Abstract
Tumor-derived mutant KRAS (v-Ki-ras-2 Kirsten rat sarcoma viral oncogene) oncoprotein is a critical driver of cancer phenotypes and a potential biomarker for many epithelial cancers. Targeted mass spectrometry analysis by multiple reaction monitoring (MRM) enables selective detection and quantitation of wild-type and mutant KRAS proteins in complex biological samples. A recently described immunoprecipitation approach (Proc. Nat. Acad. Sci. 2011, 108, 2444–2449) can be used to enrich KRAS for MRM analysis, but requires large protein inputs (2–4 mg). Here we describe sodium dodecyl sulfate-polyacrylamide gel electrophoresis-based enrichment of KRAS in a low molecular weight (20 –25 kDa) protein fraction prior to MRM analysis (GeLC-MRM). This approach reduces background proteome complexity, thus allowing mutant KRAS to be reliably quantified in low protein inputs (5–50 μg). GeLC-MRM detected KRAS mutant variants (G12D, G13D, G12V, G12S) in a panel of cancer cell lines. GeLC-MRM-analysis of wild-type and mutant was linear with respect to protein input and showed low variability across process replicates (CV = 14%). Concomitant analysis of a peptide from the highly similar HRAS and NRAS proteins enabled correction of KRAS-targeted measurements for contributions from these other proteins. KRAS peptides were also quantified in fluid from benign pancreatic cysts and pancreatic cancers at concentrations from 0.08 – 1.1 fmol/μg protein. GeLC-MRM provides a robust, sensitive approach to quantitation of mutant proteins in complex biological samples.
Keywords: KRAS, GeLC-MRM, targeted proteomics, colorectal cancer, pancreatic cyst fluid
Introduction
Although thousands of mutations have been identified in various cancers, relatively few play a causative role in tumor formation.1, 2 The v-Ki-ras-2 Kirsten rat sarcoma viral oncogene (KRAS) has been implicated as a “driver gene” and is one of the most frequently mutated genes in colorectal, lung, pancreatic and other cancers.3 KRAS is a member of the RAS GTPase protein family and missense mutations in KRAS at codons 12, 13 and 61 result in constitutive activation of the MAPK signaling pathway.2
Although KRAS mutations are readily detectable at the DNA level using PCR-based approaches, measurement of corresponding mutant protein levels offer several key advantages. Mutant proteins may serve as cancer biomarkers, as they originate at the tumor site. Mutation of KRAS is an early event in the development some cancer types, such as colorectal and pancreatic cancer.3 Quantitative measurements of wild-type and mutant protein forms also enable mechanistic studies of how mutations contribute to cancer phenotypes.
Detection of mutant proteins in complex biological samples is challenging. Activating KRAS mutations are missense (e.g., G12D, G13D) and antibodies capable of distinguishing wildtype and mutant forms have not been reported. KRAS has high sequence homology with the RAS family proteins NRAS and HRAS, further complicating antibody-based approaches. Mass spectrometry (MS)-based analyses represent the most practical strategy for mutant KRAS protein detection. However, KRAS is a relatively low abundance protein that is difficult to detect in complex biological samples, thus necessitating enrichment prior to MS analysis.
Wang and colleagues recently demonstrated that liquid-chromatography–multiple reaction monitoring (MRM) MS can be used to analyze both wild-type and mutant KRAS proteins in cancer cell lines, tumor tissues and pancreatic cyst fluid.4 They enriched KRAS by immunoprecipitation, which achieved reliable detection of wild-type and mutant KRAS from 2–4 mg input protein. Although their report provided clear proof of concept for an MRM-based approach to variant oncoprotein analysis, their approach is dependent on the availability of antibody reagents capable of immunoprecipitating the target proteins in both wild-type and variant forms.
Here we describe a simple, generally-applicable method for analysis of KRAS mutant proteins in complex biological samples. By employing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)-based enrichment of low molecular weight proteins (20–25 kDa), KRAS wild type and mutant proteins can be detected and quantified by MRM. This GeLC-MRM approach circumvents the need for antibody-based enrichment and requires only low microgram protein input. We demonstrate the application of this method to analyze KRAS proteins in cancer cell lines and pancreatic cyst fluids.
Materials and Methods
Cell culture and harvesting
All cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA) and were grown and harvested within six months of date of purchase, or grown from frozen stocks that had been made within six months of purchase. All cell lines were grown in 10% FBS and penicillin/streptomycin supplemented medium at 37 °C, 5% CO2. DLD1, COLO-205, SW480 and A549 were grown in RPMI 1640 medium, HCT 116 and HT-29 were grown in McCoy’s 5A medium, Caco-2 (20% FBS) and LS174T were grown in Minimum Essential Medium and LoVo was grown in F-12 K medium. Cells were passaged 2–3 times per week and harvested at ~80% confluency. Growth medium was aspirated, cells were washed once in 1X PBS and collected in 1X PBS. Cells were centrifuged at 300 × g for 5 min and supernatant was discarded. Cell pellets were stored at −80 °C until cell lysis was carried out.
Pancreatic Cyst Fluid collection
Informed consent was obtained from patients scheduled for surgical resection (MGH IRB Protocol #02-240). Immediately after surgical resection, the cyst fluid was aspirated through the cyst wall. Cyst fluid (0.5 mL) was aliquoted into 1.5 mL polypropylene tubes, snap frozen and stored at −80°C. The remainder of the specimen was processed in the frozen section lab according to the routine protocol. A piece of the pancreatic cyst wall was also placed in liquid nitrogen and then stored at −80°C. The pancreatic cyst was classified according to the most invasive component of the cyst.
Enrichment of low molecular weight proteins by SDS-PAGE
Cell pellets or pancreatic cyst fluid aliquots were resuspended in RIPA buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 1% NP-40) with protease inhibitors (5 μg/ml leupeptin, 5 μg/ml aprotinin), sonicated for 15 seconds and placed on ice for 20 min. Cell lysates were centrifuged at 12,000 × g for 5 min at 4 °C and the supernatant was collected. Protein concentration was determined using the bicinchoninic acid assay (Pierce, Rockford, IL). Protein samples (50 μg) were mixed with 4X NuPAGE® lithium dodecyl sulfate sample buffer (Invitrogen, Carlsbad, CA), heated for 10 min at 70°C, and separated by SDS-PAGE on a NuPAGE® Novex 10% Bis Tris gel (Invitrogen). A protein molecular weight standard (Precision Plus Protein™ Kaleidoscope Standards, Biorad, Hercules, CA) was loaded in one lane of each gel.
Electrophoresis was done at a constant 180 V for 45 min and then gels were washed once with deionized H2O, stained with SimplyBlue™ SafeStain (Invitrogen) for 1 h and destained with deionized water overnight. Five fractions per lane between the 20 kDa and 25 kDa protein markers (KRAS MW = 21.6 kDa) were excised from the gel, cut into 1 mm cubes and placed in 0.1 mL of 100 mM ammonium bicarbonate. Samples were reduced with 10 μL of 100 mM DTT for 20 min at 50 °C and alkylated with 10 μL of 200 mM iodoacetamide for 20 min at room temperature in the dark. Excess dye was removed from gel slices with 50% acetonitrile/50 mM ammonium bicarbonate and slices were dehydrated with 100% acetonitrile. The dehydrated gel pieces were evaporated to dryness in vacuo, resuspended in 0.01 μg/μL MS grade trypsin (Promega, Madison, WI) in 50 mM ammonium bicarbonate, and incubated at 37 °C overnight. Peptides were extracted with 60% acetonitrile containing 0.1% formic acid. For GeLC-MRM analysis, each fraction was evaporated separately in vacuo and then resuspended in 50 μL of 5% acetonitrile containing 0.1% formic acid. For GeLC-MRM analysis, five gel extracts were pooled, and then evaporated in vacuo. Peptides were resuspended in 50 μL of 5% acetonitrile containing 0.1% formic acid and spiked with four 13C/15N-arginine or 13C/15N-lysine (C-terminal) heavy labeled synthetic peptides (LVVVGAGGVGK, LVVVGAGDVGK, LVVVGAVGVGK, QGVEDAFYTLVR, 0.5 fmol/μL) and analyzed by LC-MRM.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis
LC-MS/MS shotgun proteomic analyses of SDS-PAGE-enriched samples were performed as described elsewhere 5. Analyses were carried out on a LTQ XL mass spectrometer (Thermo Fisher Scientific) equipped with an Eksigent NanoLC AS1 autosampler (Eksigent, Dublin, CA) and Eksigent NanoLC 1D Plus pump, nanospray source, and Xcalibur 2.0 SR2 instrument control. MS/MS spectra of the peptides were acquired by data-dependent scanning in which one full MS spectrum (mass range 400–2000 Th) was followed by five MS/MS spectra. MS/MS spectra were recorded using dynamic exclusion of previously analyzed precursors for 60 s with a repeat of 1 and a repeat duration of 1.
MS/MS scans were transcoded to mzML file format with the Scansifter algorithm.6 The resulting mzML files were searched against the Homo sapiens Ensembl protein database (derived from Ensembl genes, release 59, August 2010), which also contained the most frequently observed KRAS mutant sequences (G12D, G12V, G13D), as well as a reverse database for false discovery rate estimation. The Myrimatch algorithm (version 1.6.33) was used for all searches with precursor mass/charge (m/z) tolerance of 1.25 Th and a fragment m/z tolerance of 0.5 Th.7 The IDpicker utility (version 2.2.2) was used to assign protein identifications to the set of peptides identified by Myrimatch.8 A minimum of two peptides per protein were required for valid protein identification, with a peptide-level false discovery rate of 5%.
MRM analyses
Peptide samples were analyzed in triplicate (2 μL injection volume) on a TSQ Vantage triple quadrupole mass spectrometer (ThermoFisher Scientific, San Jose, CA) equipped with an Eksigent Ultra nanoLC solvent delivery system, autosampler and a nanospray source. The mobile phase consisted of 0.1 % formic acid in either HPLC grade water (solvent A) or 90% acetonitrile (solvent B). Sample injection was followed by a 15 min wash with 100% solvent A. The mobile phase was then programmed to 60% B over 43 min, followed by an increase to 95% B by 49 min and then held at that composition for 11 min before returning 97% solvent A. Transitions for each peptide were selected using the Skyline software package (see Table S1 for a list of peptides and corresponding precursor and product m/z values).9 Instrument parameters include Q2 gas 1.5 mTorr, scan width 0.004 Th, scan time 10 ms, and both Q1 and Q3 resolution FWHM 0.7. Standard concentration curves were generated for wildtype, G12D/G13D, G12V and NRAS/HRAS-specific peptide using synthetic peptides (New England Peptides, Gardner, MA). Stable isotope dilution was done with isotopically labeled standards (containing 13C/15N-arginine or 13C/15N-lysine) for each peptide, which were spiked into each calibration standard (0.5 fmol/μL). Concentrations for KRAS peptides were normalized to protein input and reported as fmol/μg protein.
Results
Detection of mutant KRAS peptides by LC-MS/MS shotgun proteomics
KRAS is a small (21.6 kDa) protein of relatively low abundance. In shotgun LC-MS/MS analyses of unfractionated tumor cell lines, KRAS peptides were not detected (data not shown). Others have demonstrated that gel-based enrichment enhances MS detection of specific proteins in complex biological samples.10,11,12 We used SDS-PAGE to enrich cell line protein extracts for proteins in the 20–25 kDa range (Figure 1). We analyzed 9 colon carcinoma cell lines of known KRAS genotype (three wild-type and six mutant).2 Each cell extract was run in single lane and the 20–25 kDa regions were excised (5 gel fractions per lane), the proteins were digested in-gel and the peptides from the 5 digests from each cell line were pooled for two replicate LC-MS/MS analyses. MS/MS data were searched against a human protein sequence database containing known KRAS wild-type and mutant sequences. These analyses correctly identified the KRAS variants associated with each cell line (Table 1). Only wild-type RAS peptide (LVVVGAGGVGK) was detected in KRAS wildtype cell lines (Caco-2, COLO 205, HT-29) (since this peptide is common to Ras family members N/H/K-RAS we will refer to it as wild-type RAS). Mutant KRAS peptides (G12D, G13D, G12S or G12V) were identified in KRAS mutant cell lines (DLD1, HCT 116, LoVo, LS174T, SW480, A549). Representative MS/MS spectra are provided in Figures S1–S3. At least two MS/MS spectra per cell line were observed for both wild-type and mutant peptides. This preliminary study demonstrated that SDS-PAGE-based fractionation enabled KRAS peptide detection by LC-MS/MS.
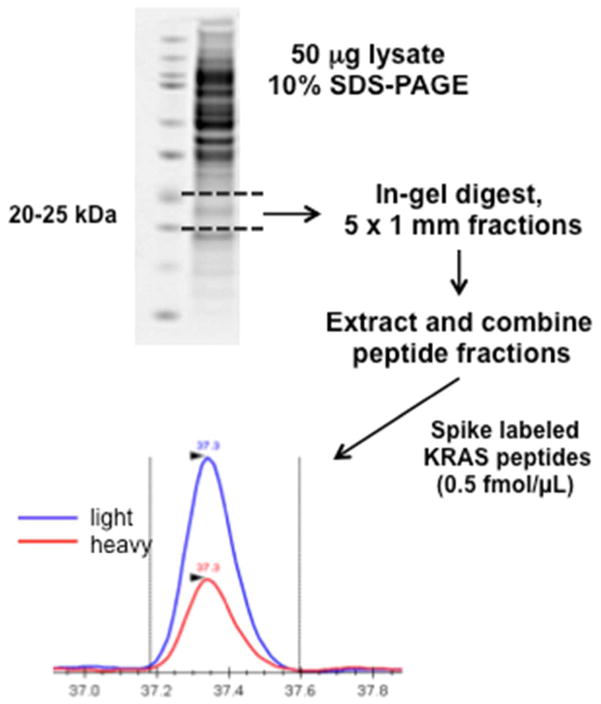
Figure 1.
GeLC-MRM work flow for detection and quantitation of wild-type RAS and mutant KRAS peptides.
Table 1.
Detection of mutant KRAS peptides in cell lines by LC-MS/MS
| Wild type cell lines | Mutant cell lines | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Caco-2 | COLO-205 | HT-29 | DLD-1 (WT/G13D) | HCT-116 (WT/G13D) | LoVo(WT/G13D) | LS174T(WT/G12D) | SW480(G12VG12V)/ | A549 (G12S/G12S) | |
| WT RAS | 9 | 3 | 4 | 3 | 4 | 5 | 8 | 5 | 5 |
| G12D | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 0 |
| G13D | 0 | 0 | 0 | 2 | 2 | 4 | 0 | 0 | 0 |
| G12V | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 |
| G12S | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
GeLC enrichment was carried out on cell lysates (50 μg) as described in “Experimental Methods” and the resulting five fractions were subject to MS/MS analysis (duplicate LC-MS/MS). MS/MS data were searched against human protein database containing common KRAS mutant sequences (2 peptide-per-protein, 5% peptide-level false discovery rate). The number of MS/MS spectra corresponding to each sequence are reported (wild-type peptide, green; mutant peptides, red).
The peptides we chose for MRM analysis included some previously used by Wang et al. 4, and others showing strong signals by LC-MS/MS. Wang et al. successfully monitored SFEDIHHYR, SFADINLYR, SFEDIHQYR peptides (specific to KRAS, NRAS and HRAS, respectively). However, in our preliminary LC-MRM analyses these peptides could not be reliably detected and were deemed not suitable for quantitation of specific RAS forms. Therefore, we monitored the NRAS/HRAS-specific peptide QGVEDAFYTLVR, which was readily detectable by LC-MRM. Since RAS proteins are relatively small and sequence homology is high among N/H/K- RAS, there were no other suitable peptides which had both strong MRM signal and RAS-specificity.
LC-MRM measurement of synthetic KRAS peptides
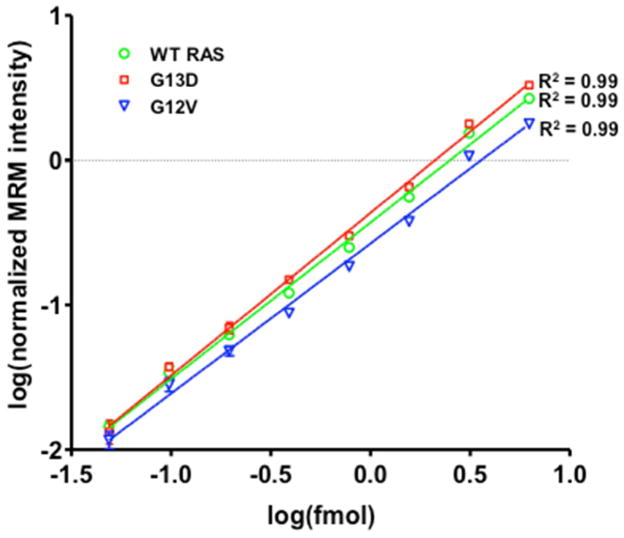
Having established that the SDS-PAGE-based method enables detection of both wild-type RAS and mutant KRAS peptides, we combined gel-based fractionation with MRM analysis to quantify KRAS protein forms in cell extracts. We first generated calibration curves for WT, G13D and G12V peptides (Figure 2). Four MRM transitions were selected for each peptide based on the most intense fragment ions observed in MRM analysis of synthetic peptides (see Table S1 for complete list of precursor/fragment ion m/z values). Calibration curve samples were made by spiking increasing amounts of unlabeled KRAS peptides (50 amol - 6 fmol) into a complex peptide background (gelC enriched peptides, 25–30 kDa) and isotope-labeled peptide standards were added to each sample at a constant amount (1 fmol/injection). Stable isotope dilution (SID) measurements showed low coefficients of variation (CV) across three replicate analyses (average CVs of 7.5%, 9.5% and 8.7% for WT, G13D and G12V respectively). Normalized peak intensities were linear with peptide input (R2 = 0.99 for wild-type RAS, G13D and G12V KRAS). Due to the highly similar MS/MS profiles of G12D and G13D peptides, a separate standard curve was not generated for G12D.
Figure 2.
LC-MRM synthetic peptide standard curves for wild-type RAS and mutant KRAS peptides. Synthetic peptides (WT, G13D, G12V) were spiked into complex peptide background (GelC-enriched peptides, 25–30 kDa) in increasing amounts (50 amol – 6 fmol). Heavy labeled versions of each peptide were added at a constant amount (0.5 fmol/μL). Summed MRM transition peak areas were normalized to heavy labeled standard peak areas.
GeLC-MRM measurement of RAS peptides in cancer cell lines
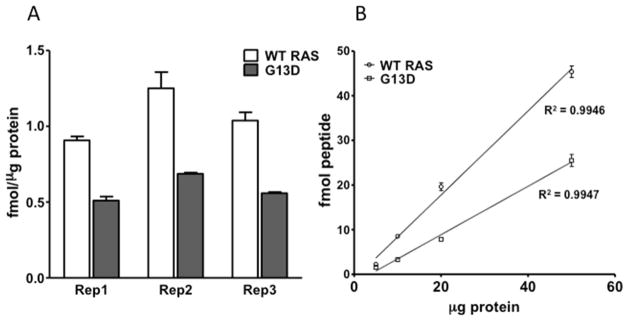
We tested the reproducibility and sensitivity of the GeLC-MRM approach to measure KRAS peptides in the colorectal cancer cell line HCT-116, which has one wild type and one mutant (G13D) KRAS allele. Three process replicates (3 lanes on the same gel, 50 μg HCT-116 protein/lane) were run in parallel and GeLC-MRM was carried out as described in Figure 1 (3 technical LC-MRM runs per sample). Measured wild-type RAS and mutant KRAS (G13D) peptide concentrations were 1.1 ± 0.17 fmol/μg protein and 0.6 ± 0.09 fmol/μg protein, respectively (Figure 3A). Across three process replicates, the GeLC-MRM approach provided reproducible measurements of peptide concentrations, with CVs of 15.2% and 13.7% for wild-type and mutant, respectively. Over 95% of detectable KRAS peptides in SDS-PAGE-fractionated HCT-116 extracts was detected in the 20–25 kDa region (Figure S4). GeLC-MRM was carried out on increasing protein amounts (5, 10, 20, 50 μg) and detection of KRAS peptides was linear with respect to protein input (R2 = 0.99 for wildtype and mutant) (Figure 3B). At protein inputs as low as 5 μg, KRAS peptides could still be reliably detected, suggesting that GeLC-MRM is a highly sensitive approach to quantify KRAS variants.
Figure 3.
GeLC-MRM performance characteristics. (A) Three GeLC-MRM process replicates were performed on HCT-116 cell lysates (three MS technical replicates each). Wildtype (white bars) and G13D mutant (grey bars) peptide concentrations were calculated from synthetic peptide standard curves and converted to fmol/μg protein units. (B) GeLC-MRM was carried out with increasing amounts of HCT-116 protein input (5, 10, 20, 50 μg, three MS technical replicates each) and total wild-type and G13D mutant peptide detected (fmol) was against protein input.
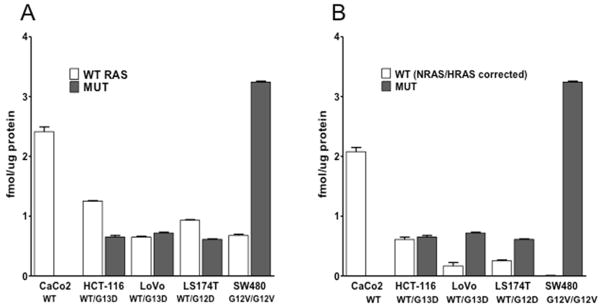
To further evaluate GeLC-MRM-based detection of KRAS peptides, we analyzed a subset of the cancer cell lines used in Table 1 (Caco-2, HCT-116, LoVo, LS174T and SW480). Due to the relatively low variation between process replicates for HCT-116 cells (Figure 3A), we analyzed a single process replicate for each cell line (3 technical MS replicates). Wild-type RAS peptide concentrations varied from 0.7 ± 0.01 fmol/μg protein in LoVo to 2.4 ± 0.08 fmol/μg protein in Caco2, whereas mutant peptide concentrations varied from 0.6 ± 0.01 fmol/μg protein in LS174T (G12D/G13D) to 3.2 ± 0.02 fmol/μg protein in SW480 (G12V) (Figure 4A). Previously reported KRAS peptide concentrations are only available for SW480 (0.8 ± 0.14 fmol/μg protein for wild-type RAS and 4.5 ± 1.05 fmol/μg protein for mutant KRAS,4) and are in general agreement with the values observed here (0.7 ± 0.02 fmol/μg protein for wild-type RAS and 3.2 ± 0.02 fmol/μg protein for mutant KRAS).
Figure 4.
GeLC-MRM quanitation of KRAS peptides in cancer cell lines. (A) Wild-type RAS (white bars) and mutant (grey bars) KRAS peptide concentrations were measured by GeLC-MRM (50 μg protein input, three technical MS replicates per sample. (B) The combined contributions of NRAS and HRAS to overall wildtype signal were calculated by measuring an NRAS/HRAS-specific peptide. These amounts were then subtracted from total wild-type concentrations, providing a NRAS/HRAS-corrected measurement of wild-type KRAS (white bars).
The peptide sequence LVVVGAGGVGK also occurs in KRAS, NRAS and HRAS. To correct for the contributions of NRAS and HRAS, we quantified a peptide specific to NRAS/HRAS, but not found in KRAS (QGVEDAFYTLVR). After subtraction of NRAS/HRAS, wild-type peptide values varied from 0.2 ± 0.06 fmol/μg protein in LoVo to 2.1 ± 0.07 fmol/μg protein in Caco-2, while SW480, which is homozygous for G12V mutant KRAS, had no detectable wild-type peptide (Figure 4B). Among KRAS wildtype and heterozygous cell lines, the relative contribution of NRAS/HRAS to wild-type signal varied considerably, with Caco-2 displaying the lowest (14%) and LoVo displaying the highest (73%).
GeLC-MRM measurement of KRAS peptides in pancreatic cyst fluids
To evaluate the ability of GeLC-MRM to quantify mutant KRAS peptides in clinical specimens, we analyzed pancreatic cyst fluid (PCF) samples collected from 15 patients who underwent surgical resection for pancreatic intraductal papillary mucinous neoplasms. Pathologic examination of the resected tissue classified the lesions as benign, carcinoma in situ and invasive carcinoma. These analyses monitored the RAS wild type, G12D/G13D and G12V peptides. While KRAS mutations are the predominant mutation among RAS genes in pancreatic cancers, there is a possibility that mutant peptides in these PCF samples are derived from NRAS or HRAS mutant proteins. Wild-type RAS was observed in all 15 patient samples and mutant KRAS was observed in 2/5 benign, 1/5 carcinoma in situ and 2/5 invasive carcinoma (Table 2). Wild-type peptide concentrations (not corrected for NRAS/HRAS) varied between 0.08 fmol/μg protein and 1.1 fmol/μg protein, while mutant peptide concentrations varied between 0.08 fmol/μg protein and 0.36 fmol/μg protein. Across all patient samples, the average wild-type peptide concentration was 0.23 fmol/μg protein and the average mutant peptide concentration was 0.17 fmol/μg protein. These values are in agreement with those reported by Wang and colleagues for analyses of three patient samples (0.13 fmol/μg protein and 0.09 fmol/μg protein for wild-type and mutant, respectively).4 Both G12D/G13D and G12V mutant peptides were detected in this set of samples. The data demonstrate that GeLC-MRM is applicable to analyze KRAS protein variants in pancreatic cyst fluids.
Table 2.
GeLC-MRM quantitation of wild-type and mutant KRAS in pancreatic cyst fluid samples
| Wild-type RAS fmol/μg protein | Mutant fmol/μg protein | |
|---|---|---|
| Benign | ||
| Patient 1 | 0.12 ± 0.001 | 0.08 ± 0.004* |
| Patient 2 | 0.16 ± f0.001 | ND |
| Patient 3 | 0.16 ± 0.004 | 0.10 ± 0.002* |
| Patient 4 | 1.11 ± 0.03 | ND |
| Patient 5 | 0.11 ± .007 | ND |
| Carcinoma in situ | ||
| Patient 6 | 0.21 ± 0.01 | 0.14 ± 0.005 * |
| Patient 7 | 0.11 ± 0.003 | ND |
| Patient 8 | 0.11 ± 0.001 | ND |
| Patient 9 | 0.18 ± 0.002 | ND |
| Patient 10 | 0.08 ± 0.002 | ND |
| Invasive carcinoma | ||
| Patient 11 | 0.10 ± 0.004 | 0.17 ± 0.05** |
| Patient 12 | 0.12 ± 0.006 | ND |
| Patient 13 | 0.08 ± 0.001 | ND |
| Patient 14 | 0.18 ± 0.003 | ND |
| Patient 15 | 0.62 ± 0.02 | 0.36 ± 0.01* |
G12D/G13D
G12V.
ND, not detected (limit of detection, 0.02 fmol/μg protein and 0.01 fmol/μg protein for G13D and G12V, respectively)
Discussion
KRAS is one of the most frequently mutated oncogenes and activating missense mutations have been identified as drivers in colorectal, pancreatic, lung and other human cancers. Analysis of KRAS and other cancer-related mutations in tumors is typically done at the DNA sequence level and this approach provides the basis for an emerging approach to personalized cancer therapies.13 Nevertheless, mutations ultimately express their effects through their effects on proteins and proteomes, so analytical approaches to measure cancer-related protein variants offer new opportunities to understand cancer biology and develop new diagnostics. A major recent advance was the demonstration by Wang and colleagues of an IP-MRM approach to analyze mutant KRAS protein in complex biological samples.4 However, the method requires antibodies capable of immunoprecipitating the target protein and thus translation to other protein targets depends on the availability of high quality antibodies. Moreover, the IP-MRM analysis required a relatively large sample protein input (2–4 mg), thus potentially limiting the types of specimens that can be analyzed. Here we use a SDS-PAGE gel-based enrichment of proteins in the molecular weight range of KRAS, which substantially reduced sample complexity and enabled quantitation of mutant and wild-type KRAS in cancer cell lines and pancreatic cyst fluids, with protein inputs of 5–50 μg. Our results for analyses of tumor cells and pancreatic cyst fluid also were concordant with the data from Wang et al.4 GeLC-MRM thus provides a general strategy for the targeted analysis of variant proteins.
Gel-based fractionation has been a widely used technique in proteomic analysis of complex protein mixtures.12,10 Here we employed a simple one-dimensional SDS-PAGE separation with gel band selection to enrich KRAS for MRM analysis. This approach provides the most readily accessible, fast and inexpensive means to enrich lower abundance proteins such as KRAS. Nevertheless, SDS-PAGE-based separations have some limitations. First, some glycosylated or other posttranslationally-modified proteins migrate over a broad molecular weight range in SDS-PAGE, thus making concentration and targeted gel band selection difficult. Second, the degree of enrichment achieved with SDS-PAGE varies considerably for different proteins. In a preliminary study, we found that gel fractionation enhanced detection of spiked proteins in a cell lysate background between 5–100 fold, compared to unfractionated samples (Zhang, H. and Liebler, D.C., unpublished data).
Our preliminary study of pancreatic cyst fluid was not meant to be a biomarker validation study, but simply to illustrate the application of the GeLC-MRM method to a relevant clinical sample type. KRAS mutations are highly prevalent in pancreatic cancers and appear at early stages of tumor development.14,15,16 Our detection of KRAS mutant proteins in pancreatic cyst fluids is consistent with the report of Wang et al., who also detected mutant KRAS proteins in premalignant pancreatic tissue and fluid.4
In conclusion, we have described an approach for the detection and quantitation of mutant KRAS in complex biological samples which provides improved sensitivity over the recently described IP-MRM method. This general approach to targeted analysis of cancer-related protein variants will be useful both in clinical biomarker investigations and in basic cancer biology studies.
Supplementary Material
Acknowledgments
We thank Dr. Robbert J.C. Slebos for helpful discussion. This work was supported in part by NIH grants U24CA126479, U24CA159988 and U01CA152647.
References
- 1.The Cancer Genome Atlas. http://cancergenome.nih.gov/
- 2.Catalogue of Somatic Mutations in Cancer. http://www.sanger.ac.uk/genetics/CGP/cosmic/
- 3.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10 (8):789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q, Chaerkady R, Wu J, Hwang HJ, Papadopoulos N, Kopelovich L, Maitra A, Matthaei H, Eshleman JR, Hruban RH, Kinzler KW, Pandey A, Vogelstein B. Mutant proteins as cancer-specific biomarkers. Proc Natl Acad Sci U S A. 2011;108 (6):2444–9. doi: 10.1073/pnas.1019203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halvey PJ, Zhang B, Coffey RJ, Liebler DC, Slebos RJ. Proteomic Consequences of a Single Gene Mutation in a Colorectal Cancer Model. J Proteome Res. 2011 doi: 10.1021/pr2009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma ZQ, Tabb DL, Burden J, Chambers MC, Cox MB, Cantrell MJ, Ham AJ, Litton MD, Oreto MR, Schultz WC, Sobecki SM, Tsui TY, Wernke GR, Liebler DC. Supporting tool suite for production proteomics. Bioinformatics. 2011;27 (22):3214–5. doi: 10.1093/bioinformatics/btr544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6 (2):654–61. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6 (9):3549–57. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26 (7):966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci U S A. 1996;93 (25):14440–5. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR, Mann M, Lodish HF. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc Natl Acad Sci U S A. 2000;97 (1):179–84. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100 (12):6940–5. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pao W, Kris MG, Iafrate AJ, Ladanyi M, Janne PA, Wistuba II, Miake-Lye R, Herbst RS, Carbone DP, Johnson BE, Lynch TJ. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res. 2009;15 (17):5317–22. doi: 10.1158/1078-0432.CCR-09-0913. [DOI] [PubMed] [Google Scholar]
- 14.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53 (4):549–54. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 15.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321 (5897):1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, Hruban RH, Maitra A, Kinzler K, Vogelstein B, Goggins M. Presence of Somatic Mutations in Most Early-Stage Pancreatic Intraepithelial Neoplasia. Gastroenterology. 2012 doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.