Abstract
Dentinogenesis imperfecta (DGI) is a hereditary defect of dentin, a calcified tissue that is the most abundant component of teeth. Most commonly, DGI is manifested as a part of osteogenesis imperfecta (OI) or the phenotype is restricted to dental findings only. In the latter case, DGI is caused by mutations in the DSPP gene, which codes for dentin sialoprotein (DSP) and dentin phosphoprotein (DPP). Although these two proteins together constitute the majority of noncollagenous proteins of the dentin, little is known about their transcriptional regulation. Here we demonstrate that mice overexpressing the Trps1 transcription factor (Col1a1-Trps1 mice) in dentin-producing cells, odontoblasts, present with severe defects of dentin formation that resemble DGI. Combined micro–computed tomography (µCT) and histological analyses revealed tooth fragility due to severe hypomineralization of dentin and a diminished dentin layer with irregular mineralization in Col1a1-Trps1 mice. Biochemical analyses of noncollagenous dentin matrix proteins demonstrated decreased levels of both DSP and DPP proteins in Col1a1-Trps1 mice. On the molecular level, we demonstrated that sustained high levels of Trps1 in odontoblasts lead to dramatic decrease of Dspp expression as a result of direct inhibition of the Dspp promoter by Trps1. During tooth development Trps1 is highly expressed in preodontoblasts, but in mature odontoblasts secreting matrix its expression significantly decreases, which suggests a Trps1 role in odontoblast development. In these studies we identified Trps1 as a potent inhibitor of Dspp expression and the subsequent mineralization of dentin. Thus, we provide novel insights into mechanisms of transcriptional dysregulation that leads to DGI. © 2012 American Society for Bone and Mineral Research.
Keywords: MINERALIZATION, DENTINOGENESIS, TRPS1, DSPP, TRANSCRIPTION
Introduction
Dentin, one of the four major components of teeth, is a calcified tissue produced by neural crest–derived cells, odontoblasts, that form a continuous single-cell layer at the periphery of the dental pulp.1 The functional odontoblast has a columnar shape with a long cellular process and its major function is to secrete matrix and regulate matrix mineralization. Polarization of odontoblasts is required for the site-specific secretion of different components of extracellular matrix, and is a key factor in specifying the site of extracellular matrix mineralization.2 Newly synthesized, unmineralized matrix is referred to as predentin. Predentin separates the odontoblast layer from the mineralized matrix, dentin. Dentin is similar to bone in its matrix protein composition3; however, unlike bone, it does not undergo remodeling and does not participate in calcium homeostasis. The molecular hallmark of dentin is a high abundance of dentin sialophosphoprotein (DSPP), which is essential for dentin development and mineralization.
The most common hereditary defect of dentin is dentinogenesis imperfecta (DGI), which is a genetically and clinically heterogeneous disorder. Type I DGI (DGI-I) is associated with forms of osteogenesis imperfecta (OI) caused by mutations in one of the type I procollagen genes, COL1A1 or COL1A2, whereas the more severe DGI-II and DGI-III are isolated dentin disorders caused by mutations in the DSPP gene.4, 5 Whereas mutations in these genes coding for the most abundant dentin proteins are responsible for the majority of DGI cases, the etiology of other genetic disorders manifesting dentin abnormalities is still unknown.6, 7
The DSPP gene encodes dentin sialophosphoprotein, a member of the Small Integrin-Binding Ligand, N-Glycosylated (SIBLING) family of extracellular matrix proteins. The SIBLING family consists of dentin matrix protein 1 (DMP1), dentin sialophosphoprotein (DSPP), bone sialoprotein (BSP), osteopontin (OPN), and matrix extracellular phosphoglycoprotein (MEPE).8 SIBLING proteins undergo extensive posttranslational modifications that include phosphorylation and glycosylation.9 In addition to these modifications, the DSPP peptide is proteolytically cleaved to form dentin sialoprotein (DSP) and dentin phosphoprotein (DPP).10 DSP and DPP are the most abundant noncollagenous dentin proteins that play distinct roles in dentin mineralization. In vivo mouse model studies have demonstrated that DSP is involved in the initiation of dentin deposition and mineralization, whereas DPP is critical for mineral maturation.11 Thus far, among the genes expressing the noncollagenous dentin proteins, only mutations in the DSPP gene have been found in inherited dentin disorders.
Tooth defects similar to the DGI-III phenotype have been demonstrated in Dspp null mice.12 The dental phenotype of Dspp null mice is characterized by: hypomineralization, an enlarged pulp cavity, a widened predentin layer, and a reduced zone of dentin. Defective tooth mineralization and dentin formation were also observed in a number of knockout and odontoblast-specific transgenic mice. In a majority of these mice, decreased Dspp levels were observed as well, underscoring the importance of transcriptional regulation, posttranslational processing, and signal transduction of DSPP for dentin formation. The expression of many signaling molecules has been detected during odontoblast differentiation and their effect on the expression of dentin-specific genes has been studied. Transforming growth factor β (TGF-β), bone morphogenic protein (BMP), fibroblast growth factor (FGF), and Wnt signaling have been demonstrated to affect the expression of Dspp and odontoblast differentiation.13–17 However, the transcription factors that mediate responses to these signals during dentinogenesis in vivo are unknown. Promoters of the Dspp and Dmp1 genes and elements driving tissue-specific expression of these genes have been well characterized. Several transcription factors, including those with well-established roles in tooth development, have been demonstrated to interact with these promoters in vitro.18–20 Interestingly, the Dspp promoter, but not the Dmp1 promoter, contains an abundance of potential binding sites for GATA transcription factors.19, 20
TRPS1 is a GATA-type transcription factor expressed during tooth development.21 In humans, mutations in the TRPS1 gene cause an autosomal dominant craniofacial and skeletal dysplasia: tricho-rhino-phalangeal syndrome (TRPS).22 TRPS patients have characteristic facies, sparse hair, and skeletal abnormalities characteristic of disturbed endochondral bone formation. In addition, TRPS patients can present with dental phenotypes that include microdontia, malocclusion, delayed tooth eruption, and supernumerary teeth.21 During early tooth development, Trps1 is highly expressed in dental mesenchyme and after cell differentiation, in dental papilla and preodontoblasts. Interestingly, Trps1 expression is significantly decreased in differentiated secretory odontoblasts, which suggests a role for Trps1 in odontoblast maturation.21 However, its role in postnatal odontoblast development and function cannot be studied using a conventional gene disruption strategy, because Trps1 homozygous mutant mice die at birth.23, 24 Previous analyses of skeletal abnormalities in Trps1 mutant mice revealed that Trps1 is involved in chondrocyte proliferation and differentiation.24–26 We and others have demonstrated that Trps1 acts as a transcriptional repressor that antagonizes Ihh signaling during endochondral bone formation.25, 26 Moreover, we have shown that the processes of chondrocyte development and mineralization of perichondrium were uncoupled in Trps1ΔGT/ΔGT mice, resulting in advanced mineralization of perichondrium. This phenotype was corrected in Trps1ΔGT/+;Runx2+/− double heterozygous mice demonstrating the genetic interaction between the Trps1 and Runx2 transcription factors in developing endochondral bones.25
In these studies, using a transgenic mouse approach, we address the role of Trps1 in mineralization. We demonstrate that sustained high expression of Trps1 in mature odontoblasts has a deleterious effect on their function. Using a combination of histological, biochemical, and molecular analyses we discovered that Trps1 directly targets the Dspp gene and represses its expression in vivo, leading to a DGI-like phenotype in transgenic mice.
Materials and Methods
Generation of Col1a1-Trps1 transgenic mice and maintenance of mouse colony
We cloned the full-length V5-tagged Trps1 cDNA under the 2.3-kb fragment of the collagen type I promoter in a vector containing a tyrosinase expression cassette.27 Mice were maintained on an FVB/N background and housed in the Baylor College of Medicine Transgenic Mouse Facility according to standards of the Center of Comparative Medicine in full compliance with all applicable federal and state guidelines. Transgenic mice were identified by eye and fur color, and the genotypes were confirmed in the offspring of founder mice by PCR with primers specific for the Trps1 cDNA (gcagcatatgccagcatctttg) and WPRE sequence (actgacaattccgtggtgttgtcg). Transgenic mice were fed with a soft diet produced by milling of the standard mouse diet (Teklad, Harlan Laboratories Inc., Indianapolis, IN, USA). All studies were performed with prior approval of Institutional Animal Care and Use Committee.
Micro–computed tomography analyses
We fixed mouse heads in formalin for 48 hours followed by 70% ethanol washes and scanned using ScanCo micro–computed tomography (µCT)-40 instrument with 16-µm resolution. Images were reconstructed and analyzed using ScanCo software. All cross-section images of incisors were taken from the region located 0.8 mm posterior to the anterior terminus of the bone dorsal to the incisor.
Histology, immunohistochemistry, and RNA in situ hybridization
We fixed mouse heads in 4% paraformaldehyde. For P7 mice and older, heads were decalcified in 10% EDTA prior to paraffin embedding; 7-µm sections were stained with hematoxylin and eosin (H&E) according to standard protocols. Undecalcified tissues were embedded in methyl methacrylate; 5-µm sections were stained with either von Kossa reagent or Alizarin Red according to standard protocols. Immunohistochemistry (IHC) was done using enzymatic antigen retrieval and antibodies: anti-Trps1 (Abnova, Taipei, Taiwan, 1:50 dilution), anti-osteocalcin (Abcam, Cambridge, UK, 1:200 dilution), and anti-TNAP (Abcam, 1:100 dilution); and aminoethyl carbazole (AEC) chromogen detection (Invitrogen, Carlsbad, CA, USA). In situ hybridization (ISH) was performed as described.28 The generation of the Trps1 probe was as described.25 The Dspp probe was generated as described for the rat Dspp.29 The Dmp1 probe was generated by PCR amplification with primers specific for Dmp1 cDNA (F: aagactgtcattctccttgtg, R: cccgtactcttagagagtcc) and subsequent cloning of the PCR product into the EcoRV site of the pBluescriptK/S(+) plasmid. The antisense and sense Dmp1 probes were generated by in vitro transcription with T3 and T7 polymerases, respectively.
Analyses of SIBLING proteins
We performed extraction and analyses of dentin matrix proteins as described in detail.30, 31 Briefly, proteins were extracted from the dentin of 3-month-old males (n = 8 each group). For each analyzed genotype total protein extracts were combined and equal amounts of proteins from the wild-type (WT) and transgenic dentin were used in the comparative analyses. After removal of collagen fraction, noncollagenous proteins were separated into a total of 120 sequential elution fractions by a Q-sepharose ion-exchange column connected to a fast liquid protein chromatography (FLPC) system. Fractions 31 to 55 containing different types of the SIBLING proteins were analyzed on an SDS-PAGE gel and stained with Stains-All to visualize the proteins. This standardized procedure allows for comparison of the relative levels of SIBLING proteins in the analyzed samples.30–32 DSP protein was detected by Western blot with monoclonal anti-DSP 2G7.3 antibodies.33
Generation of stable cell lines using piggyBac transposon system
PiggyBac-expressing vector pCMV-piggyBac and transposon constructs were kindly provided by Dr. Mathew Wilson34 (Baylor College of Medicine). MDPC-23 cells were transfected with 1:5 ratio of transposase-expressing:transposon plasmids using Lipofectamine 2000 according to the manufacturer's protocol. Puromycin selection (5 µg/mL) of transposon-positive cells was begun 48 hours after transfection.
RNA extraction, cDNA synthesis, and qPCR for gene expression and chromatin immunoprecipitation
We extracted RNA using Trizol according to the manufacturer's protocol. One microgram (1 µg) of total RNA after DNase I treatment was converted to cDNA with a SuperScript III Reverse Transcriptase kit (Invitrogen). Quantitative PCR assays were performed using a LightCycler and FastStart DNA Master SYBR Green reaction mix (Roche, Indianapolis, IN, USA).
Chromatin immunoprecipitation assay
We performed chromatin immunoprecipitation (ChIP) using a Chromatin Immunoprecipitation Assay Kit (Millipore, Billerica, MA, USA) according to the manufacturer's protocol. V5 antibodies (Invitrogen) were used to immunoprecipitate V5-tagged Trps1-DNA complexes and normal mouse immunoglobulin G (IgG) (Santa Cruz, Santa Cruz, CA, USA) was used as a specificity control. Primers for ChIP analyses were designed to detect immunoprecipitated chromatin containing either one potential GATA binding site (fragments A and B) or two sites (fragment C). GAPDH was used as a specificity control. Primer sequences are available upon request. The ChIP assay was performed in triplicate.
Luciferase reporter Assay
We generated luciferase reporter constructs by PCR amplification of the mouse Dspp promoter fragments with the following forward primers F1: caggtagaactccatgagtttcag, F2: ctatggaagtgcagcttcgagg, F3: gttcaggcaagtactttttcc, F4: tacctcaggaatgataggggtc; and a reverse (R) primer: tgagagtggcacactgtcctg. PCR products were cloned into the SmaI restriction site of pGL3 basic vector (Promega). Point mutations in selected putative GATA consensus elements were generated by site-specific mutagenesis (QuikChange II Site-Directed Mutagenesis Kit; Stratagene, La Jolla, CA, USA) using the following mutagenesis oligonucleotides: M1 tgtacctcaggaataataggggtcttaaatagcc, M2 gatatatcacactgattatatatatttaagacacaaaa, M3 ctttaaacccccacataagggatcctaagcagtgattg, M4 cagggatcctaagcagttattggttgagaaaattatc. Reporter constructs were cotransfected with a plasmid expressing renilla luciferase (phRL-null; Promega, Madison, WI, USA). Luciferase activity was assayed 48 hours after transfection.
Statistical analysis
Statistical analyses of the weight of the 3-week-old and 4-week-old mice were done using one-way ANOVA and comparison to control (WT mice) was made using Dunnett's test. For the 5-week-old and 6-week-old mice, ANOVA on ranks was performed because the data were not normally distributed, and the Dunnett's test was used to compare the various groups to the control (WT mice). All other statistical analyses were done using paired t test.
Results
Generation and characterization of Col1a1-Trps1 transgenic mice
In our previous studies on the role of the Trps1 transcriptional repressor during endochondral bone formation, we demonstrated that Trps1 deficiency results in advanced mineralization of perichondrium. These data suggest that Trps1 acts as a negative regulator of mineralization. To address the role of the Trps1 transcription factor in the mineralization process in vivo, we generated transgenic mice expressing V5-tagged Trps1 from a cell-type specific Col1a1 promoter (Col1a1-Trps1 mice) that specifically drives transgene expression in osteoblasts and odontoblasts.35 To simplify phenotypic discrimination of transgenic animals by fur and eye color we included a marker gene–expressing cassette in our transgenic construct (Fig. 1A). This additional cassette contains the tyrosinase cDNA expressed from its endogenous promoter independently of the Col1a1-Trps1 expression.27 The tyrosinase minigene is also expressed in odontoblasts, resulting in pigment deposition in these cells; however, it does not impair dentin formation (Supplemental Fig. S1).
Fig. 1.
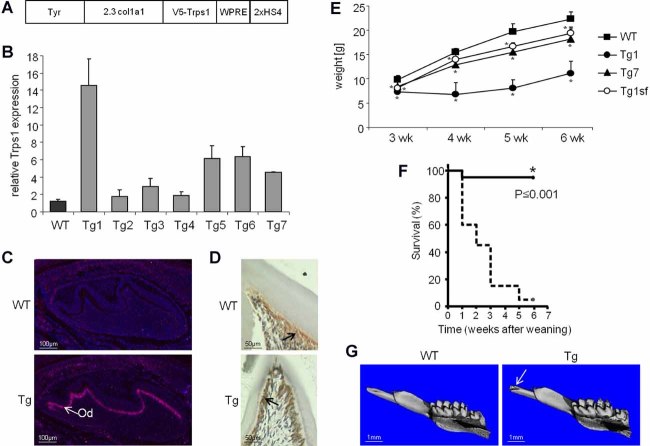
Gross characterization of Col1a1-Trps1 transgenic mice. (A) Schematic of the transgenic construct: Tyr = tyrosinase cassette containing tyrosinase promoter and cDNA for coat/eye color identification of transgenic animals27; 2.3 col1a1 = 2.3 kb fragment of the Col1a1 promoter; V5-Trps1 = V5-tagged Trps1 cDNA; WPRE = woodchuck hepatitis virus posttranscriptional regulatory element; 2xHS4 = 2 copies of HS4 chromatin insulator. (B) Analyses of Trps1 expression in calvarium of WT and Col1a1-Trps1 mice by qRT-PCR (P2). Bars represent mean value of relative Trps1 expression in WT mice and seven transgenic lines (Tg) normalized to GAPDH expression (n = 3). The value of Trps1 expression in WT animals was arbitrarily set to 1. Error bars represent standard deviations. (C) Analyses of Trps1 expression in teeth of WT and Col1a1-Trps1 mice (P2) by RNA in situ hybridization. Arrows indicate Trps1 expression in transgenic odontoblasts (Od). (D) Analyses of Trps1 expression in teeth of WT and Col1a1-Trps1 mice (P14) by immunohistochemistry. Arrows indicate Trps1 in odontoblasts. (E) Weight curves of WT and Col1a1-Trps1 mice (males) demonstrate growth retardation of transgenic animals. The phenotype is more severe in the transgenic line Tg1 that overexpresses Trps1 at higher levels than the Tg7 line. The growth retardation is corrected by providing soft food diet (open circles: Tg1 transgenic males fed with soft food). Data are shown as mean (n = 10 for each group except Tg1 4wk [4 weeks old], n = 6; Tg1 5wk [5 weeks old] and Tg1 6wk [6 weeks old], n = 4), error bars represent standard deviation, *p ≤ 0.05. (F) Kaplan-Mayer survival curve demonstrating postweaning lethality of Col1a1-Trps1 mice (n = 20 each group). The experiment was terminated at 6 weeks after weaning. (G) µCT images showing broken incisors (arrow) in Col1a1-Trps1 mice (3 weeks old).
We first analyzed all transgenic lines for levels of Trps1 expression in calvaria by quantitative RT-PCR on mRNA isolated from P2 mice (Fig. 1B). For detailed analyses of dental and skeletal phenotype we selected two lines (with different overexpression levels of Trps1) from a total of seven transgenic lines generated. The Tg1 line is the higher overexpressing line with Trps1 mRNA levels in calvaria 15 times higher than in WT littermates. As a second line we chose line Tg7 with lower levels of the Trps1 overexpression (4.5 times more than WT littermates). The phenotype of these transgenic lines is similar and the severity of abnormalities is directly correlated with the level of transgene overexpression. Col1a1-Trps1 mice are normal at birth and show no apparent defects during the first 3 weeks of life. However, shortly after weaning they present with a severe growth retardation phenotype and demonstrate a high incidence of lethality (Fig. 1E, F; Supplemental Fig. S2).
Additionally, for the Tg1 line we confirmed the transgene overexpression in odontoblasts using RNA ISH and IHC (Fig. 1C, D). Analyses of Trps1 expression at the protein level revealed that Trps1 persists at low levels throughout the life of odontoblasts in WT mice. Thus, in WT mice Trps1 is highly expressed in developing odontoblasts, whereas in secretory odontoblasts its expression is maintained at low levels. In contrast, in transgenic mice high expression of Trps1 is sustained in functional odontoblasts.
Overexpression of Trps1 in odontoblasts causes defective dentin formation
The most common cause of the postweaning growth retardation and lethality is problems in feeding. Considering that our transgene is expressed in teeth and bone, we performed gross analyses of the craniofacial skeleton using µCT to assess potential problems with mineralization. µCT analyses of 3-week-old mice revealed that all Col1a1-Trps1 mice presented with fractured incisors at the time of weaning (Fig. 1G). Additionally, cross-sectional images demonstrated dramatically reduced layers of dentin in both incisors and molars of transgenic versus age-matched WT littermates (Fig. 2A), which resulted in the enlarged dental pulp cavity. These results suggested that the growth retardation phenotype of Col1a1-Trps1 mice might be caused by difficulties with mastication. To compensate for this, we provided Col1a1-Trps1 mice with a soft food diet starting upon weaning, and continued throughout the life of the animals. Indeed, by reducing masticatory stresses on dentition, we were able to restore the normal lifespan of Col1a1-Trps1 mice. In addition, there was no significant difference in the total body weight of WT and transgenic animals fed with soft food (Fig. 1E; Supplemental Fig. S2). These results indicate that severe malnutrition due to tooth fragility was the underlying cause of postweaning growth retardation and mortality seen in Col1a1-Trps1 mice.
Fig. 2.
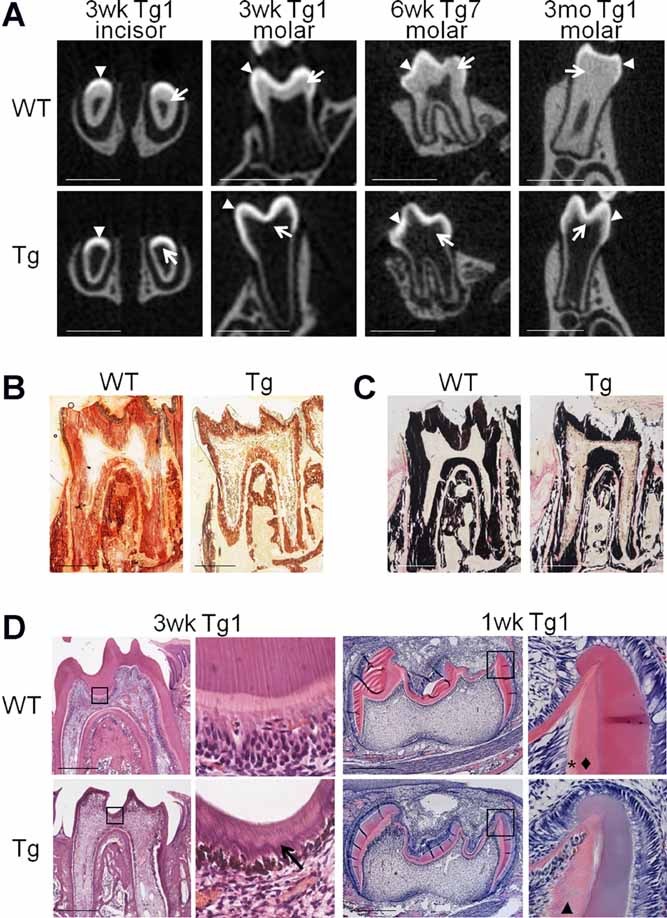
Defective dentin formation in Col1a1-Trps1 mice. (A) µCT images (cross sections) of WT and transgenic teeth (Tg1 and Tg7 line) demonstrating a diminished dentin layer in transgenic mice. Arrowheads: enamel, arrows: dentin. Transgenic animals were fed with soft food to assure proper nutrition. Scale bars = 1 mm. (B) Alizarin red staining of undecalcified molar sections (3 weeks old) demonstrating reduced dentin layer in transgenic teeth. Scale bars = 500 µm. (C) Von Kossa staining of undecalcified molar sections (3 weeks old) demonstrating reduced dentin layer in transgenic teeth. Scale bars = 500 µm. (D) Histological analyses of molars showing abnormal dentin formation in transgenic mice. In the dentin of 3-week-old (3wk) transgenic animals (erupted teeth) an irregular mineralization front is observed (arrow). In the 1-week-old (1wk) WT animals (unerupted teeth) two distinctive layers of predentin and dentin are present (asterisk and diamond, respectively), while in transgenic animals only one layer is present (arrowhead). Black boxes indicate areas magnified on the images to the right. Scale bars = 500 µm.
Although the soft food diet rescued postweaning growth retardation and lethality in Col1a1-Trps1 mice, the notably abnormal dentin phenotype did not improve, indicating that dentin formation was not correctable over time in animals expressing Trps1 in secretory odontoblasts. This was confirmed by µCT analyses of teeth from 6-week-old and 3-month-old Col1a1-Trps1 mice fed with soft food showing a significantly reduced dentin layer in transgenic animals compared to WT littermates (Fig. 2A). The defective dentin phenotype was further confirmed by histological analyses of molars. Alizarin Red and von Kossa staining of undecalcified teeth demonstrated a reduced layer of mineralized dentin in Col1a1-Trps1 mice compared to WT littermates (Fig. 2B, C). Notably less dentin in transgenic mice was also demonstrated on decalcified tissue sections (Fig. 2D). Additionally, at higher magnification of histological images the irregular mineralization front is clearly evident in the teeth of transgenic animals, suggesting that expression of Trps1 in mature odontoblasts affects not only matrix secretion, but also matrix mineralization. Disturbed dentin mineralization can be readily seen in unerupted teeth. In WT P7 molars, distinctive layers of predentin and dentin are apparent, whereas in the teeth of transgenic animals, this organized predentin-dentin layered structure is diminished (Fig. 2D). In summary, overexpression of Trps1 in secretory odontoblasts disturbs dentin formation, which leads to dramatically reduced dentin layers and tooth fragility. Concomitantly, µCT analyses of bone showed no significant differences between WT and transgenic mice (Supplemental Fig. S3). Thus, the phenotype of Col1a1-Trps1 mice appears to recapitulate features of dentinogenesis imperfecta in humans, though the mechanism was not yet clear.
Decreased levels of major noncollagenous dentin matrix proteins in Col1a1-Trps1 mice
In contrast to DGI-I, in which both bone and dentin are affected, presentation of DGI-II and DGI-III is restricted to dental abnormalities. These forms of DGI are caused by mutations in the DSPP gene. The DSPP gene encodes a protein that is proteolytically processed to form DSP and DPP, which are the most abundant noncollagenous proteins in the dentin matrix. To understand the molecular basis of abnormal dentin formation in Col1a1-Trps1 mice, we compared the relative abundance of DSP, DPP, and other SIBLING proteins in the dentin matrices between WT and transgenic animals.30–32 These analyses showed dramatically reduced levels of DPP in transgenic animals (Fig. 3A). At the same time, levels of the N-terminal fragment of DMP1 showed no apparent differences as evaluated by Stains-All staining. Since DSP is less sensitive to Stains-All staining, we compared levels of DSP between WT and transgenic mice by Western blot. These analyses also showed dramatically decreased levels of DSP (Fig. 3B).
Fig. 3.
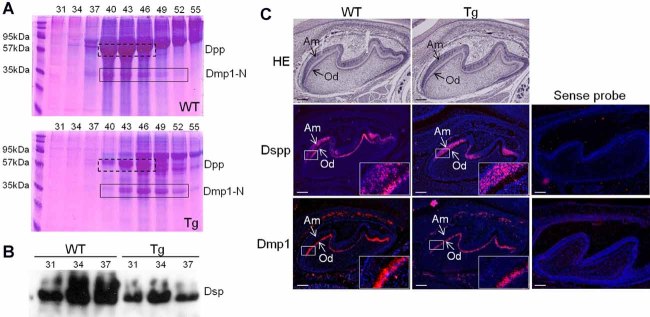
Decreased levels of DSP and DPP proteins, and Dspp gene expression in odontoblast of Col1a1-Trps1 mice. (A) Detection of SIBLING proteins isolated from dentin of 8-week-old WT and transgenic mice. Proteins were extracted from dentin and separated into 120 fractions using ion-exchange chromatography. Selected fractions containing SIBLING proteins were analyzed on an SDS-PAGE gel. SIBLING proteins were visualized by Stains-All. Numbers above the gel image indicate the chromatographic protein fractions analyzed on the gel; these fractions contained DPP and DMP1 fragments. The dashed-line box marks DPP protein, the solid-line box marks the N-terminal fragment of DMP1 protein. In transgenic mice a total level of DPP is decreased in comparison with WT mice, whereas levels of DMP1 show no difference. (B) Western blot with DSP antibodies shows decreased levels of DSP in transgenic dentin. (C) RNA in situ hybridization with probes specific for Dspp and Dmp1 (antisense molars, P2; sense molars P4). The boxed areas are magnified on the images in the right low corner of each respective image. In WT animals Dspp is expressed in both odontoblasts and ameloblasts, while in Col1a1-Trps1 mice Dspp expression in odontoblasts is significantly reduced. There is no difference in expression of Dmp1 between WT and transgenic odontoblasts. Od = odontoblasts; Am = ameloblasts. Scale bars = 100 µm.
To evaluate whether sustained high expression of Trps1 in secretory odontoblasts affects their differentiation status, we compared expression of other odontoblast markers in WT and transgenic teeth.36 Tissue nonspecific alkaline phosphatase (TNAP), which is expressed in preodontoblasts as well as in mature odontoblast in WT mice, was not changed in Col1a1-Trps1 teeth. More importantly, osteocalcin (OC), which is expressed in late odontoblasts, showed no apparent differences in expression between WT and transgenic teeth either (Fig. 4). In summary, analyses of dentin matrix proteins demonstrated that overexpression of Trps1 in mature odontoblasts leads specifically to decreased levels of the major noncollagenous dentin matrix proteins, DSP and DPP, without affecting the maturation of odontoblasts, because osteocalcin and DMP1 (markers of differentiated odontoblasts) remain unchanged.
Fig. 4.
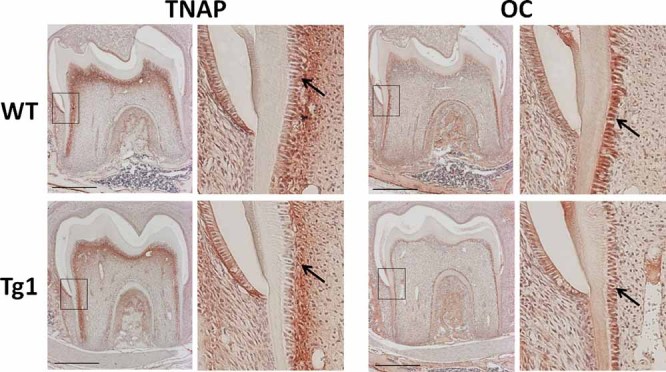
Immunohistochemical analysis of odontoblast markers in WT and Col1a1-Trps1 molars (P14). Tissue nonspecific alkaline phosphatase (TNAP) and osteocalcin (OC) are detected both in WT and Col1a1-Trps1 (Tg1) odontoblasts (arrows). Black boxes indicate area magnified on the images to the right of each respective image. Scale bars = 500 µm.
Trps1 represses Dspp expression through direct interaction with its promoter
Because both DSP and DPP proteins are expressed from the same gene, we next compared Dspp expression between WT and Col1a1-Trps1 transgenic teeth. We did this by RNA ISH on unerupted molars from P2 mice. As expected, in WT mice the Dspp transcripts were detected in odontoblasts and also in ameloblasts, where Dspp is transiently expressed (Fig. 3C). In contrast, Dspp expression in odontoblasts of transgenic mice was dramatically reduced but it was unchanged in ameloblasts. We also compared expression of the Dmp1 gene. The RNA ISH results were consistent with the protein analyses and no differences were observed in the expression of Dmp1 between WT and transgenic odontoblasts (Fig. 3C). These results demonstrate that overexpression of Trps1 in odontoblasts specifically represses expression of the Dspp gene in a cell autonomous fashion, suggesting that the Dspp gene might be a direct target of the Trps1 transcription factor.
To test whether Trps1 can repress Dspp by directly regulating its promoter, we first screened the mouse Dspp promoter for consensus elements of GATA transcription factors. We performed computational analyses of the 1.5-kb fragment of the Dspp promoter and 5′ untranslated region (5′UTR) using the TFSEARCH program, ver. 3.1.37 These analyses identified 11 consensus binding sites for GATA transcription factors in the Dspp promoter (Fig. 5A). Although GATA binding sites are fairly common among gene promoters, their relatively high abundance in the Dspp promoter suggests that GATA transcription factors play an important role in regulation of Dspp expression. To test whether Trps1 can directly bind to four GATA consensus elements that are the most proximal to the transcriptional start site, we performed ChIP assays in the MDPC-23 mouse odontoblastic cell line38 that stably expresses V5-tagged Trps1. We immunoprecipitated DNA-protein complexes using V5 antibodies, and performed quantitative PCR to evaluate the relative occupancy of Trps1 on the selected GATA elements in the Dspp promoter. Three independent ChIP experiments demonstrated an average enrichment of 2-fold to 3.5-fold for the analyzed Dspp promoter fragments in the DNA immunoprecipitated from cells expressing V5-Trps1 as compared with the control cell line lacking exogenous Trps1 (Fig. 5A). There was no difference in the abundance of DNA fragments between these cell lines for the Gapdh reference gene. ChIP assay with control antibodies (normal mouse IgG) showed no differences, providing additional support for specific binding of Trps1 to the Dspp promoter (Fig. 5A).
Fig. 5.
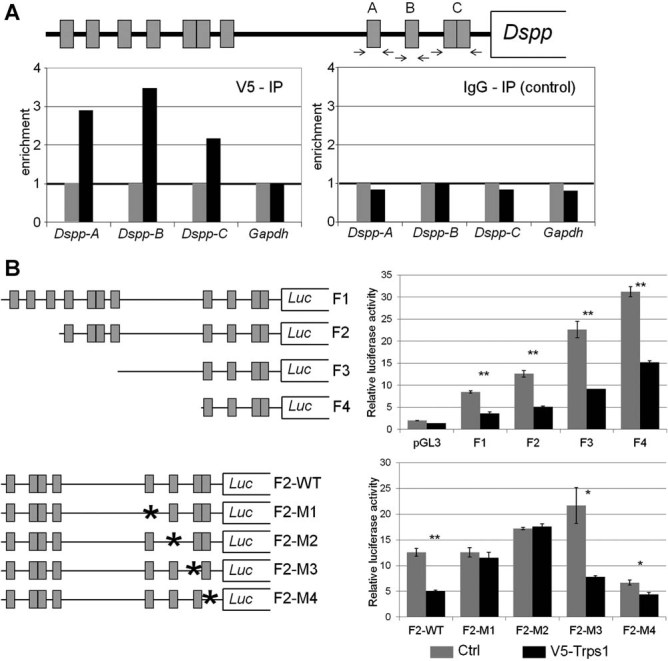
Role of putative GATA consensus elements in Trps1-mediated repression of the Dspp. (A) Interaction of Trps1 with putative GATA consensus sites in the mouse Dspp promoter analyzed by ChIP assay. Top: Schematic representation of the location of GATA elements (gray boxes) in the 1.5-kb fragment of the Dspp promoter. Arrows indicate localization of qPCR primers. Bottom: Results of qPCR on DNA immunoprecipitated with V5 antibodies (left panel) and control IgG antibodies (right panel) from the odontoblastic cell line stably overexpressing V5-tagged Trps1 (black bars) versus control cell line (gray bars). The values for the control cell line were arbitrarily set as 1 to better demonstrate the enrichment of the Dspp promoter sequences in the DNA immunoprecipitated from the cell line expressing V5-tagged Trps1. (B) Comparison of the Dspp promoter-luciferase reporter activity in control MDPC-23 cell line (Crtl, gray bars) and MDPC-23 cell line overexpressing V5-tagged Trps1 (V5-Trps1, black bars). Top: Schematics of Dspp-luciferase reporter constructs with different fragments of the Dspp promoter and their relative activities in MDPC-23 cell lines. Bottom: Schematic of the F2 Dspp-luciferase reporter constructs carrying single point mutations (asterisks) disrupting putative GATA consensus elements and their relative activity. Error bars represent the standard deviations of three independent experiments. Statistical significance was calculated using paired t test, *p ≤ 0.05, **p ≤ 0.01.
To address the role of these GATA consensus elements in Trps1-mediated repression of the Dspp, we took an in vitro approach. First, we generated four reporter constructs (plasmids F1, F2, F3, and F4), in which firefly luciferase was expressed from various fragments of the mouse Dspp promoter (Fig. 5B). Control and V5-Trps1 expressing MDPC-23 stable cell lines were transfected with each of these constructs and a renilla luciferase-expressing plasmid to normalize for the transfection efficiency. Analyses of the reporter activity detected a 2-fold reduction in the activity of each reporter in MDPC-23 cells overexpressing Trps1 in comparison with control cells. Furthermore, using the F2 Dspp reporter, we generated four additional reporter constructs carrying single point mutations in the putative GATA consensus elements that we analyzed for the Trps1 binding by ChIP (Fig. 5). Comparison of the mutant F2 Dspp reporters activity in control and Trps1 overexpressing MDPC-23 cells demonstrated that eliminating the potential Trps1 binding site in either fragment A (construct F2-M1) or B (construct F2-M2) results in an increase in the reporter activity in V5-Trps1 MDPC-23 cells to levels comparable with control cells (Fig. 5B). This suggests that these putative GATA consensus binding sites are required for Trps1-mediated repression of the F2 Dspp reporter.
Collectively, these results demonstrate that Trps1 can directly bind GATA consensus sites in the Dspp promoter and this activity results in the repression of Dspp expression in vivo.
Discussion
In our previous studies focused on the role of the Trps1 transcription factor in endochondral bone development, we uncovered that mice mutated for the Trps1 gene present with an advanced mineralization of perichondrium phenotype.25 This suggests that Trps1 is a negative regulator of mineralization. To understand the function of Trps1 in mineralizing tissues we generated Col1a1-Trps1 transgenic mice overexpressing Trps1 specifically in osteoblasts and odontoblasts. Analyses of these mice demonstrated that Trps1 acts as a tissue-specific repressor of mineralization, with a strong effect on dentin, but with little influence on bone. Further, we determined that this context-dependent activity is specified by direct repression of the Dspp gene by the Trps1 transcription factor. As a result, the dentin matrix in Col1a1-Trps1 mice exhibits dramatically reduced levels of DSP and DPP proteins, and, consequently, severely impaired mineralization. Hence, the phenotype of Col1a1-Trps1 mice recapitulates main features of Dspp knockout (KO) mice and the human genetic disorder DGI.
Interestingly, the bone tissue of Col1a1-Trps1 mice is not significantly affected, implying the tissue-specific effect of Trps1 on mineralization. This specificity might be determined by different pools of target genes in osteoblasts and odontoblasts. Alternatively, Trps1 may repress genes critical for dentinogenesis but not for bone formation. Although dentin is similar to bone in terms of the extent of its mineralization and the expression of many of the same transcription factors and matrix proteins, high expression of the Dspp gene characterizes dentin as a unique mesenchyme-derived mineralized tissue.39–41 More importantly, the key role of Dspp in dentin formation is underscored by severe dentin impairment in cases where the DSPP gene is disrupted. For comparison, mutations in type I procollagen genes, COL1A1 or COL1A2, affect both bone and dentin, demonstrating the requirement of collagen type I in both tissues. Thus, the downregulation of Dspp maybe be the major factor determining the tissue-specific Trps1-mediated repression of mineralization. Another important difference between bone and dentin is that dentin does not undergo remodeling, thus the quantity of dentin solely reflects odontoblast activity, whereas overall bone mass is a net result of the opposing activities of osteoblasts and osteoclasts. However, in our studies we did not measure the dynamic parameters of bone remodeling; therefore, we cannot exclude that the osteoblast activity is affected by Trps1 overexpression.
Recently, Piscopo and colleagues42 showed that Trps1 binds elements in the osteocalcin promoter and represses osteocalcin promoter-driven reporter expression in various osteogenic cell lines. In addition, they demonstrated that osteocalcin expression is inversely correlated with levels of Trps1 in bone marrow stromal cells undergoing osteogenic differentiation. Considering these data, and the fact that osteocalcin is also a marker of mature odontoblasts, we compared expression of osteocalcin in WT and Col1a1-Trps1 teeth using immunohistochemistry. However, we did not observe significant differences between WT and transgenic mice, indicating that overexpression of Trps1 does not affect osteocalcin expression in vivo. Additionally, these data, together with unchanged DMP1 expression, suggest that overexpression of Trps1 does not cause the arrest of odontoblast differentiation; rather the dental phenotype of Col1a1-Trps1 mice results from impaired odontoblast function.
Consistent with significantly reduced levels of DSPP, dentin abnormalities observed in Col1a1-Trps1 mice are similar to those reported for Dspp KO mice, a mouse model of DGI. Dspp deficiency caused either by genetic disruption of the gene or by Trps1-mediated repression of Dspp expression results in enlarged pulp chambers and reduced dentin layers that are evident on µCT images of teeth. Moreover, at the histological level, decreased dentin and an irregular mineralization front are apparent in both mouse models, which further confirms defective dentin formation. In contrast to Dspp KO mice, we have not observed calcospherites or sporadic unmineralized areas in dentin of Col1a1-Trps1 mice. This might be due to residual Dspp expression in transgenic mice, because, unlike Dspp KO mice in which expression of the gene is completely abolished, Col1a1-Trps1 transgenic mice express very low levels of DSPP that accumulates in dentin over time. Alternatively, it is possible that Trps1 overexpression in secretory odontoblasts may also affect expression of other genes, resulting in the phenotype of Col1a1-Trps1 transgenic mice differing from the phenotype of Dspp KO mice.
DGI can be phenocopied in genetically manipulated mice by either mutating the Dspp gene or by diminishing Dspp expression indirectly by dysregulating other genes. The latter animal models provide valuable insight into the molecular mechanisms controlling the expression of this highly tissue-specific gene. In spite of its critical role in dentin formation, little is known about factors that determine spatial and temporal expression of Dspp in vivo. Animal studies have previously demonstrated that activation of TGF-β signaling represses the Dspp gene. Although TGF-β signaling is necessary for odontoblast differentiation,17, 43 overexpression of TGF-β1 in secretory odontoblasts results in decreased Dspp expression and the DGI phenotype in transgenic mice.13 Additional studies in odontoblastic cells have revealed that Smad3 mediates this inhibitory effect of TGF-β on the Dspp gene16; however, transcription factors that directly repress Dspp in response to TGF-β are unknown. Whether or not Trps1 is the mediator of TGF-β signaling in odontoblasts, remains to be determined. Trps1 is indeed a good candidate, given its high expression in dental mesenchyme and preodontoblasts in comparison with mature odontoblasts, in which it is expressed at low levels. This suggests that, akin to TGF-β signaling, Trps1 is involved in odontoblast differentiation, whereas in mature odontoblasts it suppresses dentin formation. In addition, the unusually high number of GATA elements in the Dspp promoter suggests an important role for this family of transcription factors in its regulation. Dramatically reduced levels of Dspp expression in odontoblasts overexpressing Trps1 demonstrate that this GATA-type transcription factor is dominant over the activity of other transcription factors stimulating Dspp expression. These transcription factors remain to be determined, because, in spite of extensive in vitro analyses of various transcription factors as candidate regulators of Dspp expression, none has been validated in vivo.15, 41, 44–46
Our transgenic mouse approach, in which high Trps1 expression is sustained in secretory odontoblasts, may help to understand the molecular pathology underlying dentin defects caused by transcriptional dysregulation. Such a mechanism has been identified in Ambras syndrome, a rare disorder caused by a translocation with a break point in the Trps1 promoter, leading to the aberrant pattern of Trps1 expression.47 Unlike TRPS patients that are characterized by sparse slow-growing hair, the most prominent feature of Ambras syndrome is extensive growth of hair over the entire body (hypertrichosis). Interestingly, although not described in detail, Ambras patients also present with a dental phenotype, indicating that dysregulation of the spatial and/or temporal pattern of Trps1 expression results in dental abnormalities in humans. Whether the aberrant expression pattern of Trps1 contributes to other disorders with dental presentations remains to be determined. For example, the genetic cause of spondylometaphyseal dysplasia with DGI (odontochondrodysplasia [ODCD]) is still unknown.48 Interestingly, in this disorder both endochondral bone formation and dentinogenesis are affected, pointing to involvement of a gene that functions in both tissues. In our studies, we identified the Trps1 transcription factor, expressed during endochondral bone as well as tooth development, as a potent inhibitor of dentin formation, thus it is plausible that Trps1 and a gene responsible for ODCD function in the same pathway. Our results not only identified the Dspp gene as the first Trps1 target gene in odontoblasts, but also shed light on the in vivo regulation of this gene, which is critical for dentin formation. Thus, revealing the Trps1 molecular network in odontoblasts will certainly contribute to greater understanding of the molecular mechanisms of dentinogenesis and associated human dental disorders.
Acknowledgments
We thank Dr. Mathew Wilson (Baylor College of Medicine) for kindly providing the transposon construct and the piggyBac-expressing plasmid. We thank Tony Winters, Elda Munivez, Yuqing Chen, Margaretha Guenther, and the UAB Center for Metabolic Bone Disease Comparative Pathology Core for excellent technical assistance. This work was supported by the University of Alabama at Birmingham Faculty Development Grant Program (to DN) and Rolanette and Berdon Lawrence Bone Disease Program of Texas (to DN), National Institutes of Health (R03AR057128 to DN; DE016990 and HD22657 to BL; DE005092 to CQ), and the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (HD024064 to BL).
Authors' roles: Study design: DN, CQ, and BL. Study conduct: DN, YS, IM, BD, and TB. Data collection: DN, YS, BD, and TB. Data analysis: DN, YS, IM, and BD. Data interpretation: DN, YS, IM, RDS, CQ, and BL. Drafting manuscript: DN, YS, CQ, and TB. Revising manuscript content: DN, RDS, CQ, and BL. Approving final version of manuscript: DN, YS, IM, TB, RDS, CQ, and BL. DN takes responsibility for the integrity of the data analysis.
Disclosures
All authors state that they have no conflicts of interest.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.D'Souza DR, Qin C. Development of the pulpdentin complex. In: Hargreaves KM, Goodis HE, Tay FR, editors. Seltzer and Bender's dental pulp. 2nd ed. Chicago: Quintessence; 2012. pp. 1–26. [Google Scholar]
- 2.Arana-Chavez VE, Massa LF. Odontoblasts: the cells forming and maintaining dentine. Int J Biochem Cell Biol. 2004;8:1367–73. doi: 10.1016/j.biocel.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Embery G, Hall R, Waddington R, Septier D, Goldberg M. Proteoglycans in dentinogenesis. Crit Rev Oral Biol Med. 2001;4:331–49. doi: 10.1177/10454411010120040401. [DOI] [PubMed] [Google Scholar]
- 4.Barron MJ, McDonnell ST, Mackie I, Dixon MJ. Hereditary dentine disorders: dentinogenesis imperfecta and dentine dysplasia. Orphanet J Rare Dis. 2008:31. doi: 10.1186/1750-1172-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu JC, Simmer JP. Developmental biology and genetics of dental malformations. Orthod Craniofac Res. 2007;2:45–52. doi: 10.1111/j.1601-6343.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg M, Septier D, Oldberg A, Young MF, Ameye LG. Fibromodulin-deficient mice display impaired collagen fibrillogenesis in predentin as well as altered dentin mineralization and enamel formation. J Histochem Cytochem. 2006;5:525–37. doi: 10.1369/jhc.5A6650.2005. [DOI] [PubMed] [Google Scholar]
- 7.Kim JW, Simmer JP. Hereditary dentin defects. J Dent Res. 2007;5:392–9. doi: 10.1177/154405910708600502. [DOI] [PubMed] [Google Scholar]
- 8.Qin C, Brunn JC, Jones J, George A, Ramachandran A, Gorski JP, Butler WT. A comparative study of sialic acid-rich proteins in rat bone and dentin. Eur J Oral Sci. 2001;2:133–41. doi: 10.1034/j.1600-0722.2001.00001.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Chen L, Jahangiri A, Chen B, Wu Y, Chuang HH, Qin C, MacDougall M. Expression and processing of small integrin-binding ligand N-linked glycoproteins in mouse odontoblastic cells. Arch Oral Biol. 2008;9:879–89. doi: 10.1016/j.archoralbio.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin C, Brunn JC, Cook RG, Orkiszewski RS, Malone JP, Veis A, Butler WT. Evidence for the proteolytic processing of dentin matrix protein 1. Identification and characterization of processed fragments and cleavage sites. J Biol Chem. 2003;36:34700–8. doi: 10.1074/jbc.M305315200. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki S, Sreenath T, Haruyama N, Honeycutt C, Terse A, Cho A, Kohler T, Muller R, Goldberg M, Kulkarni AB. Dentin sialoprotein and dentin phosphoprotein have distinct roles in dentin mineralization. Matrix Biol. 2009;4:221–9. doi: 10.1016/j.matbio.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sreenath T, Thyagarajan T, Hall B, Longenecker G, D'Souza R, Hong S, Wright JT, MacDougall M, Sauk J, Kulkarni AB. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem. 2003;27:24874–80. doi: 10.1074/jbc.M303908200. [DOI] [PubMed] [Google Scholar]
- 13.Thyagarajan T, Sreenath T, Cho A, Wright JT, Kulkarni AB. Reduced expression of dentin sialophosphoprotein is associated with dysplastic dentin in mice overexpressing transforming growth factor-beta 1 in teeth. J Biol Chem. 2001;14:11016–20. doi: 10.1074/jbc.M010502200. [DOI] [PubMed] [Google Scholar]
- 14.Yamashiro T, Zheng L, Shitaku Y, Saito M, Tsubakimoto T, Takada K, Takano-Yamamoto T, Thesleff I. Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation. 2007;5:452–62. doi: 10.1111/j.1432-0436.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen S, Gluhak-Heinrich J, Martinez M, Li T, Wu Y, Chuang HH, Chen L, Dong J, Gay I, MacDougall M. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J Biol Chem. 2008;28:19359–70. doi: 10.1074/jbc.M709492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He WX, Niu ZY, Zhao SL, Jin WL, Gao J, Smith AJ. TGF-beta activated Smad signalling leads to a Smad3-mediated down-regulation of DSPP in an odontoblast cell line. Arch Oral Biol. 2004;11:911–8. doi: 10.1016/j.archoralbio.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Oka S, Oka K, Xu X, Sasaki T, Bringas P, Jr, Chai Y. Cell autonomous requirement for TGF-beta signaling during odontoblast differentiation and dentin matrix formation. Mech Dev. 2007;6:409–15. doi: 10.1016/j.mod.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, Unterbrink A, Kadapakkam S, Dong J, Gu TT, Dickson J, Chuang HH, MacDougall M. Regulation of the cell type-specific dentin sialophosphoprotein gene expression in mouse odontoblasts by a novel transcription repressor and an activator CCAAT-binding factor. J Biol Chem. 2004;40:42182–91. doi: 10.1074/jbc.M402476200. [DOI] [PubMed] [Google Scholar]
- 19.Chen S, Gu TT, Sreenath T, Kulkarni AB, Karsenty G, MacDougall M. Spatial expression of Cbfa1/Runx2 isoforms in teeth and characterization of binding sites in the DSPP gene. Connect Tissue Res. 2002;2–3:338–44. doi: 10.1080/03008200290000691. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Inozentseva-Clayton N, Dong J, Gu TT, MacDougall M. Binding of two nuclear factors to a novel silencer element in human dentin matrix protein 1 (DMP1) promoter regulates the cell type-specific DMP1 gene expression. J Cell Biochem. 2004;2:332–49. doi: 10.1002/jcb.20051. [DOI] [PubMed] [Google Scholar]
- 21.Kantaputra P, Miletich I, Ludecke HJ, Suzuki EY, Praphanphoj V, Shivdasani R, Wuelling M, Vortkamp A, Napierala D, Sharpe PT. Tricho-rhino-phalangeal syndrome with supernumerary teeth. J Dent Res. 2008;11:1027–31. doi: 10.1177/154405910808701102. [DOI] [PubMed] [Google Scholar]
- 22.Momeni P, Glockner G, Schmidt O, von Holtum D, Albrecht B, Gillessen-Kaesbach G, Hennekam R, Meinecke P, Zabel B, Rosenthal A, Horsthemke B, Ludecke HJ. Mutations in a new gene, encoding a zinc-finger protein, cause tricho-rhino-phalangeal syndrome type I. Nat Genet. 2000;1:71–4. doi: 10.1038/71717. [DOI] [PubMed] [Google Scholar]
- 23.Malik TH, Von Stechow D, Bronson RT, Shivdasani RA. Deletion of the GATA domain of TRPS1 causes an absence of facial hair and provides new insights into the bone disorder in inherited tricho-rhino-phalangeal syndromes. Mol Cell Biol. 2002;24:8592–600. doi: 10.1128/MCB.22.24.8592-8600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suemoto H, Muragaki Y, Nishioka K, Sato M, Ooshima A, Itoh S, Hatamura I, Ozaki M, Braun A, Gustafsson E, Fassler R. Trps1 regulates proliferation and apoptosis of chondrocytes through Stat3 signaling. Dev Biol. 2007;2:572–81. doi: 10.1016/j.ydbio.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Napierala D, Sam K, Morello R, Zheng Q, Munivez E, Shivdasani RA, Lee B. Uncoupling of chondrocyte differentiation and perichondrial mineralization underlies the skeletal dysplasia in tricho-rhino-phalangeal syndrome. Hum Mol Genet. 2008;14:2244–54. doi: 10.1093/hmg/ddn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wuelling M, Kaiser FJ, Buelens LA, Braunholz D, Shivdasani RA, Depping R, Vortkamp A. Trps1, a regulator of chondrocyte proliferation and differentiation, interacts with the activator form of Gli3. Dev Biol. 2009;1:40–53. doi: 10.1016/j.ydbio.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Zhou G, Zheng Q, Engin F, Munivez E, Chen Y, Sebald E, Krakow D, Lee B. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci U S A. 2006;50:19004–9. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morello R, Zhou G, Dreyer SD, Harvey SJ, Ninomiya Y, Thorner PS, Miner JH, Cole W, Winterpacht A, Zabel B, Oberg KC, Lee B. Regulation of glomerular basement membrane collagen expression by LMX1B contributes to renal disease in nail patella syndrome. Nat Genet. 2001;2:205–8. doi: 10.1038/84853. [DOI] [PubMed] [Google Scholar]
- 29.Baba O, Qin C, Brunn JC, Jones JE, Wygant JN, McIntyre BW, Butler WT. Detection of dentin sialoprotein in rat periodontium. Eur J Oral Sci. 2004;2:163–70. doi: 10.1111/j.0909-8836.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- 30.Huang B, Sun Y, Maciejewska I, Qin D, Peng T, McIntyre B, Wygant J, Butler WT, Qin C. Distribution of SIBLING proteins in the organic and inorganic phases of rat dentin and bone. Eur J Oral Sci. 2008;2:104–12. doi: 10.1111/j.1600-0722.2008.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, Gandhi V, Prasad M, Yu W, Wang X, Zhu Q, Feng JQ, Hinton RJ, Qin C. Distribution of small integrin-binding ligand, N-linked glycoproteins (SIBLING) in the condylar cartilage of rat mandible. Int J Oral Maxillofac Surg. 2011;3:272–81. doi: 10.1016/j.ijom.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y, Lu Y, Chen L, Gao T, D'Souza R, Feng JQ, Qin C. DMP1 processing is essential to dentin and jaw formation. J Dent Res. 2011;5:619–24. doi: 10.1177/0022034510397839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin C, Brunn JC, Baba O, Wygant JN, McIntyre BW, Butler WT. Dentin sialoprotein isoforms: detection and characterization of a high molecular weight dentin sialoprotein. Eur J Oral Sci. 2003;3:235–42. doi: 10.1034/j.1600-0722.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 34.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;1:139–45. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 35.Rossert J, Eberspaecher H, de Crombrugghe B. Separate cis-acting DNA elements of the mouse pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J Cell Biol. 1995;5:1421–32. doi: 10.1083/jcb.129.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narayanan K, Srinivas R, Ramachandran A, Hao J, Quinn B, George A. Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1. Proc Natl Acad Sci U S A. 2001;8:4516–21. doi: 10.1073/pnas.081075198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;1:362–7. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanks CT, Fang D, Sun Z, Edwards CA, Butler WT. Dentin-specific proteins in MDPC-23 cell line. Eur J Oral Sci. 1998:260–6. doi: 10.1111/j.1600-0722.1998.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 39.Butler WT, Brunn JC, Qin C. Dentin extracellular matrix (ECM) proteins: comparison to bone ECM and contribution to dynamics of dentinogenesis. Connect Tissue Res. 2003:171–8. [PubMed] [Google Scholar]
- 40.D'Souza RN, Cavender A, Sunavala G, Alvarez J, Ohshima T, Kulkarni AB, MacDougall M. Gene expression patterns of murine dentin matrix protein 1 (Dmp1) and dentin sialophosphoprotein (DSPP) suggest distinct developmental functions in vivo. J Bone Miner Res. 1997;12:2040–9. doi: 10.1359/jbmr.1997.12.12.2040. [DOI] [PubMed] [Google Scholar]
- 41.Galler KM, Yasue A, Cavender AC, Bialek P, Karsenty G, D'Souza RN. A novel role for Twist-1 in pulp homeostasis. J Dent Res. 2007;10:951–5. doi: 10.1177/154405910708601007. [DOI] [PubMed] [Google Scholar]
- 42.Piscopo DM, Johansen EB, Derynck R. Identification of the GATA factor TRPS1 as a repressor of the osteocalcin promoter. J Biol Chem. 2009;46:31690–703. doi: 10.1074/jbc.M109.052316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Souza RN, Happonen RP, Flanders KC, Butler WT. Histochemical localization of transforming growth factor-beta 1 in developing rat molars using antibodies to different epitopes. J Biol Buccale. 1990;4:299–306. [PubMed] [Google Scholar]
- 44.Chen S, Rani S, Wu Y, Unterbrink A, Gu TT, Gluhak-Heinrich J, Chuang HH, MacDougall M. Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J Biol Chem. 2005;33:29717–27. doi: 10.1074/jbc.M502929200. [DOI] [PubMed] [Google Scholar]
- 45.Narayanan K, Ramachandran A, Peterson MC, Hao J, Kolsto AB, Friedman AD, George A. The CCAAT enhancer-binding protein (C/EBP)beta and Nrf1 interact to regulate dentin sialophosphoprotein (DSPP) gene expression during odontoblast differentiation. J Biol Chem. 2004;44:45423–32. doi: 10.1074/jbc.M405031200. [DOI] [PubMed] [Google Scholar]
- 46.Cho YD, Yoon WJ, Woo KM, Baek JH, Park JC, Ryoo HM. The canonical BMP signaling pathway plays a crucial part in stimulation of dentin sialophosphoprotein expression by BMP-2. J Biol Chem. 2010;47:36369–76. doi: 10.1074/jbc.M110.103093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fantauzzo KA, Tadin-Strapps M, You Y, Mentzer SE, Baumeister FA, Cianfarani S, Van Maldergem L, Warburton D, Sundberg JP, Christiano AM. A position effect on TRPS1 is associated with Ambras syndrome in humans and the Koala phenotype in mice. Hum Mol Genet. 2008;22:3539–51. doi: 10.1093/hmg/ddn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldblatt J, Carman P, Sprague P. Unique dwarfing, spondylometaphyseal skeletal dysplasia, with joint laxity and dentinogenesis imperfecta. Am J Med Genet. 1991;2:170–2. doi: 10.1002/ajmg.1320390211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.