TFIIH in receptor-mediated transcription [17], [18]. Furthermore, data from S. cerevisiae studies demonstrate a role for Rad2 (the ERCC5 homolog) in RNA polymerase II-mediated transcription [19]. In addition, ERCC5 is thought to have a possible role in the removal of oxidative damage by BER and possibly other pathways [20], [21]. Numerous studies using various tumor cell lines or tissues indicates that ERCC5 plays a key role in carcinogenesis and that its deficiency leads to DNA repair defects, genomic instability, failure of modulation of gene transcription [22]–[26]. Genetic disorders resulting from mutations in the ERCC5 gene, such as xeroderma pigmentosum (XP), Cockayne syndrome (CS), and tri-chothiodystrophy (TTD), underscore the biological importance of this gene [14], and most of these syndromes follow a recessive genetic model, in which heterozygotes are unaffected, but mutant homozygotes manifest the disease [27].
The ERCC5 gene is located on chromosome 13q22-q33, consists of 15 exons that range from 61 to 1074 bp and 14 introns that range from 250 to 5763 bp, and spans 32 kb [28]. To date, at least 73 non-synonymous SNPs (nsSNPs) in the ERCC5 coding region have been identified (http://www.ncbi.nlm.nih.gov/SNP/), and 24 SNPs in the gene region have been studied for their association with cancer risk (Table S1), of which only four were nsSNPs (Figure S1). For example, the Asp1104His polymorphism (rs17655 G>C) is common [minor allele frequency (MAF) >0.05] and regarded as a tagger, which was most frequently investigated for its association with cancer risk. Interestingly, relatively few nsSNPs are present in the eight NER core genes, suggesting the conservativeness of these genes for their biological importance.
In a published meta-analysis, a total of 12 SNPs of the eight NER core genes, including 6 nsSNPs, have been investigated for the associations with cancer risks [29]–[44], among which 5 nsSNPs were found to be associated either with risk of a specific cancer or risk of the overall cancer risks mostly under recessive genetic models (Table S2), but no published meta-analysis has summarized all reported studies of the Asp1104His polymorphism in association with risk of all cancer types. It is biologically plausible that the Asp1104His polymorphism, causing a change from aspartate to histidine at codon 1104 in ERCC5 protein, may result in an alteration of the gene function, thus likely altering risk of developing cancers, possibly following a recessive genetic model.
To date, although a number of studies have been performed to investigate the association between the ERCC5 Asp1104His polymorphism and cancer risk, the evidence regarding the role of SNPs of the ERCC5 gene as a genetic marker for cancer risk is conflicting, partially because of the possible lack of a main effect of the SNP on risk of any single type of cancer, a possibly low penetrance or weak effect, or the relatively small sample size in each of published studies. Therefore, we performed a meta-analysis to identify statistical evidence for an association between the ERCC5 Asp1104His polymorphism and cancer risk using all published data to date.
Materials and Methods
Identification and Eligibility of Relevant Studies
We searched two electronic databases (MEDLINE and EMBASE) for all relevant articles with the following terms: “ERCC5” or “XPG”, “DNA repair”, “polymorphism” or “variant”, “case-control”, “risk”, “association”, and “cancer” or “carcinoma” or “neoplasm” or “malignance” (last search was updated on Sept 1, 2011). References of the retrieved articles or reviews on this topic were also manually screened for additional relevant eligible studies.
We defined inclusion criteria as follows: written in English or Chinese; case-control design; sufficient information for estimating odds ratio (OR) and their 95% confidence interval (CI); observed genotype frequencies in the controls in agreement with Hardy-Weinberg equilibrium (HWE). Abstracts and unpublished reports were not considered. Investigations in subjects with family history or cancer-prone disposition were also excluded. Meanwhile, if studies had overlapping subjects, we selected the most recent study that included the largest number of individuals in the publications. Additionally, we also checked for minor allelic frequency (MAF) among studies by different genotype frequencies in ethnic groups based on Hapmap or dbSNP frequencies reported for the different ethnic groups, and the datasets were excluded if they had a very high probability of inaccurate reported.
Data Extraction
Two investigators (Zhu ML and Wang MY) independently extracted the following information from each study: the first author, year of publication, country of origin, ethnicity, cancer type, source of controls (population-based, hospital-based, family-based and mixed controls), genotyping method, number of genotyped cases and controls, numbers of genotypes for ERCC5 Asp1104His (rs17655) in cases and controls, and main findings. For studies including subjects of different ethnic groups, we extracted data separately for each ethnic group and categorized as Caucasian, Asian, African and others. When a study did not state what ethic groups were included or if it was impossible to separate participants according to the data presented, we termed the sample as ‘others’.
Correlation Analysis of ERCC5 mRNA Expression
We provide biological plausibility of the studied SNP, we downloaded the Asp1104His genotyping data from the HapMap phase II release 23 data set consisting of 3.96 million SNP genotypes from 270 individuals from four populations (CEU: 90 Utah residents from northern and western Europe; CHB: 45 unrelated Han Chinese in Beijing; JPT: 45 unrelated Japanese in Tokyo; YRI: 90 Yoruba in Ibadan, Nigeria) (http://www.sanger.ac.uk/humgen/hapmap3) [45]. The data on ERCC5 mRNA expression levels from EBV-transformed B lymphoblastoid cell lines from the same 270 HapMap individuals were available online (http://app3.titan.uio.no/biotools/tool.php?app=snpexp) as well [46], [47]. Then, we conducted linear regression model-trend test for assessing the correlation between Asp1104His and ERCC5 mRNA expression for different populations.
Statistical Methods
We assessed the association between the ERCC5 Asp1104His polymorphism and cancer risk by crude ORs and 95% CIs. Then, we calculated the pooled ORs and 95% CIs under an assumption of a recessive genetic model (His/His vs. Asp/Asp or His/His vs. Asp/His + Asp/Asp). In addition, we performed stratification analyses by cancer type (if one cancer type contained less than three studies, it was merged into the ‘other cancers’ group), ethnicity, source of controls, study design and sample size (<500, 500–1000, and >1000).
We evaluated the between-study heterogeneity by using the Chi square-based Q-test, which was considered significant if P<0.10. Values from single studies were combined using models of both random effects (DerSimonian and Laird method 1986) [48] and fixed effects (Mantel–Haenszel method) [49]. When P value of the heterogeneity test was >0.10, the fixed-effects model was used, which indicates no significant heterogeneity of the effect size across all studies; otherwise, the random-effects model was more appropriate, which tends to provide wider CIs, when the results of the constituent studies differ among themselves. To evaluate the effect of individual studies on overall risk of cancers, we conducted sensitivity analyses by excluding each study individually and recalculating the ORs and 95% CI. We also used the inverted funnel plot and the Egger’s test to examine the potential influence of publication bias (linear regression analysis) [50]. HWE among controls for each study was examined by the Pearson's goodness-of-fit chi-square test. The boxplot presentation and trend tests were performed with Statistical Analysis System software (v.9.1 SAS Institute, Cary, NC) All other statistical analyses were carried out with STATA software, version 11.0 (Stata Corporation, College Station, TX). All P values were two-sided with a significance level of 0.05, unless specified otherwise.
Results
Study Characteristics
We identified a total of 74 relevant publications after initial screening. Among these, 62 publications had met the inclusion criteria and were subjected to further examination. We excluded 8 publications because they did not present detailed genotyping information [51]–[58]. We also excluded 3 publications because they included the overlapped data with those included in the analysis [59], [60], [61]. Furthermore, we removed 7 publications because their genotype distributions among the controls deviated from HWE [62]–[68]. Therefore, our final data pooling consisted of 44 publications [69]–[112] with a total of 23490 cancer cases and 27168 controls, of which there were actual 49 case-control studies, because 5 publications provided more than one individual study (Figure S2). These 49 studies included 9 breast cancer studies, 8 skin cancer studies, 5 lung cancer studies, 5 bladder cancer studies, 3 head and neck cancer studies, 3 colorectal cancer studies, 3 non-Hodgkin lymphoma studies, and 13 studies of other cancers. Of these, there were 27 hospital-based studies, 20 population-based studies, 1 family-based study, and 1 study with mixed controls. In addition, 29 of 49 studies were conducted in Caucasians, 4 were conducted in African-Americans, 6 were conducted in Asians, and the remaining 10 were conducted in other ethnic groups. Several genotyping methods were used, including the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP), which was the most frequently used method, TaqMan, sequencing, Illumina, SNaPshot, SNPlex and Mass spectrometry, but two publications did not provide information about genotyping methods. Additionally, all studies were in keep with HapMap or dbSNP frequencies reported for the different ethnic groups (Table S3).
Quantitative Synthesis
When all eligible studies were pooled into one dataset for the meta-analysis, we found no statistical association between the ERCC5 Asp1104His polymorphism and overall cancer risk under the recessive genetic models: His/His vs. Asp/Asp: OR = 0.99, 95% CI: 0.92–1.06 or His/His vs. Asp/His + Asp/Asp: OR = 0.98, 95% CI: 0.93–1.03.
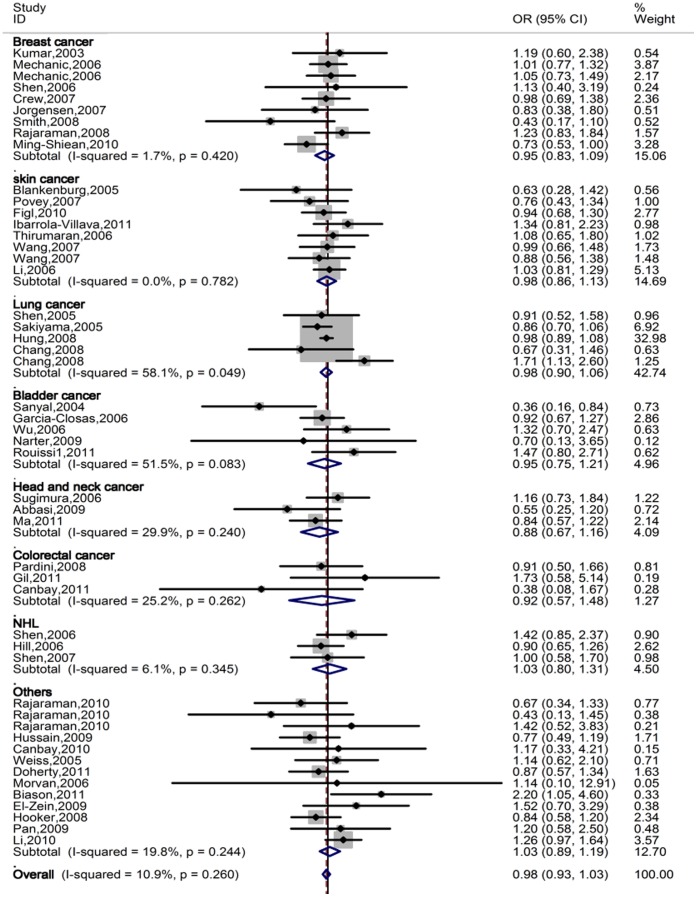
In the stratified analysis by ethnicity, we did not observe any association between the polymorphism and cancer risk in the recessive genetic models, neither and had the similar results in the stratified analyses by tumor type, source of controls, and sample size in cases ( Table 1 , Figure 2 ).
Table 1. Meta-analysis of the association between the ERCC5 Asp1104His polymorphism and cancer risk under the XP recessive genetic model for 49 studies.
| Variables | No. of studies | No. of subjects Cases/controls | His/His vs. Asp/Asp | His/His vs. Asp/His+Asp/Asp | ||||
| OR (95%CI)c | P OR a | P het b | OR (95%CI)c | P OR a | P het b | |||
| All | 49 | 23490/27168 | 0.99 (0.92–1.06) | 0.707 | 0.242 | 0.98 (0.93–1.03) | 0.393 | 0.260 |
| Cancer type | ||||||||
| Breast | 9 | 4915/5277 | 0.99 (0.85–1.15) | 0.882 | 0.362 | 0.95 (0.83–1.09) | 0.497 | 0.420 |
| Skin | 8 | 3909/4263 | 0.95 (0.78–1.16) | 0.622 | 0.586 | 0.98 (0.86–1.13) | 0.809 | 0.782 |
| Lung | 5 | 4702/5654 | 0.99 (0.86–1.15) | 0.937 | 0.032 | 0.98 (0.90–1.06) | 0.566 | 0.049 |
| Bladder | 5 | 2304/2253 | 0.94 (0.74–1.20) | 0.621 | 0.112 | 0.95 (0.75–1.21) | 0.686 | 0.083 |
| Head and neck | 3 | 1429/1954 | 0.90 (0.67–1.22) | 0.510 | 0.194 | 0.88 (0.67–1.16) | 0.364 | 0.240 |
| Colorectal | 3 | 743/879 | 0.97 (0.59–1.58) | 0.900 | 0.372 | 0.92 (0.57–1.48) | 0.720 | 0.262 |
| NHL | 3 | 2105/1957 | 1.07 (0.83–1.38) | 0.594 | 0.238 | 1.03 (0.80–1.32) | 0.839 | 0.345 |
| Others | 13 | 3383/4931 | 1.02 (0.86–1.21) | 0.850 | 0.384 | 1.03 (0.89–1.19) | 0.696 | 0.244 |
| Ethnicity | ||||||||
| Caucasian | 29 | 13316/15586 | 0.99 (0.89–1.10) | 0.814 | 0.287 | 0.98 (0.90–1.08) | 0.709 | 0.452 |
| African-American | 4 | 1318/1330 | 1.11 (0.89–1.38) | 0.365 | 0.020 | 1.03 (0.86–1.24) | 0.747 | 0.016 |
| Asian | 6 | 2314/2443 | 0.90 (0.76–1.06) | 0.216 | 0.292 | 0.93 (0.82–1.06) | 0.262 | 0.091 |
| Others | 10 | 6542/7809 | 1.00 (0.88–1.15) | 0.982 | 0.878 | 0.98 (0.90–1.07) | 0.679 | 0.852 |
| Source of controls | ||||||||
| Hospital | 27 | 9787/11586 | 0.95 (0.85–1.05) | 0.298 | 0.186 | 0.97 (0.89–1.05) | 0.426 | 0.129 |
| Population | 20 | 10333/11150 | 1.02 (0.91–1.14) | 0.732 | 0.325 | 0.99 (0.89–1.09) | 0.795 | 0.427 |
| Others | 2 | 3370/4432 | 1.02 (0.85–1.23) | 0.810 | 0.853 | 0.98 (0.90–1.08) | 0.706 | 0.795 |
| Sample size | ||||||||
| <500 | 33 | 7469/10388 | 0.98 (0.87–1.11) | 0.755 | 0.055 | 0.96 (0.86–1.06) | 0.396 | 0.067 |
| 500–1000 | 11 | 8170/7890 | 1.02 (0.90–1.15) | 0.781 | 0.704 | 1.01 (0.91–1.11) | 0.923 | 0.649 |
| >1000 | 5 | 7851/8890 | 0.96 (0.84–1.09) | 0.516 | 0.893 | 0.97 (0.90–1.05) | 0.474 | 0.916 |
P value of the Z-test for odds ration test.
P value of the Q-test for heterogeneity test.
Fixed effects model.
Figure 2. Forest plot of overall cancer risk of different cancer types associated with ERCC5 Asp1104His polymorphism (His/His vs. Asp/His + Asp/Asp) by the fixed effects for each of the 44 published studies.
For each study, the estimates of OR and its 95% CI were plotted with a box and a horizontal line. The symbol filled diamond indicates pooled OR and its 95% CI.
Heterogeneity and Sensitivity Analyses
There were no between-study heterogeneity among overall studies of the ERCC5 Asp1104His polymorphism in the recessive genetic models (χ2 = 54.45, P = 0.242 for heterogeneity test and χ2 = 53.86, P = 0.260 for heterogeneity test for His/His vs. Asp/Asp and His/His vs. Asp/His + Asp/Asp, respectively). In the sensitivity analyses, the influence of each study on the pooled OR was checked by repeating the meta-analysis while omitting each study, one at a time. This procedure validated the stability of our results. Furthermore, the inclusion of 7 studies, whose genotype distributions among the controls deviated from HWE, affected between-study heterogeneity for His/His vs. Asp/Asp (χ2 = 72.21, P = 0.060), but did not influence the result of the meta-analysis significantly: His/His vs. Asp/Asp: OR = 1.00, 95% CI: 0.93–1.09. His/His vs. Asp/His + Asp/Asp: OR = 0.98, 95% CI: 0.93–1.03.
Publication Bias
We conducted Begg’s funnel plot and Egger’s test to access the publication bias of all included studies. The shape of the funnel plot seemed symmetrical (Figure S3), suggesting that there was no obvious publication bias. Furthermore, the Egger’s test further provided statistical evidence that there was no significant publication bias in this meta-analysis (the Egger’s test: His/His vs. Asp/Asp: P = 0.897, His/His vs. Asp/His + Asp/Asp: P = 0.749).
Correlation Analysis for ERCC5 mRNA Expression and Asp1104His Genotypes
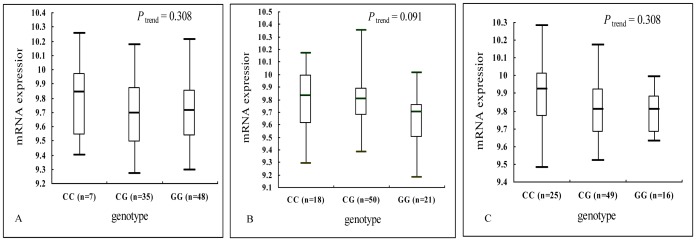
In the genotype-phenotype correlation analysis using the lymphoblastoid cell lines derived from peripheral lymphocytes from 270 people, we found no trend for the allele effect on ERCC5 mRNA expression for Europeans (P trend = 0.308), Asians (P trend = 0.091) and Africans (P trend = 0.308) ( Figure 3 ).
Figure 3. Correlation between Asp1104His and ERCC5 mRNA expression for different populations. A.
: CEU, 90 Utah residents from northern and western Europe; B: Asians, 45 unrelated Han Chinese in Beijing (CHB) and 45 unrelated Japanese in Tokyo (JPT); C: YRI, 90 Yoruba in Ibadan, Nigeria.
Discussion
On the basis of eligible 49 case-control studies with a total of 23490 cancer cases and 27168 controls, our meta-analysis comprehensively evaluated the association between the ERCC5 Asp1104 His polymorphism and risk of different types of cancers, and we did not find statistical evidence for such an association in the recessive genetic models as shown in the XP syndrome. Similarly, in subgroup analyses, we consistently showed no statistical association between the polymorphism and cancer risk in any of the subgroups. Furthermore, the observed null associations were supported by the data that the variant genotypes of the SNP were not associated with mRNA expression levels of ERCC5 in the lymphoblastoid cell lines.
DNA repair deregulation is a crucial factor in the multistep process of carcinogenesis, and the ERCC5 gene is a vital component of the DNA repair machinery. It has been observed that deficiency of ERCC5 may result in severe autosomal recessive diseases including XP, CS and TTD [14] characterized by solar hypersensitivity of the skin, high predisposition for developing cancers (mainly epithelial and melanoma) on areas exposed to sunlight. Furthermore, studies have suggested that ERCC5 SNPs are associated with development of some cancers, such as breast cancer [44] and smoking-related cancers [23], [24]. These suggest a possible link between the ERCC5 function and development of cancer. The biological mechanisms of the ERCC5 gene in carcinogenesis may be complicated, among which nsSNPs, leading to an amino acid change in the protein product and modulating the individual DNA repair capacity phenotype [113], [114], may account for some of the known genetic variations related to risk of cancers. However, our meta-analysis suggests that there is no statistical evidence for an association between the ERCC5 Asp1104His polymorphism and overall cancer risk, which is consistent with the previous two meta-analyses conducted in breast cancer and melanoma, respectively. The former included some studies departure from HWE in the control population, and the latter only contained three studies. Although we excluded the inappropriate studies and expanded the sample size, the null results were not altered. Furthermore, as far as our knowledge is concerned, none of the SNPs in NER have ever been identified as susceptibility locus in the published genome-wide association studies (GWAS) for common diseases including cancers based on common SNPs, which are similar to our results. This is a challenge to the theory of common variants and common diseases [115]. It is likely that, as NER genes are considered susceptibility genes, the role of NER variants in cancer development may be dependent on the degree of exposures that cause damage to DNA. Therefore, without detailed information about such exposures for further adjustment or stratification, the results of the observed associations may be either biased or masked. For example, XP patients can drastically reduce risk of developing skin cancer by avoiding exposure to sunlight. Another possibility is that the common variants are unlikely to have a significant biological effect. For common variants, in most cases, the disease-associated variant itself is unlikely to be functionally relevant [115]. The third possibility is that the genetic risk of cancer conferred by the common variants is very modest and the penetrance of the variants is very small, which means that even if the polymorphism is crucial for carcinogenesis, extremely large-scale evidence would be necessary to establish with high confidence the presence of specific associations. The inclusion of rare variants and larger samples in future genome-wide association studies would help to reveal low-penetrance susceptibility loci that are more likely to be associated with cancer risk.
Furthermore, we did not observe biological evidence for the effect of this SNP on the gene expression in terms of mRNA levels, which biological support for the result of no association. Although a sequence homology-based tool predicted this ERCC5 polymorphism to be a deleterious substitution [116], and computational algorithms by SIFT and SNPs3D tools aslo identified the polymorphism with some functional implications (http://compbio.cs.queensu.ca/F-SNP/), such a potentially functional relevance have not been validated experimentally to date. Diversities of variant-related risk associations in various kinds of cancer may result from different mechanisms of carcinogenesis among different cancer types. Although some studies have discovered some sequence variants in the region of chromosome 5p15.33 and 8q24 that are associated with risk of different cancer types [117]–[122], it is still uncertain whether the same polymorphism may have non-specific effect on different types of cancer. Therefore, further functional studies should be undertaken to explore the mechanism underlying the variant-related associations with cancer risk.
It would be hard to interpret results, if significant heterogeneity were present. However, in this meta-analysis, we did not find any obvious heterogeneity and publish bias across studies. Nevertheless, some limitations should be addressed. Firstly, although funnel plot and Egger’s test show no publication bias, selection bias could have occurred because only studies published in English and Chinese were included. Secondly, because the reference groups were not uniformly defined, some selected population-based controls and some used hospital-based cancer-free controls, non-differential misclassification bias is possible; in addition, some control groups may not be representative of the general population. Thirdly, our results were based on unadjusted OR estimates, because not all published studies presented adjusted ORs or when they did, the ORs were not adjusted by the same potential confounders, such as age, sex and exposure variables. Thus, more comprehensive individual datasets are needed to allow for the adjustment by some co-variants and further evaluation of potential gene-environmental interactions for susceptibility to cancer. Fourthly, although the sample size of our study was relatively large, the statistical power was still limited in the analyses of subgroups with small sample sizes, particularly when the effect size is small. Therefore, studies with larger sample sizes with sufficient large subgroups should be undertaken to validate our findings.
In summary, our meta-analysis shows that the ERCC5 Asp1104His polymorphism appeared to be unlikely to confer susceptibility to cancers. Further well-designed studies with larger sample sizes will be necessary to validate the findings in the present meta-analysis.
Supporting Information
ERCC5 gene map labeled with nine SNPs have been studied for associations with cancer risk. (A) Six SNPs are located in the coding region, among which four are nsSNPs, of which two are synonymous SNPs; two SNPs are located in the 5′ untranslated region, and one SNP is located in the 3′ untranslated region; six SNPs are tagging SNPs. (B) Nine SNPs with a minor allelic frequency in different populations obtained from the dbSNP database.
(TIF)
Flow chart of included studies for this meta-analysis.
(TIF)
Funnel plot analysis to detect publication bias for ERCC5 Asp1104His under the recessive genetic models (A, His/His vs. Asp/Asp and B, His/His vs. Asp/His + Asp/Asp) for all 44 studies. Each point represents an individual study for the indicated association.
(TIF)
Summary of 24 SNPs of the ERCC5/XPG gene that have been studied for their associations with cancer risk.
(DOCX)
Summary of Studied SNPs in the eight NER genes reviewed in all published meta-analysis.
(DOCX)
Characteristics of the 44 references included in the meta-analysis for ERCC5 Asp1104His.
(DOCX)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by a grant from “China’s Thousand Talents Program” Recruitment at Fudan University, a grant from the Ministry of Health (201002007) and the National Natural Science Foundation of China (81101808). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 2.Wakasugi M, Reardon JT, Sancar A. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem. 1997;272:16030–16034. doi: 10.1074/jbc.272.25.16030. [DOI] [PubMed] [Google Scholar]
- 3.O’Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 4.Scherly D, Nouspikel T, Corlet J, Ucla C, Bairoch A, et al. Complementation of the DNA repair defect in xeroderma pigmentosum group G cells by a human cDNA related to yeast RAD2. Nature. 1993;363:182–185. doi: 10.1038/363182a0. [DOI] [PubMed] [Google Scholar]
- 5.Wakasugi M, Reardon JT, Sancar A. The Non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. Journal of Biological Chemistry. 1997;272:16030–16034. doi: 10.1074/jbc.272.25.16030. [DOI] [PubMed] [Google Scholar]
- 6.Constantinou A, Gunz D, Evans E, Lalle P, Bates PA, et al. Conserved residues of human XPG protein important for nuclease activity and function in nucleotide excision repair. J Biol Chem. 1999;274:5637–5648. doi: 10.1074/jbc.274.9.5637. [DOI] [PubMed] [Google Scholar]
- 7.Iyer N, Reagan MS, Wu KJ, Canagarajah B, Friedberg EC. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry. 1996;35:2157–2167. doi: 10.1021/bi9524124. [DOI] [PubMed] [Google Scholar]
- 8.Araujo SJ, Nigg EA, Wood RD. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Molecular and Cellular Biology. 2001;21:2281–2291. doi: 10.1128/MCB.21.7.2281-2291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorel F, Constantinou A, Dunand-Sauthier I, Nouspikel T, Lalle P, et al. Definition of a short region of XPG necessary for TFIIH interaction and stable recruitment to sites of UV damage. Mol Cell Biol. 2004;24:10670–10680. doi: 10.1128/MCB.24.24.10670-10680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He ZG, Henricksen LA, Wold MS, Ingles CJ. Rpa Involvement in the Damage-Recognition and Incision Steps of Nucleotide Excision-Repair. Nature. 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 11.Dunand-Sauthier I, Hohl M, Thorel F, Jaquier-Gubler P, Clarkson SG, et al. The spacer region of XPG mediates recruitment to nucleotide excision repair complexes and determines substrate specificity. Journal of Biological Chemistry. 2005;280:7030–7037. doi: 10.1074/jbc.M412228200. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair. 2009;8:544–556. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. Journal of Biological Chemistry. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 14.Fagbemi AF, Orelli B, Scharer OD. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair. 2011;10:722–729. doi: 10.1016/j.dnarep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanawalt PC. Controlling the efficiency of excision repair. Mutat Res. 2001;485:3–13. doi: 10.1016/s0921-8777(00)00071-9. [DOI] [PubMed] [Google Scholar]
- 16.Emmert S, Slor H, Busch DB, Batko S, Albert RB, et al. Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J Invest Dermatol. 2002;118:972–982. doi: 10.1046/j.1523-1747.2002.01782.x. [DOI] [PubMed] [Google Scholar]
- 17.Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, et al. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: implications for Cockayne syndrome in XP-G/CS patients. Mol Cell. 2007;26:231–243. doi: 10.1016/j.molcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Scharer OD. Hot topics in DNA repair: the molecular basis for different disease states caused by mutations in TFIIH and XPG. DNA Repair (Amst) 2008;7:339–344. doi: 10.1016/j.dnarep.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SK, Yu SL, Prakash L, Prakash S. Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription. implications for Cockayne syndrome. Cell. 2002;109:823–834. doi: 10.1016/s0092-8674(02)00795-x. [DOI] [PubMed] [Google Scholar]
- 20.Klungland A, Hoss M, Gunz D, Constantinou A, Clarkson SG, et al. Base excision repair of oxidative DNA damage activated by XPG protein. Molecular Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 21.Bessho T. Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repairenzyme thymine glycol DNA glycosylase. Nucleic Acids Res. 1999;27:979–983. doi: 10.1093/nar/27.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koeppel F, Poindessous V, Lazar V, Raymond E, Sarasin A, et al. Irofulven cytotoxicity depends on transcription-coupled nucleotide excision repair and is correlated with XPG expression in solid tumor cells. Clinical Cancer Research. 2004;10:5604–5613. doi: 10.1158/1078-0432.CCR-04-0442. [DOI] [PubMed] [Google Scholar]
- 23.Cheng L, Sturgis EM, Eicher SA, Spitz MR, Wei Q. Expression of nucleotide excision repair genes and the risk for squamous cell carcinoma of the head and neck. Cancer. 2002;94:393–397. doi: 10.1002/cncr.10231. [DOI] [PubMed] [Google Scholar]
- 24.Cheng L, Spitz MR, Hong WK, Wei Q. Reduced expression levels of nucleotide excision repair genes in lung cancer: a case-control analysis. Carcinogenesis. 2000;21:1527–1530. [PubMed] [Google Scholar]
- 25.Bartolucci R, Wei J, Sanchez JJ, Perez-Roca L, Chaib I, et al. XPG mRNA expression levels modulate prognosis in resected non-small-cell lung cancer in conjunction with BRCA1 and ERCC1 expression. Clin Lung Cancer. 2009;10:47–52. doi: 10.3816/CLC.2009.n.007. [DOI] [PubMed] [Google Scholar]
- 26.Walsh CS, Ogawa S, Karahashi H, Scoles DR, Pavelka JC, et al. ERCC5 is a novel biomarker of ovarian cancer prognosis. Journal of Clinical Oncology. 2008;26:2952–2958. doi: 10.1200/JCO.2007.13.5806. [DOI] [PubMed] [Google Scholar]
- 27.Kraemer KH, Lee MM, Andrews AD, Lambert WC. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch Dermatol. 1994;130:1018–1021. [PubMed] [Google Scholar]
- 28.Emmert S, Schneider TD, Khan SG, Kraemer KH. The human XPG gene: gene architecture, alternative splicing and single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:1443–1452. doi: 10.1093/nar/29.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Gu S, Wu Q, Li Y, Fu X, et al. No association of ERCC1 C8092A and T19007C polymorphisms to cancer risk: a meta-analysis. European journal of human genetics : EJHG. 2007;15:967–973. doi: 10.1038/sj.ejhg.5201855. [DOI] [PubMed] [Google Scholar]
- 30.Cao C, Zhang Y-M, Wang R, Sun S-F, Chen Z-B, et al. Excision repair cross complementation group 1 polymorphisms and lung cancer risk: a meta-analysis. Chinese Medical Journal. 2011;124:2203–2208. [PubMed] [Google Scholar]
- 31.Vineis P, Manuguerra M, Kavvoura FK, Guarrera S, Allione A, et al. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. Journal of the National Cancer Institute. 2009;101:24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 32.Kiyohara C, Takayama K, Nakanishi Y. Lung cancer risk and genetic polymorphisms in DNA repair pathways: a meta-analysis. Journal of nucleic acids. 2010;2010:701760. doi: 10.4061/2010/701760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qian B, Zhang H, Zhang L, Zhou X, Yu H, et al. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung cancer (Amsterdam, Netherlands) 2011;73:138–146. doi: 10.1016/j.lungcan.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 34.Qiu L, Wang Z, Shi X, Wang Z. Associations between XPC polymorphisms and risk of cancers: A meta-analysis. European journal of cancer (Oxford, England : 1990) 2008;44:2241–2253. doi: 10.1016/j.ejca.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 35.Stern MC, Lin J, Figueroa JD, Kelsey KT, Kiltie AE, et al. Polymorphisms in DNA repair genes, smoking, and bladder cancer risk: findings from the international consortium of bladder cancer. Cancer Research. 2009;69:6857–6864. doi: 10.1158/0008-5472.CAN-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng W, Cong X-F, Cai W-H, Yang S, Mao C, et al. Current evidences on XPC polymorphisms and breast cancer susceptibility: a meta-analysis. Breast cancer research and treatment. 2011;128:811–815. doi: 10.1007/s10549-011-1369-6. [DOI] [PubMed] [Google Scholar]
- 37.Wang F, Chang D, Hu F-l, Sui H, Han B, et al. DNA repair gene XPD polymorphisms and cancer risk: a meta-analysis based on 56 case-control studies. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:507–517. doi: 10.1158/1055-9965.EPI-07-2507. [DOI] [PubMed] [Google Scholar]
- 38.Li C, Jiang Z, Liu X. XPD Lys(751)Gln and Asp (312)Asn polymorphisms and bladder cancer risk: a meta-analysis. Molecular biology reports. 2009;37:301–309. doi: 10.1007/s11033-009-9693-1. [DOI] [PubMed] [Google Scholar]
- 39.Mocellin S, Verdi D, Nitti D. DNA repair gene polymorphisms and risk of cutaneous melanoma: a systematic review and meta-analysis. Carcinogenesis. 2009;30:1735–1743. doi: 10.1093/carcin/bgp207. [DOI] [PubMed] [Google Scholar]
- 40.Pabalan N, Francisco-Pabalan O, Sung L, Jarjanazi H, Ozcelik H (2010 ) Meta-analysis of two ERCC2 (XPD) polymorphisms, Asp312Asn and Lys751Gln, in breast cancer. Breast cancer research and treatment. 124:531–541. doi: 10.1007/s10549-010-0863-6. [DOI] [PubMed] [Google Scholar]
- 41.Qiu L-X, Yao L, Zhang J, Zhu X-D, Zhao X-M, et al. XPD Lys751Gln polymorphism and breast cancer susceptibility: a meta-analysis involving 28,709 subjects. Breast cancer research and treatment. 2010;124:229–235. doi: 10.1007/s10549-010-0813-3. [DOI] [PubMed] [Google Scholar]
- 42.Chen B, Zhou Y, Yang P, Wu X-T. ERCC2 Lys751Gln and Asp312Asn polymorphisms and gastric cancer risk: a meta-analysis. Journal of cancer research and clinical oncology. 2011;137:939–946. doi: 10.1007/s00432-010-0956-y. [DOI] [PubMed] [Google Scholar]
- 43.Yuan L, Cui D, Zhao E-J, Jia C-Z, Wang L-D, et al. XPD Lys751Gln polymorphism and esophageal cancer risk: a meta-analysis involving 2288 cases and 4096 controls. World journal of gastroenterology : WJG. 2011;17:2343–2348. doi: 10.3748/wjg.v17.i18.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding D-P, He X-F, Zhang Y (2011 ) Lack of association between XPG Asp1104His and XPF Arg415Gln polymorphism and breast cancer risk: a meta-analysis of case-control studies. Breast cancer research and treatment. 129:203–209. doi: 10.1007/s10549-011-1447-9. [DOI] [PubMed] [Google Scholar]
- 45.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 46.Holm K, Melum E, Franke A, Karlsen TH. SNPexp - A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC bioinformatics. 2010;11:600–604. doi: 10.1186/1471-2105-11-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazlsy C, et al. Relative impact of nucleotide and copy number variation on gene phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 49.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 50.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. British Medical Journal. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen M, Kamat AM, Huang M, Grossman HB, Dinney CP, et al. High-order interactions among genetic polymorphisms in nucleotide excision repair pathway genes and smoking in modulating bladder cancer risk. Carcinogenesis. 2007;28:2160–2165. doi: 10.1093/carcin/bgm167. [DOI] [PubMed] [Google Scholar]
- 52.Cui Y, Morgenstern H, Greenland S, Tashkin DP, Mao J, et al. Polymorphism of Xeroderma Pigmentosum group G and the risk of lung cancer and squamous cell carcinomas of the oropharynx, larynx and esophagus. Int J Cancer. 2006;118:714–720. doi: 10.1002/ijc.21413. [DOI] [PubMed] [Google Scholar]
- 53.Hung RJ, Baragatti M, Thomas D, McKay J, Szeszenia-Dabrowska N, et al. Inherited predisposition of lung cancer: A hierarchical modeling approach to DNA repair and cell cycle control pathways. Cancer Epidemiology Biomarkers & Prevention. 2007;16:2736–2744. doi: 10.1158/1055-9965.EPI-07-0494. [DOI] [PubMed] [Google Scholar]
- 54.Joshi AD, Corral R, Siegmund KD, Haile RW, Le Marchand L, et al. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009;30:472–479. doi: 10.1093/carcin/bgn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu YH, Scheurer ME, El-Zein R, Cao YM, Do KA, et al. Association and Interactions between DNA Repair Gene Polymorphisms and Adult Glioma. Cancer Epidemiology Biomarkers & Prevention. 2009;18:204–214. doi: 10.1158/1055-9965.EPI-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michiels S, Danoy P, Dessen P, Bera A, Boulet T, et al. Polymorphism discovery in 62 DNA repair genes and haplotype associations with risks for lung and head and neck cancers. Carcinogenesis. 2007;28:1731–1739. doi: 10.1093/carcin/bgm111. [DOI] [PubMed] [Google Scholar]
- 57.Mort R, Mo L, McEwan C, Melton DW. Lack of involvement of nucleotide excision repair gene polymorphisms in colorectal cancer. British Journal of Cancer. 2003;89:333–337. doi: 10.1038/sj.bjc.6601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen H, Ding Q, Fang ZJ, Xia GW, Fang J. Population study of genetic polymorphisms and superficial bladder cancer risk in Han-Chinese smokers in Shanghai. International Urology and Nephrology. 2009;41:855–864. doi: 10.1007/s11255-009-9560-y. [DOI] [PubMed] [Google Scholar]
- 59.An J, Liu Z, Hu Z, Li G, Wang LE, et al. Potentially functional single nucleotide polymorphisms in the core nucleotide excision repair genes and risk of squamous cell carcinoma of the head and neck. Cancer Epidemiol Biomarkers Prev. 2007;16:1633–1638. doi: 10.1158/1055-9965.EPI-07-0252. [DOI] [PubMed] [Google Scholar]
- 60.Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Chen C, et al. Nucleotide excision repair genotype and the incidence of endometrial cancer: Effect of other risk factors on the association. Gynecologic Oncology. 2006;103:891–896. doi: 10.1016/j.ygyno.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 61.Rouissi K, Ouerhani S, Hamrita B, Bougatef K, Marrakchi R, et al. Smoking and polymorphisms in xenobiotic metabolism and DNA repair genes are additive risk factors affecting bladder cancer in Northern Tunisia. Pathology and Oncology Research. 2011;17:879–886. doi: 10.1007/s12253-011-9398-3. [DOI] [PubMed] [Google Scholar]
- 62.Berhane N, Sobti RC, Mahdi SA. DNA repair genes polymorphism (XPG and XRCC1) and association of prostate cancer in a north Indian population. Molecular biology reports. 2012;39:2471–2479. doi: 10.1007/s11033-011-0998-5. [DOI] [PubMed] [Google Scholar]
- 63.Goncalves FT, Francisco G, de Souza SP, Luiz OC, Festa-Neto C, et al. European ancestry and polymorphisms in DNA repair genes modify the risk of melanoma: A case-control study in a high UV index region in Brazil. Journal of Dermatological Science. 2011;64:59–66. doi: 10.1016/j.jdermsci.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 64.He X, Ye F, Zhang J, Cheng Q, Shen J, et al. Susceptibility of XRCC3, XPD, and XPG genetic variants to cervical carcinoma. Pathobiology. 2008;75:356–363. doi: 10.1159/000164220. [DOI] [PubMed] [Google Scholar]
- 65.Jeon HS, Kim KM, Park SH, Lee SY, Choi JE, et al. Relationship between XPG codon 1104 polymorphism and risk of primary lung cancer. Carcinogenesis. 2003;24:1677–1681. doi: 10.1093/carcin/bgg120. [DOI] [PubMed] [Google Scholar]
- 66.McKean-Cowdin R, Barnholtz-Sloan J, Inskip PD, Ruder AM, Butler M, et al. Associations between Polymorphisms in DNA Repair Genes and Glioblastoma. Cancer Epidemiology Biomarkers & Prevention. 2009;18:1118–1126. doi: 10.1158/1055-9965.EPI-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, et al. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132–2138. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wen SX, Tang PZ, Zhang XM, Zhao D, Guo YL, et al. Association between genetic polymorphism in xeroderma pigmentosum G gene and risks of laryngeal and hypopharyngeal carcinomas. Acta Academiae Medicinae Sinicae. 2006;28:703–706. [PubMed] [Google Scholar]
- 69.Kumar R, Hoglund L, Zhao C, Forsti A, Snellman E, et al. Single nucleotide polymorphisms in the XPG gene: determination of role in DNA repair and breast cancer risk. Int J Cancer. 2003;103:671–675. doi: 10.1002/ijc.10870. [DOI] [PubMed] [Google Scholar]
- 70.Mechanic LE, Millikan RC, Player J, de Cotret AR, Winkel S, et al. Polymorphisms in nucleotide excision repair genes, smoking and breast cancer in African Americans and whites: a population-based case-control study. Carcinogenesis. 2006;27:1377–1385. doi: 10.1093/carcin/bgi330. [DOI] [PubMed] [Google Scholar]
- 71.Shen J, Desai M, Agrawal M, Kennedy DO, Senie RT, et al. Polymorphisms in nucleotide excision repair genes and DNA repair capacity phenotype in sisters discordant for breast cancer. Cancer Epidemiology Biomarkers and Prevention. 2006;15:1614–1619. doi: 10.1158/1055-9965.EPI-06-0218. [DOI] [PubMed] [Google Scholar]
- 72.Crew KD, Gammon MD, Terry MB, Fang FZ, Zablotska LB, et al. Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiology Biomarkers and Prevention. 2007;16:2033–2041. doi: 10.1158/1055-9965.EPI-07-0096. [DOI] [PubMed] [Google Scholar]
- 73.Jorgensen TJ, Visvanathan K, Ruczinski I, Thuita L, Hoffman S, et al. Breast cancer risk is not associated with polymorphic forms of xeroderma pigmentosum genes in a cohort of women from Washington County, Maryland. Breast Cancer Res Treat. 2007;101:65–71. doi: 10.1007/s10549-006-9263-3. [DOI] [PubMed] [Google Scholar]
- 74.Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, et al. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132–2138. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rajaraman P, Bhatti P, Doody MM, Simon SL, Weinstock RM, et al. Nucleotide excision repair polymorphisms may modify ionizing radiation-related breast cancer risk in US radiologic technologists. International Journal of Cancer. 2008;123:2713–2716. doi: 10.1002/ijc.23779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hsu MS, Yu JC, Wang HW, Chen ST, Hsiung CN, et al. Synergistic Effects of Polymorphisms in DNA Repair Genes and Endogenous Estrogen Exposure on Female Breast Cancer Risk. Annals of Surgical Oncology. 2010;17:760–771. doi: 10.1245/s10434-009-0802-0. [DOI] [PubMed] [Google Scholar]
- 77.Blankenburg S, Konig IR, Moessner R, Laspe P, Thoms KM, et al. No association between three xeroderma pigmentosum group C and one group G gene polymorphisms and risk of cutaneous melanoma. Eur J Hum Genet. 2005;13:253–255. doi: 10.1038/sj.ejhg.5201296. [DOI] [PubMed] [Google Scholar]
- 78.Li C, Hu Z, Liu Z, Wang LE, Strom SS, et al. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:2526–2532. doi: 10.1158/1055-9965.EPI-06-0672. [DOI] [PubMed] [Google Scholar]
- 79.Povey JE, Darakhshan F, Robertson K, Bisset Y, Mekky M, et al. DNA repair gene polymorphisms and genetic predisposition to cutaneous melanoma. Carcinogenesis. 2007;28:1087–1093. doi: 10.1093/carcin/bgl257. [DOI] [PubMed] [Google Scholar]
- 80.Figl A, Scherer D, Nagore E, Bermejo JL, Botella-Estrada R, et al. Single-nucleotide polymorphisms in DNA-repair genes and cutaneous melanoma. Mutation Research - Genetic Toxicology and Environmental Mutagenesis. 2010;702:8–16. doi: 10.1016/j.mrgentox.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 81.Ibarrola-Villava M, Pena-Chilet M, Fernandez LP, Aviles JA, Mayor M, et al. Genetic polymorphisms in DNA repair and oxidative stress pathways associated with malignant melanoma susceptibility. European Journal of Cancer. 2011. [DOI] [PubMed]
- 82.Thirumaran RK, Bermejo JL, Rudnai P, Gurzau E, Koppova K, et al. Single nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skin. Carcinogenesis. 2006;27:1676–1681. doi: 10.1093/carcin/bgi381. [DOI] [PubMed] [Google Scholar]
- 83.Wang LE, Li C, Strom SS, Goldberg LH, Brewster A, et al. Repair capacity for UV light induced DNA damage associated with risk of nonmelanoma skin cancer and tumor progression. Clin Cancer Res. 2007;13:6532–6539. doi: 10.1158/1078-0432.CCR-07-0969. [DOI] [PubMed] [Google Scholar]
- 84.Sanyal S, Festa F, Sakano S, Zhang Z, Steineck G, et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- 85.Garcia-Closas M, Malats N, Real FX, Welch R, Kogevinas M, et al. Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:536–542. doi: 10.1158/1055-9965.EPI-05-0749. [DOI] [PubMed] [Google Scholar]
- 86.Wu X, Gu J, Grossman HB, Amos CI, Etzel C, et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78:464–479. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Narter KF, Ergen A, Agachan B, Gormus U, Timirci O, et al. Bladder cancer and polymorphisms of DNA repair genes (XRCC1, XRCC3, XPD, XPG, APE1, hOGG1). Anticancer Res. 2009;29:1389–1393. [PubMed] [Google Scholar]
- 88.Rouissi K, Ouerhani S, Hamrita B, Bougatef K, Marrakchi R, et al. Smoking and Polymorphisms in Xenobiotic Metabolism and DNA Repair Genes are Additive Risk Factors Affecting Bladder Cancer in Northern Tunisia. Pathol Oncol Res. 2011. [DOI] [PubMed]
- 89.Shen M, Berndt SI, Rothman N, Demarini DM, Mumford JL, et al. Polymorphisms in the DNA nucleotide excision repair genes and lung cancer risk in Xuan Wei, China. Int J Cancer. 2005;116:768–773. doi: 10.1002/ijc.21117. [DOI] [PubMed] [Google Scholar]
- 90.Sakiyama T, Kohno T, Mimaki S, Ohta T, Yanagitani N, et al. Association of amino acid substitution polymorphisms in DNA repair genes TP53, POLI, REV1 and LIG4 with lung cancer risk. Int J Cancer. 2005;114:730–737. doi: 10.1002/ijc.20790. [DOI] [PubMed] [Google Scholar]
- 91.Hung RJ, Christiani DC, Risch A, Popanda O, Haugen A, et al. International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol Biomarkers Prev. 2008;17:3081–3089. doi: 10.1158/1055-9965.EPI-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, et al. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer. 2008;123:2095–2104. doi: 10.1002/ijc.23801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sugimura T, Kumimoto H, Tohnai I, Fukui T, Matsuo K, et al. Gene-environment interaction involved in oral carcinogenesis: Molecular epidemiological study for metabolic and DNA repair gene polymorphisms. Journal of Oral Pathology and Medicine. 2006;35:11–18. doi: 10.1111/j.1600-0714.2005.00364.x. [DOI] [PubMed] [Google Scholar]
- 94.Abbasi R, Ramroth H, Becher H, Dietz A, Schmezer P, et al. Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int J Cancer. 2009;125:1431–1439. doi: 10.1002/ijc.24442. [DOI] [PubMed] [Google Scholar]
- 95.Hongxia M, Hongping Y, Zhensheng L, Li EW, Erich MS, et al. Polymorphisms of XPG/ERCC5 and risk of squamous cell carcinoma of the head and neck. Pharmacogenetics and Genomics. 2011.
- 96.Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, et al. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008;638:146–153. doi: 10.1016/j.mrfmmm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 97.Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA, et al. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual’s susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2011. [DOI] [PubMed]
- 98.Canbay E, Cakmakoglu B, Zeybek U, Sozen S, Cacina C, et al. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin. 2011;27:1295–1302. doi: 10.1185/03007995.2011.573544. [DOI] [PubMed] [Google Scholar]
- 99.Shen M, Zheng T, Lan Q, Zhang Y, Zahm SH, et al. Polymorphisms in DNA repair genes and risk of non-Hodgkin lymphoma among women in Connecticut. Hum Genet. 2006;119:659–668. doi: 10.1007/s00439-006-0177-2. [DOI] [PubMed] [Google Scholar]
- 100.Hill DA, Wang SS, Cerhan JR, Davis S, Cozen W, et al. Risk of non-Hodgkin lymphoma (NHL) in relation to germline variation in DNA repair and related genes. Blood. 2006;108:3161–3167. doi: 10.1182/blood-2005-01-026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shen M, Purdue MP, Kricker A, Lan Q, Grulich AE, et al. Polymorphisms in DNA repair genes and risk of non-Hodgkin’s lymphoma in New South Wales, Australia. Haematologica. 2007;92:1180–1185. doi: 10.3324/haematol.11324. [DOI] [PubMed] [Google Scholar]
- 102.Hussain SK, Mu LN, Cai L, Chang SC, Park SL, et al. Genetic variation in immune regulation and DNA repair pathways and stomach cancer in China. Cancer Epidemiol Biomarkers Prev. 2009;18:2304–2309. doi: 10.1158/1055-9965.EPI-09-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Canbay E, Agachan B, Gulluoglu M, Isbir T, Balik E, et al. Possible associations of APE1 polymorphism with susceptibility and HOGG1 polymorphism with prognosis in gastric cancer. Anticancer Res. 2010;30:1359–1364. [PubMed] [Google Scholar]
- 104.Weiss JM, Weiss NS, Ulrich CM, Doherty JA, Voigt LF, et al. Interindividual variation in nucleotide excision repair genes and risk of endometrial cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2524–2530. doi: 10.1158/1055-9965.EPI-05-0414. [DOI] [PubMed] [Google Scholar]
- 105.Doherty JA, Weiss NS, Fish S, Fan W, Loomis MM, et al. Polymorphisms in Nucleotide Excision Repair Genes and Endometrial Cancer Risk. Cancer Epidemiol Biomarkers Prev. 2011. [DOI] [PMC free article] [PubMed]
- 106.Hooker S, Bonilla C, Akereyeni F, Ahaghotu C, Kittles RA. NAT2 and NER genetic variants and sporadic prostate cancer susceptibility in African Americans. Prostate Cancer Prostatic Dis. 2008;11:349–356. doi: 10.1038/sj.pcan.4501027. [DOI] [PubMed] [Google Scholar]
- 107.Le Morvan V, Longy M, Bonaiti-Pellie C, Bui B, Houede N, et al. Genetic polymorphisms of the XPG and XPD nucleotide excision repair genes in sarcoma patients. Int J Cancer. 2006;119:1732–1735. doi: 10.1002/ijc.22009. [DOI] [PubMed] [Google Scholar]
- 108.Biason P, Hattinger CM, Innocenti F, Talamini R, Alberghini M, et al. Nucleotide excision repair gene variants and association with survival in osteosarcoma patients treated with neoadjuvant chemotherapy. Pharmacogenomics J. 2011. [DOI] [PMC free article] [PubMed]
- 109.El-Zein R, Monroy CM, Etzel CJ, Cortes AC, Xing Y, et al. Genetic polymorphisms in DNA repair genes as modulators of Hodgkin disease risk. Cancer. 2009;115:1651–1659. doi: 10.1002/cncr.24205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rajaraman P, Hutchinson A, Wichner S, Black PM, Fine HA, et al. DNA repair gene polymorphisms and risk of adult meningioma, glioma, and acoustic neuroma. Neuro Oncol. 2010;12:37–48. doi: 10.1093/neuonc/nop012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pan J, Lin J, Izzo JG, Liu Y, Xing J, et al. Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway Carcinogenesis. 2009;30:785–792. doi: 10.1093/carcin/bgp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li LM, Zeng XY, Ji L, Fan XJ, Li YQ, et al. [Association of XPC and XPG polymorphisms with the risk of hepatocellular carcinoma]. Zhonghua Gan Zang Bing Za Zhi. 2010;18:271–275. doi: 10.3760/cma.j.issn.1007-3418.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 113.Vodicka P, Stetina R, Polakova V, Tulupova E, Naccarati A, et al. Association of DNA repair polymorphisms with DNA repair functional outcomes in healthy human subjects. Carcinogenesis. 2007;28:657–664. doi: 10.1093/carcin/bgl187. [DOI] [PubMed] [Google Scholar]
- 114.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 115.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nature genetics. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nature genetics. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 119.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nature genetics. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 120.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nature genetics. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nature genetics. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 122.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KKH, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nature genetics. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ERCC5 gene map labeled with nine SNPs have been studied for associations with cancer risk. (A) Six SNPs are located in the coding region, among which four are nsSNPs, of which two are synonymous SNPs; two SNPs are located in the 5′ untranslated region, and one SNP is located in the 3′ untranslated region; six SNPs are tagging SNPs. (B) Nine SNPs with a minor allelic frequency in different populations obtained from the dbSNP database.
(TIF)
Flow chart of included studies for this meta-analysis.
(TIF)
Funnel plot analysis to detect publication bias for ERCC5 Asp1104His under the recessive genetic models (A, His/His vs. Asp/Asp and B, His/His vs. Asp/His + Asp/Asp) for all 44 studies. Each point represents an individual study for the indicated association.
(TIF)
Summary of 24 SNPs of the ERCC5/XPG gene that have been studied for their associations with cancer risk.
(DOCX)
Summary of Studied SNPs in the eight NER genes reviewed in all published meta-analysis.
(DOCX)
Characteristics of the 44 references included in the meta-analysis for ERCC5 Asp1104His.
(DOCX)