Abstract
Background and Aims
In the genus Anemone two small groups of taxa occur with the highest ploidy levels 2n = 6x = 48, belonging to the closely related clades: the montane/alpine Baldensis clade and the more temperate Multifida clade. To understand the formation of polyploids within these groups, the evolution of allohexaploid A. baldensis (AABBDD, 2n = 6x = 48) from Europe and allotetraploid Anemone multifida (BBDD, 2n = 4x = 32) from America was analysed.
Methods
Internal transcribed spacer and non-transcribed spacer sequences were used as molecular markers for phylogenetic analyses. Cytogenetic studies, including genomic in situ hybridization with genomic DNA of potential parental species as probe, fluorescence in situ hybridization with 5S and 18S rDNA as probes and 18S rDNA restriction analyses, were used to identify the parental origin of chromosomes and to study genomic changes following polyploidization.
Key Results
This study shows that A. multifida (BBDD, 2n= 4x = 32) and A. baldensis (AABBDD, 2n = 6x = 48) are allopolyploids originating from the crosses of diploid members of the Multifida (donor of the A and B subgenomes) and Baldensis groups (donor of the D subgenome). The A and B subgenomes are closely related to the genomes of A. sylvestris, A. virginiana and A. cylindrica, indicating that these species or their progeny might be the ancestral donors of the B subgenome of A. multifida and A and B subgenomes of A. baldensis. Both polyploids have undergone genomic changes such as interchromosomal translocation affecting B and D subgenomes and changes at rDNA sites. Anemone multifida has lost the 35S rDNA loci characteristic of the maternal donor (B subgenome) and maintained only the rDNA loci of the paternal donor (D subgenome).
Conclusions
It is proposed that A. multifida and A. baldensis probably had a common ancestor and their evolution was facilitated by vegetation changes during the Quaternary, resulting in their present disjunctive distribution.
Keywords: 5S rDNA intergenic spacer, Anemone, fluorescence in situ hybridization, genomic in situ hybridization, intergenomic translocation, ITS, polyploidy
INTRODUCTION
Recent investigations have shown that all seed plants and angiosperms encountered polyploidy or whole-genome duplication events throughout their evolutionary history (Jiao et al., 2011). Polyploidization is accompanied by rapid and dynamic genetic and epigenetic changes, as well as changes in gene expression and phenotypic variation (Cifuentes et al., 2010; Gaeta and Pires, 2010; Parisod et al., 2010). Changes in genome structure range from gene conversion, sequence loss or gain, sequence amplification or reduction, to all known forms of chromosomal rearrangements (Lim et al., 2007a, b; Weiss-Schneeweiss et al., 2007; Książcyk et al., 2011). Many hybrids and polyploids suffer from numerous rDNA rearrangements, including repeat and locus loss (Lim et al., 2007a, b; Weiss-Schneeweiss et al., 2007; Kotseruba et al., 2010; Książcyk et al., 2011), interlocus recombination and repeat replacement (Kovarik et al., 2005). The mechanism and consequences of these changes remain largely unknown. Recent studies of Tragopogon and Brassica polyploids have provided evidence that homeologous pairing and recombination could be a key mechanism for genome restructuring (Lim et al., 2008; Gaeta and Pires, 2010). Szadkowski et al. (2010) showed that the very first meiosis of somatically doubled resynthesized Brassica napus already acts as a genome blender, with many of the genetic exchanges between different subgenomes transmitted to the progeny. Moreover, polyploidy formation pathways are shown to have an impact on genetic rearrangements in resynthesized Brassica napus as the meiosis of the F1 interspecific hybrid generated more gametes with recombined chromosomes than did meiosis of the plant produced by somatic doubling (Szadkowski et al., 2011).
The genus Anemone sensu stricto consists of approx. 150 species of perennial, low-growing herbs of worldwide distribution and with considerable diversity in morphology (Tamura, 1995). Two basic chromosome numbers (n = 7 and n = 8), substantial karyotype divergence involving rDNA and heterochromatin distribution, and the existence of different ploidy levels are prominent features of Anemone genome evolution (Baumberger, 1970; Mlinarec et al., 2012). However, polyploidy is unevenly distributed in Anemone and is restricted to a few lineages. The most common polyploids are tetraploids, while hexaploidy is rare (Gajewski, 1946; Heimburger, 1961). The only three hexaploids recognized (A. baldensis, A. lithophila and A. drummondii) are found among the Baldensis group with the disjunct areas in the Arctic and the mountains of southern Europe, one the Earth's most polyploidy-rich areas (Brochmann et al., 2004).
Allotetraploid A. multifida (2n = 4x = 32) is representative of the Multifida group, a small group of species from North America (A. cylindrica, A. virginiana and A. multifida) and one species disjunctly distributed in Europe (A. sylvestris). Anemone multifida is widely distributed, ranging from the north-eastern part of North America to Alaska and southward to New Mexico and California, and reappearing again in the mountains of southern Chile and Argentina. Anemone sylvestris is native to meadows and dry deciduous woodlands of central and western Europe. Allohexaploid A. baldensis (2n = 6x = 48) is representative of the Baldensis group, a small group of species from the arctic and alpine tundra of North America (A. parviflora, A. lithophila, A. drummondii) and one species disjunctly distributed in Europe (A. baldensis). Anemone baldensis occurs in the Pyrenees, Alps, Dinnarids and Carpathian mountains of Europe. Hybrids between members of the Multifida group are readily obtained (Heimburger, 1962), suggesting a close relationship between them. Hexaploid A. jancziewskii, obtained experimentally from the crossing of A. multifida with A. sylvestris, has existed for more than 120 years in several European botanical gardens (Janczewski, 1892; Gajewski, 1946).
Studies on the origin and evolution of the polyploid Anemone species and their relationships with the diploid taxa have been initiated only recently (Mlinarec et al., 2012). Cytogenetic studies to date involve analysis of chromosome number and karyotype as well as rDNA and heterochromatin characterization (Heimburger, 1959; Baumberger, 1970; Marks and Schweizer, 1974; Mlinarec et al., 2006, 2009, 2012). Based on karyotype morphology and additional phylogenetic analyses, Mlinarec et al. (2012) proposed a hybrid origin for the hexaploid A. baldensis with the diploid A. sylvestris as putative progenitor. Similarly, incongruence between the chloroplast and nuclear DNA phylogenetic trees indicated a possible hybridogenous genomic constitution of A. multifida, with the presumed parental species among the members of the Baldensis and Multifida groups (Meyer et al., 2010).
To confirm the allopolyploid origin of tetraploid A. multifida (2n = 4x = 32) and hexaploid A. baldensis (2n = 6x = 48) and elucidate the parental origin of their chromosomes among species from the Baldensis and Multifida groups, we performed, phylogenetic analyses using internal transcribed spacer (ITS) and non-transcribed spacer (NTS) sequences as molecular markers, cytogenetic studies including genomic in situ hybridization (GISH) with genomic DNAs of potential parental species as probe, fluorescence in situ hybridization (FISH) with 5S and 18S rDNA as probe and 18S rDNA restriction analyses. We identified three subgenomes (AABBDD) in A. baldensis and two (BBDD) in A. multifida. The A and B subgenomes are closely related to the genomes of A. sylvestris, A. virginiana and A. cylindrica, indicating that these species or their progeny might be the ancestral donors of the B subgenome of A. multifida and A and B subgenomes of A. baldensis. The D subgenome of A. multifida and A. baldensis probably originated from the Baldensis group.
MATERIALS AND METHODS
Plant material
Information on all plant material used is given in Table 1. Plants were grown in pots in the Botanical Garden of the University of Zagreb. All species were identified based on morphological and karyological characteristics. For karyological studies, actively growing root-tip meristems were pretreated with 0·05 % colchicine (Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) for 4 h at room temperature, fixed in a solution of ethanol and acetic acid (3 : 1) for 24 h at –20 °C, and stored in 70 % ethanol at –20 °C until use. For cloning as well as for GISH probes, high-quality genomic DNA was isolated from young leaves using the Qiagen mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions.
Table 1.
Plant material, accession numbers and chromosome number (2n) for Anemone
| Taxon | Accession no. | Locality | 2n |
|---|---|---|---|
| Anemone baldensis L. | 698G | Hungary, Vácrátót, 1989, Botanical Garden of the University of Zagreb | 48 |
| Anemone cylindrica A. Gray | 6559B | Germany, seed from the Botanical Garden Chemnitz, Botanical garden of the University of Zagreb | 16 |
| Anemone multifida Poir. | 12427 | Germany, seed from Botanical Garden Chemnitz, Botanical garden of the University of Zagreb | 32 |
| Anemone sylvestris L. | 1451B | Croatia, Čučerje, Medvednica, Botanical Garden of the University of Zagreb | 16 |
| Anemone virginiana L. | 11838B | Germany, seed from the Botanical Garden Chemnitz, origin: Canada, Quebec, Country Deux-Montaignes, Oka, 2005, Botanical Garden of the University of Zagreb | 16 |
PCR amplification and cloning
PCR amplifications of the entire spacer region with the partial 5S rDNA gene as well as ITS region of 35S rDNA were carried out in a 50 µL reaction mixture containing 10 ng template DNA, 0·4 µm of each primer, 200 µm dNTPs, 2·5 U GoTaq DNA Polymerase and corresponding 1× (1·5 mm MgCl2) Green Reaction Buffer (Promega Corp., Madison, WI, USA), using the primer pairs described previously (Besendorfer et al., 2005; Puizina et al., 2008). After an initial denaturing step at 94 °C for 3 min, amplification was carried out using 30 cycles consisting of denaturation at 94 °C for 1 min, annealing at 54 °C for 10 s and primer extension at 72 °C for 1 min, with a final extension at 72 °C for 10 min. Cloning and transformation were carried out using the InsTAclone PCR Cloning Kit (Fermentas GmbH, Germany) or pGEM-T Easy Vector System (Promega) according to the manufacturers' instructions. Sequencing was carried out by Macrogen Inc. (Seoul, Korea).
Sequence alignment and phylogenetic analysis
For the polyploid taxa we employed 20 NTS and 16 ITS clones of A. baldensis and 15 NTS and ten ITS clones of A. multifida, while for diploid taxa (A. sylvestris, A. virginiana, A. cylindrica) we employed 3–6 NTS clones and 1–3 ITS sequences. ITS sequences of Anemone parviflora (FJ639887), A. sylvestris (FJ639894), A. drummondii (AY055404), A. multifida (AY055405), A. virginiana (FJ639898) and A. lithophila (FJ639883) were mined from the GenBank databse using keywords (Anemone, ITS). These sequences are the result of direct sequencing (Meyer et al., 2010), while NTS and ITS sequences obtained in this study are the result of cloning and their accession numbers are indicated in Fig. 1. Sequences were aligned with Clustal_x v1·81 (Thompson et al., 1997).
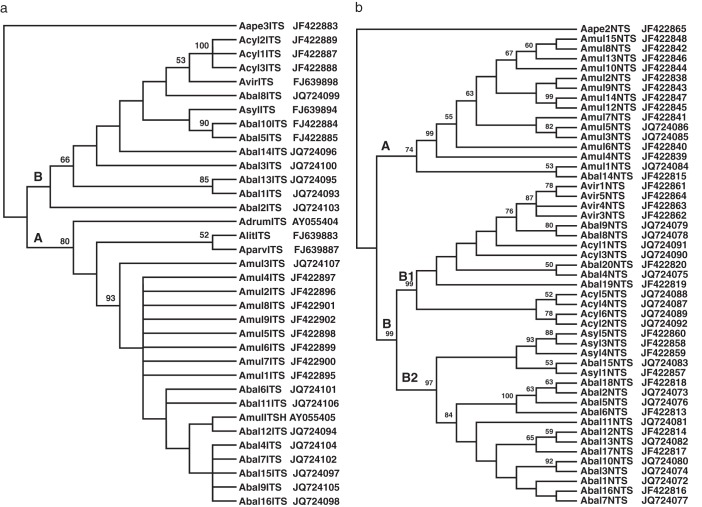
Fig. 1.
Phylogenetic relationships among cloned ITS and NTS sequences of A. baldensis (Abal), A. cylindrica (Acyl), A. drummondii (Adrum), A. lithophila (Alit), A. multifida (Amul), A. parviflora (Aparv), A. sylvestris (Asyl) and A. virginiana (Avir): (a) 50 % majority-rule consensus tree of the 1603 most-parsimonious trees based on ITS sequences, (b) 50 % majority-rule consensus tree of the 478 most-parsimonious trees based on NTS sequences. The A clade (Baldensis clade) and B clade (Multifida clade) are indicated. Bootstrap support values (>50 %) are shown above nodes. Sequences of A. apennina were used as outgroup. GenBank sequence accession numbers are indicated next to the name of the particular clone.
Phylogenetic signal in each dataset was determined from the tree-length distribution of 1000 000 trees, using the g1-statistic (Hillis and Huelsenbeck, 1992). An unweighted maximum-parsimony (MP) analysis was conducted using PAUP* 4·0b10 (Swofford, 2003). Heuristic searches were performed with 1000 random addition sequence replicates using tree bisection reconnection (TBR) branch swapping. Gaps were treated as missing data. Bootstrap support values (Felsenstein, 1985) from 1000 replicates were calculated using the heuristic search options as above except random addition sequence with 100 replicates. As outgroup taxa we employed the ITS (JF422883) and NTS (JF422865) sequences of the closely related Mediterranean species A. apennina.
Chromosome preparation and FISH
Chromosome preparations for FISH and GISH were as described by Mlinarec et al. (2006). FISH and GISH experiments were performed according to Mlinarec et al. (2012). Clone pTa794, containing the complete 410-bp BamHI fragment of the 5S rRNA gene and the spacer region of wheat (Gerlach and Dyer, 1980), was used as the 5S rDNA probe. The 2·4-kb HindIII fragment of the partial 18S rRNA gene and ITS1 from Cucurbita pepo, cloned into pUC19 (Torres-Ruiz and Hemleben, 1994), was used as the 35S rDNA probe. The genomic and rDNA probes were directly labelled with Cy3-dCTP (Amersham, GE Healthcare, Little Chalfont, UK) and FITC-dUTP (Roche Diagnostics GmbH, Mannheim, Germany) by using a nick-translation kit according to the manufacturer's instructions (Roche). After overnight hybridization, slides were given a stringent wash in 0·1× saline sodium citrate resulting in DNA duplexes with an estimated >85 % sequence identity. The preparations were mounted in antifade buffer Vectashield (Vector Laboratories, Peterborough, UK) containing DAPI counterstain (2 µg mL−1) and stored at 4 °C. Signals were visualized and photographs captured on an Olympus BX51 microscope, equipped with a highly sensitive Olympus DP70 digital camera. Images were uniformly processed using Adobe Photoshop for colour contrast and brightness. An average of ten well-spread metaphases was analysed for each individual. Three individuals per taxon were analysed.
Chromosome analyses and construction of ideograms
Acetocarmine staining was performed according to standard protocols (Mlinarec et al., 2006). Chromosome measurements were made on three well-spread chromosome plates of each of three individuals. For chromosome classification, the nomenclature of Levan et al. (1964) was followed. Chromosomes were identified according to type, total length, arm ratio, fluorochrome banding pattern and GISH hybridization signal, and were ranked in order of increasing length.
Southern hybridization
Genomic DNA (gDNA) was isolated from 150 mg fresh tissue of five Anemone species: A. cylindrica, A. virginiana, A. sylvestris, A. multifida and A. baldensis using the GenElute Plant Genomic DNA Miniprep Kit (Sigma-Aldrich, St. Louis, MO, USA). gDNA (2·7 µg) was digested overnight with EcoRV (New England Biolabs, Ipswich, MA, USA). Digested gDNA (1·35 µg) was loaded per lane on a 1 % (w/v) agarose gel and electrophoretically separated for several hours at 100 V. DNA was blotted onto a positively charged nylon membrane (Roche, Basel, Schwitzerland) for 1 h by using a Model 785 Vacuum Blotter (BioRad, Hercules, CA, USA). Crosslinking was performed for 3 min (0·24 J cm−2) on a UVlink CL508M crosslinker (Uvitec, Cambridge, UK). The 2·4-kb HindIII fragment of the partial 18S rRNA gene and ITS1 from Cucurbita pepo, cloned into pUC19 (Torres-Ruiz and Hemleben, 1994), was used as a template to generate the DIG-dUTP-labelled probe by using the DIG DNA Labelling and Detection Kit (Roche) according to the manufacturer in combination with the universal M13-forward and M13-reverse primers. The membrane was prehybridized for 1 h and hybridized overnight at 42 °C. Subsequently, the membrane was washed at room temperature and at 68 °C according to the manufacturer's instructions. DIG was detected using the Anti-DIG-AP, Fab fragments (Roche). The hybridized probe was visualized directly on the membrane by applying the NBT/BCIP solution (Roche) according to the manufacturer's instructions. Strongest signal was found after overnight incubation.
RESULTS
Phylogenetic analysis of ITS and NTS sequences
Phylogenetic analyses were performed with ITS and NTS sequences to determine if they can be useful in evaluation of possible cases of hybridization among different Anemone species (Fig. 1). In addition to A. baldensis and A. multifida, we included in phylogenetic analyses their putative parental species A. sylvestris, A. virginiana and A. cylindrica (members of the Multifida clade) as well as A. parviflora, A. drummondii and A. lithophila (members of the Baldensis clade). Anemone apennina was used as outgroup to root the NTS and ITS trees.
For ITS, the alignment of 35 sequences had 549 characters, of which 434 were constant, 60 were parsimony-uninformative and 55 were parsimony-informative. The length–frequency distribution from 1000 000 random trees showed a strong left skew (g1 = –0·38) as compared with the critical value of g1 = –0·13 (at P = 0·01) for 25 taxa and 50 characters, indicating the presence of highly significant phylogenetic signal in the dataset. The heuristic search resulted in 1603 equally parsimonious trees with a length of 168 steps (consistency index (CI) = 0·72, retention index (RI) = 0·89, rescaled consistency index (RC) = 0·64).
The NTS dataset included 49 sequences that comprised 538 characters, of which 215 were constant, 138 were parsimony-uninformative and 138 were parsimony-informative. The distribution of lengths for the 1000 000 random trees evaluated was strongly skewed to the left (g1 = –0·53) compared with the critical value of g1 = –0·12 (at P = 0·01) for 25 taxa and 100 characters (Hillis and Huelsenbeck, 1992). This g1 value indicates that the data are significantly more structured than are random data and implies the presence of strong phylogenetic signal in the NTS dataset. An unweighted parsimony analysis yielded 478 most-parsimonious trees of length 708 steps (CI = 0·68, RI = 0·84, RC = 0·57).
Phylogenetic analyses separated the sequences in either the ITS or the NTS tree into two divergent clades: A and B. All sequences originating from A. multifida only derived from within clade A, while those generated from A. baldensis fell within the two distinct clades A and B (Fig. 1).
In Fig. 1(a), the ITS tree displays a clear rake-like branching pattern containing a number of unresolved polytomies, suggesting a low rate of sequence divergence. Some of the ITS sequences of A. baldensis are associated with those of A. parviflora, A. drummondii and A. lythophila in clade A [bootstrap support (BS) = 80 %], named the Baldensis clade, whereas the other A. baldensis sequences are closely related to ITS sequences obtained from the diploid taxa of the Multifida group (A. sylvestris, A. cylindrica and A. virginiana) named the Multifida clade (clade B). Two of the A. baldensis clones (Abal5ITS and Abal10ITS) showed close affinities to the sequence AsylITS derived from A. sylvestris, whereas another (Abal8ITS) is rather close to those of A. virginiana and A. cylindrica. Interestingly, all the ITS clones isolated from the tetraploid A. multifida formed a well-supported monophyletic group (BS = 93 %) together with the A. baldensis ITS sequences within the Baldensis clade (clade A).
In the NTS tree (Fig. 1b), the data confirm the genomic sequence heterogeneity of A. baldensis, as observed for ITS sequences. NTS sequences are distributed in two main clades. One clone (Abal14NTS) is related to the A. multifida clones in a distinct clade (BS = 74 %) corresponding to the previous Baldensis clade A. All the other NTS clones fall in the Multifida clade B (BS = 99 %). Within the latter, the A. baldensis clones are separated into two subsets belonging each to two well-supported sister subclades (B1 and B2). One A. baldensis NTS subset is clearly embedded with A. cylindrica and A. virginiana in subclade B1 (BS = 99 %), while the other is associated with A. sylvestris in subclade B2 (BS = 97 %).
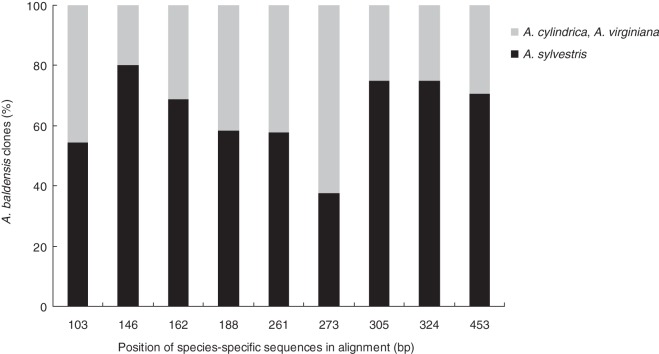
To gain further insight from the NTS sequences we more accurately examined sequence alignment of cloned NTS sequences from the five studied Anemone species. Analysis of NTS sequences revealed 12 species-specific nucleotide positions for which the sequence origin could be associated with the corresponding Anemone species (Supplementary Data Fig. S1). In agreement with the NTS tree, species-specific nucleotide positions in A. multifida NTS clones did not reveal similarity to any other analysed clones of other Anemone species. Only one A. multifida clone, Amul1NTS, showed similarity with one A. baldensis clone, Abal14NTS, but only in five of 12 species-specific sequences (ACTC at position 103, TTGTTC at position 146, GGGTAATG at position 162, GGCTCTCA at position 273 and GGC at position 324) (Supplementary Data Fig. S1). By contrast, species-specific positions identical to either A. sylvestris, A. virginiana or A. cylindrica were identified in the NTS clones of A. baldensis. The majority of 20 analysed A. baldensis clones possessed species-specific positions that were identical to A. sylvestris (36–78 % identity depending on the position), while some were identical to A. cylindrica and/or A. virginiana (Fig. 2). Note that the NTS sequences of A. cylindrica and A. virginiana were almost identical (sharing 11 of 12 species-specific sequences) and differed from A. sylvestris in ten of 12 species-specific positions (Supplementary Data Fig. S1).
Fig. 2.
The proportion of A. baldensis NTS clones that possess species-specific sequences identical to A. sylvestris, A. virginiana and A. cylindrica. The species-specific sequences are shown in Supplementary Data Fig. S1.
These phylogenetic analyses proved to be useful in revealing the polyploid origin of A. baldensis, suggesting that this allohexaploid originates from crosses between the members of the Multifida and Baldensis clades. The highly supported association between the NTS sequences of A. baldensis and those from A. sylvestris, A. cylindrica or A. virginiana suggests that all three species could have been implicated in the origin of A. baldensis. Furthermore, although the DNA sequence data did not reveal the hybrid origin of A. multifida, a strong association of the ITS sequences with the Baldensis group suggests that the potential candidate for one parental species comes from this group.
GISH in A. multifida and A. baldensis
To determine the origin of the Anemone polyploids, labelled gDNA of presumed parental species was hybridized to mitotic chromosomes of A. multifida (2n = 4x = 32) and A. baldensis (2n = 6x = 48). GISH of A. multifida root-tip metaphases revealed cross-hybridization of A. virginiana (Fig. 3a, d), A. sylvestris (Fig. 3c) and A. cylindrica (data not shown) gDNAs to only one chromosome set of A. multifida. The labelled chromosome set was designed as the B chromosome set in A. multifida (Fig. 3d).
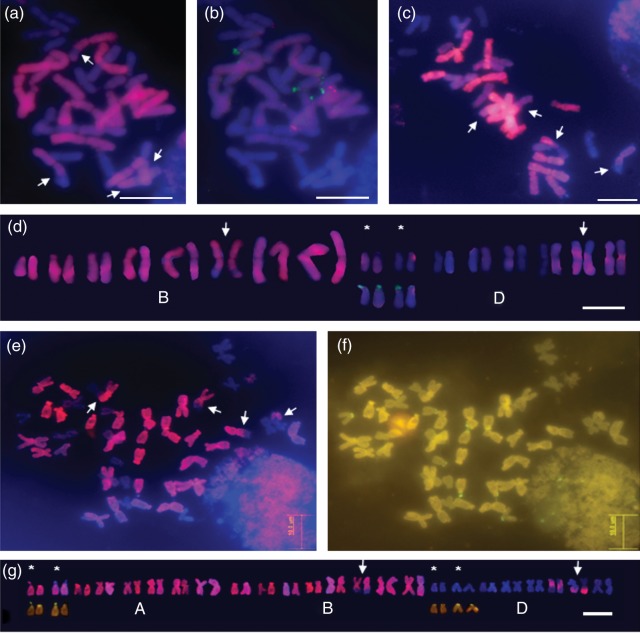
Fig. 3.
Karyotype analyses of A. multifida (a–d) and A. baldensis (e–g). GISH on mitotic chromosomes of A. multifida with labelled gDNA of A. virginiana (a, d; in red) and A. sylvestris (c; in red). GISH on mitotic chromosomes of A. baldensis with labelled gDNA of A. sylvestris (e, g; in red). The unidentified subgenome shows DAPI-specific staining (in blue). Intergenomic translocation is marked with arrows. 35S rDNA bearing chromosome pairs are marked with asterisks. The chromosome sets of the corresponding subgenomes are marked A, B and D. FISH on A. multifida (b) and A. baldensis (f) mitotic chromosomes with differentially labelled 35S rDNA (in green) and/or 5S rDNA (in red). Scale bars = 10 µm.
GISH analysis in A. baldensis was more complex due to the presence of three chromosome sets distinguishable by size (Mlinarec et al., 2012). GISH of A. baldensis root-tip metaphases showed cross-hybridization of A. sylvestris (Fig. 3e, g), A. virginiana (data not shown) and A. cylindrica (data not shown) gDNAs to two chromosome sets of A. baldensis. One labelled chromosome set had chromosomes of similar size, morphology and rDNA FISH pattern to those of A. sylvestris and was identified as A. sylvestris-like or the A chromosome set (Fig. 3g). The other labelled chromosome set was most similar to A. cylindrica and A. virginiana (A. cylindrica and A. virginiana have chromosomes of similar size, morphology and rDNA loci position; Mlinarec et al., 2012) and was designed as the B chromosome set in A. baldensis.
Karyotype analysis of A. multifida and A. baldensis showed that the unidentified chromosome set that showed DAPI-specific staining consists of chromosomes of similar size, morphology and rDNA FISH pattern (Table 2; see Fig. 5 below). The unidentified chromosome set was designed as the D chromosome set in both polyploid species.
Table 2.
Chromosome measurements of A. baldensis and A. multifida
| Chromosome |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | |||||||||
| A. baldensis, 2n = 48 | 0·8 | 1·1 | 0·9 | 2·1 | 2·8 | 3·4 | 3·7 | 4·7 | ||||||||
| 4·0 | 4·2 | 4·3 | 4·5 | 3·6 | 3·9 | 4·3 | 4·8 | |||||||||
| A | 5·0 | 4·8 | 3·8 | 5·3 | 4·8 | 5·2 | 2·1 | 6·6 | 1·3 | 6·4 | 1·2 | 7·3 | 1·2 | 8·0 | 1·0 | 9·5 |
| 1·0 | 1·2 | 1·3 | 2·1 | 3·9 | 4·0 | 4·5 | 4·7 | |||||||||
| 4·5 | 4·7 | 4·9 | 3·9 | 4·3 | 4·5 | 5·2 | 5·7 | |||||||||
| B | 4·5 | 5·5 | 4·0 | 5·9 | 3·8 | 6·2 | 1·9 | 6 | 1·1 | 8·2 | 1·1 | 8·5 | 1·2 | 9·7 | 1·2 | 10·4 |
| 0·8 | 1·0 | 1·0 | 1·9 | 2·5 | 3 | 3·2 | 3·6 | |||||||||
| 3·3 | 3·3 | 3·4 | 3·3 | 3·3 | 4·0 | 4·1 | 4·2 | |||||||||
| D | 5·1 | 4·1 | 3·3 | 4·3 | 3·4 | 4·4 | 1·7 | 5·2 | 1·3 | 5·8 | 1·3 | 7·0 | 1·3 | 7·3 | 1·2 | 7·8 |
| A. multifida, 2n = 32 | 0·7 | 1·0 | 1·3 | 2·4 | 3·8 | 4·4 | 4·9 | 5·1 | ||||||||
| 5·0 | 5·3 | 5·4 | 5·0 | 4·3 | 4·7 | 5·8 | 5·9 | |||||||||
| B | 7·1 | 5·7 | 5·3 | 6·3 | 4·2 | 6·7 | 2·1 | 7·4 | 1·1 | 8·1 | 1·1 | 9·1 | 1·2 | 10·7 | 1·2 | 11·0 |
| 0·8 | 1·1 | 1·0 | 1·9 | 2·6 | 3·1 | 3·5 | 3·3 | |||||||||
| 3·3 | 3·3 | 3·5 | 3·2 | 3·3 | 4·1 | 3·9 | 4·5 | |||||||||
| D | 5·1 | 4·1 | 3·0 | 4·4 | 3·5 | 4·5 | 1·7 | 5·1 | 1·3 | 5·9 | 1·3 | 7·2 | 1·1 | 7·4 | 1·4 | 7·8 |
The first measurement in each column refers to the short arms; the second, to the long arms; and the third, to the totals. Arm ratios appear at the left. Note that the D chromosome set of A. baldensis and A. multifida are of similar size and morphology. Chromosome measurements of A. multifida are taken from Heimburger (1959).
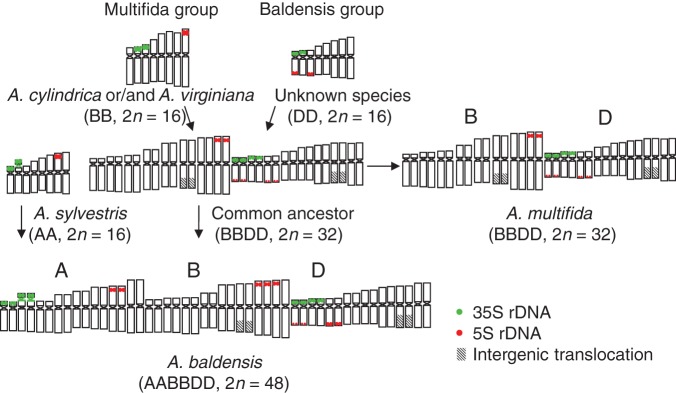
Fig. 5.
Scheme of the evolutionary steps involved in the formation of polyploids A. multifida and A. baldensis. Ideogram of A. sylvestris, A. cylindrica, A. virginiana and 5S rDNA loci distribution in A. baldensis are taken from Mlinarec et al. (2012).
To test the affinity between the D subgenomes of A. multifida and A. baldensis, labelled gDNA of A. multifida was hybridized to mitotic chromosomes of A. baldensis. Interestingly, GISH of A. baldensis root-tip metaphases revealed hybridization of A. multifida genomic DNA to all three chromosome sets of A. baldensis (Supplementary Data Fig. S2).
Thus, the cytogenetic data obtained in this study are in agreement with the ITS and NTS phylogeny, confirming that A. multifida and A. baldensis are allopolyploids. The B chromosome set in A. multifida and the A and B chromosome sets in A. baldensis are closely related to A. sylvestris, A. cylindrica or A. virginiana. The similarity between the complete genomes of A. multifida and A. baldensis, as revealed by GISH, suggest that these two polyploids, in addition to the A and B subgenome, also have the D subgenome in common. On the basis of these observations, we propose that A. multifida and A. baldensis originate from similar progenitor species or species in closely related taxa.
Genomic changes following polyploidization
In allotetraploid A. multifida rDNA from two parental genomes is expected, whereas in allohexaploid A. baldensis rDNA from three parental genomes is expected. Surprisingly, detailed ITS sequence analysis revealed only one group of ITS sequences in A. multifida and only two groups of ITS sequences in A. baldensis. Furthermore, FISH revealed that two pairs of 35S rDNA sites instead of four were detected in A. multifida and four instead of six were detected in A. baldensis (Fig. 3b, f). Subsequent GISH revealed that the 35S rDNA sequences assigned to the B subgenome of A. multifida were not detectable (Fig. 3d, g, asterisk) indicating that the major proportion of 35S rDNA sequences of the B subgenome have been lost in A. multifida.
We also observed that in A. baldensis and A. multifida, two pairs of homeologous chromosomes carry terminal reciprocal translocation, one pair belonging to the B subgenome and the other to the D subgenome. Homeologous translocation occurred among similar homeologous linkage groups B6–D7 in both A. multifida (Fig. 3a, c, d, arrows) and A. baldensis (Fig. 3e, g, arrows). It was observed in all three tested individuals of A. baldensis and A. multifida, again suggesting that A. baldensis and A. multifida originate from similar progenitor species or species in closely related taxa.
Southern blot analysis
As we have shown by GISH analysis that the 35S rDNA of the Multifida-origin subgenome (the B subgenome) was missing in A. multifida, we wanted to confirm this by Southern blot analysis (Fig. 4). gDNAs of A. cylindrica, A. virginiana, A. sylvestris, A. multifida and A. baldensis were restricted and hybridized with the 18S rDNA subunit probe (Fig. 4). The quantity of gDNA loaded into each lane was approximately the same for each species (Fig. 4a). Species of the Multifida group (A. cylindrica, A. virginiana and A. sylvestris) generated a similar pattern of bands, having one strong species-specific band of approx. 4 kb and two smaller bands corresponding to approx. 3 and approx. 2·5 kb. Interestingly, these bands were completely absent in A. multifida, supporting our previous conclusion that the major proportion of the 35S rDNA from the B subgenome has been lost in A. multifida. In addition, two bands corresponding to 7 and 10 kb were detected in A. multifida which were also present in A. baldensis (Fig. 4b, asterisks). As A. multifida and A. baldensis share the D subgenome, which is not present in species of the Multifida group, these larger bands probably originate from that subgenome. Furthermore, the remaining bands in A. baldensis (Fig. 4b, arrows) were of similar size to those in the lanes of species of the Multifida group, suggesting that they originate from the Multifida-origin subgenome (the A and B subgenomes). The hybridization signals of these bands in A. baldensis were of similar intensity to those in the lanes of species of the Multifida group. Given that A. baldensis has inherited two chromosome sets from species of the Multifida group, the similar intensity between these signals indicates a subsequent loss of the 35S rDNA loci of the A or B subgenomes in A. baldensis, in agreement with the FISH results.
Fig. 4.
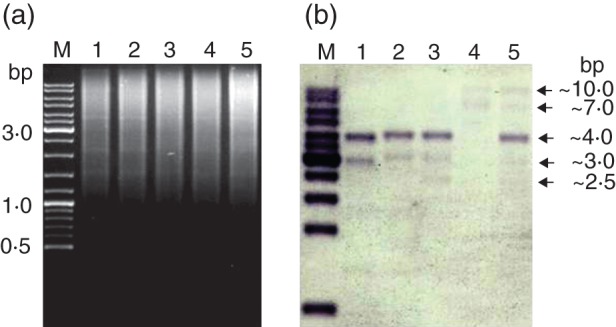
Genomic restriction digests (a) and Southern blotting analyses (b) of 1, A. cylindrica; 2, A. virginiana; 3, A. sylvestris; 4, A. multifida; and 5, A. baldensis with restriction endonuclease EcoRV and probed with 18S rDNA. Asterisks indicate the 35S rDNA of the Baldensis-origin subgenome (the D subgenome), while arrows indicate 35S rDNA of the Multifida-origin subgenome (the A and B subgenomes) in A. baldensis and A. multifida.
Phenotype similarities between A. baldensis and A. multifida and their potential parental species A. sylvestris, A. cylindrica and A. virginiana
As polyploids often show new phenotypic characteristics not present in their diploid progenitors we compared some morphological characteristics of A. baldensis and A. multifida with their putative parental species A. sylvestris, A. cylindrica and A. virginiana. On the basis of leaf morphology, A. baldensis represents an intermediate phenotype between those of A. multifida and A. sylvestris, A. cylindrica and A. virginiana, while achene fruits of A. baldensis resemble those of A. multifida and A. sylvestris, although they are considerably larger than those of both A. multifida and A. sylvestris (Supplementary Data Fig. S3).
DISCUSSION
Origin of A. multifida and A. baldensis
The results of this study show that A. multifida (2n = 4x = 32) and A. baldensis (2n = 6x = 48) are allopolyploids composed of two (BBDD) and three subgenomes (AABBDD), respectively. In the case of A. baldensis, GISH showed cross-hybridization of gDNAs of A. sylvestris, A. virginiana and A. cylindrica to its A and B chromosome sets despite the fact that these chromosome sets are distinguishable by size and morphology (Mlinarec et al., 2012). Such cross-hybridization to A. baldensis is probably a result of small intergenomic divergence between members of the Multifida group. That the members of the Multifida clade occur in similar habitats and are interfertile (Heimburger, 1962) provide further support for their relatedness. However, sequence analyses were useful in distinguishing between the two chromosome sets of the Multifida origin in A. baldensis. The clustering of NTS sequences of the Multifida origin into two sublades (B1 and B2) suggests that sequences in each subclade could be matched with the sequences of their potential parental species (Kotseruba et al., 2010). Therefore, clustering of NTS sequences of A. baldensis with those of A. sylvestris within the B2 subclade indicates that A. sylvestris might be involved in the origin of A. baldensis. The karyotype similarity between the A chromosome set and the chromosome set of A. sylvestris indicates that the latter may be a potential donor of the A chromosome set to A. baldensis. Accordingly, clustering of NTS sequences of A. baldensis with those of both A. cylindrica and A. virginiana within the B1 subclade suggests that both species are also involved in the origin of A. baldensis. Therefore, the B chromosome set of A. baldensis could originate from the common ancestor of A. cylindrica and A. virginiana or it could be of homoploid origin involving A. cylindrica and A. virginiana as parental species. Both hypotheses are equally possible and could be resolved by the use of single-copy nuclear genes in phylogenetic analysis (Kelly et al., 2010). Thus, DNA analytical data support our hypothesis that all three species (A. sylvestris, A. cylindrica or A. virginiana) or their progeny could be the ancestral donors of the A and B chromosome sets in A. baldensis. This is further supported by the finding that the NTS clones of A. baldensis contain species-specific sequences identical to either A. sylvestris, A. cylindrica or A. virginiana.
In A. multifida, the ITS sequences did not help in revealing the origin of the B subgenome as the major proportion of the B subgenome-type 35S rDNA repeats have been lost in this species. rDNA loci loss has been reported in many hybrids and allopolyploids such as Nicotiana tabaccum (Lim et al., 2000), Iris versicolor (Lim et al., 2007b), grass Zingeria kochi (Kotseruba et al., 2010) and Brassica napus (Książcyk et al., 2011). In addition, we obtained incongruent results from the molecular and cytogenetic analyses of NTS sequences of A. multifida. We expected that the major 5S rDNA locus from the B subgenome would have NTS sequences associated with the Multifida group because the B subgenome originates from this group of species. However, all clones derived from A. multifida formed a single well-supported clade that differed significantly from the clones of the Multifida group. This finding suggests that the 5S rDNA underwent interlocus homogenization in A. multifida.
Previous phylogenetic studies which used plastid DNA sequences as molecular markers place A. multifida within the Multifida clade (Meyer et al., 2010). This suggests that in A. multifida the Multifida-origin subgenome (the B subgenome) is maternal. Accordingly, we concluded that in the same species, the Baldensis-origin subgenome (the D subgenome) is paternal. Loss of 35S rDNA loci from the maternal parent (donor of the B genome) and maintenance in the paternal parent (donor of the D subgenome) suggest that in A. multifida the maternal genome evolves more rapidly than the paternal one. Although the nucleo-cytoplasmic interaction hypothesis of Gill (1991) predicts that the paternal genome should evolve more rapidly than the maternal one, there are more and more examples of genetic changes targeted at the maternal genome donor (this study; Lim et al., 2000; Clarkson et al., 2005; Guggisberg et al., 2008).
Here we observed an interesting correlation between A. multifida and A. baldensis. Both species share genomic changes such as intergenomic translocation between B and D subgenomes and loss of the maternal-type 35S rDNA. Intergenomic reciprocal recombination, observed in A. multifida and A. baldensis, suggests that chromosome pairing between homeologous chromosomes took place. Accumulating evidence indicates that chromosome pairing between homeologous chromosomes is commonplace in polyploids, notably in neopolyploids such as Brassica napus (Nicolas et al., 2007, 2009; Szadkowski et al., 2011) and synthetic allotetraploid tobacco (Petit et al., 2010). The similarity between the complete genomes of A. multifida and A. baldensis, as shown on both molecular and cytogenetic level, implies that these two polyploids, in addition to the A and B subgenome, also have the D subgenome in common. However, the origin of the D subgenome remains unclear. Its karyotype is unique among diploid Anemone species investigated so far regarding chromosome size and position of 5S rDNA (Mlinarec et al., 2012). However, similar karyological characteristics found in the sister genus Pulsatilla (Mlinarec et al., 2012) suggest that the Pulsatilla sp. could be the genome donors to the D chromosome set of A. multifida and A. baldensis. However, we consider this hypothesis to be unlikely because no molecular marker used so far places A. multifida and A. baldensis close to the Pulsatilla clade. By contrast, phylogenetic analyses showed that A. multifida and A. baldensis are closely related to the Multifida and Baldensis groups (this study; Meyer et al., 2010). This suggests that one of the diploid members of the Baldensis group or its progeny might be the ancestral donors of the D chromosome set to A. multifida and A. baldensis. Anemone parviflora, as the only extant diploid candidate for the genome donor, does not appear likely as a progenitor because no A. parviflora-like ITS sequences were found in both polyploids (this study). Alternatively, the possibility that the genome donor of the D subgenome was closely related to the members of the Baldensis group, but went extinct due to dramatic climatic change, cannot be ruled out. Future phylogenetic and karyological studies that include all species of the Baldensis group are expected to reveal the origin of the D subgenome as well as the origin of two other hexaploids, A. drummondii and A. lithophila.
Biogeographical implications
We propose here that the New World species A. multifida and Old World species A. baldensis have either the same parental species or parental species in closely related taxa (as shown in a model in Fig. 5). How and when their parental species came into contact is a question that can be tackled on palaeoclimatic grounds.
The early Quaternary Arctic flora was recruited from survivors of the arcto-Tertiary forests, combined with immigrants from temperate mountain ranges (Murray, 1995). One such immigrant, the progenitor of the Multifida clade and probably the most similar to extant A. virginiana and A. cylindrica, immigrated into the Arctic area in the early Quaternary. Soon after, it crossed with the likely progenitor of the Baldensis clade (Fig. 5). Successive cycles of divergent evolution among populations isolated in different glacial refugia and migration into deglaciated terrain led to the origin of two allotetraploid taxa, A. multifida (2n = 4x = 32) and A. baldensis (2n = 4x = 32), with their distribution centres in North America and Europe, respectively. Indeed, tetraploid A. baldensis has been reported in the Arctic area (Baumberger, 1970) but these reports are relatively old and should be taken with caution. In Europe, tetraploid A. baldensis (2n = 4x = 32) could have crossed with diploid A. sylvestris (2n = 2x = 16), resulting in the formation of hexaploid A. baldensis (2n = 6x = 48) (Fig. 5). This hypothesis is also supported by leaf morphology. Anemone multifida has a characteristic leaf shape, which is unique among Anemone species, while A. baldensis is of intermediary type between A. multifida and A. sylvestris regarding leaf morphology (Supplementary Data Fig. S3). ITS sequence analysis showed that the genome of A. sylvestris was involved in the formation of A. baldensis before the Pleistocene glaciations. A mutation rate of roughly one mutation in the ITS region every 100 000 years (Kay et al., 2006) suggests that hexaploid A. baldensis originated between 3 and 2·5 Mya. Range contraction/expansion cycles during the Quaternary led to subsequent migration of hexaploid A. baldensis southward in Europe. In the same period, A. multifida could have spread across North America and then migrated to South America along with the glacial range expansion. Postglacially, i.e. during the last 10 000–15 000 years, A. baldensis and A. multifida have probably achieved their present disjunctive distribution in the mountains of southern Europe and South America, respectively.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank D. Mihelj for maintaining the plant material. We are indebted to D. Papeš for ongoing support. This work was funded by the Ministry of Science, Education and Sport of the Republic of Croatia, grant no. 119-1191196-1201.
LITERATURE CITED
- Baumberger H. Chromosomenzahlbestimmungen und karyotypanalysen bei den Gattungen Anemone, Hepatica und Pulsatilla. Berichte der Schweizerischen Botanischen Gesellschaft. 1970;80:17–95. [Google Scholar]
- Besendorfer V, Krajačić-Sokol I, Jelenić SP, et al. Two classes of 5S rDNA unit arrays of the silver fir, Abies alba Mill.: structure, localization and evolution. Theoretical and Applied Genetics. 2005;110:730–741. doi: 10.1007/s00122-004-1899-y. [DOI] [PubMed] [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, et al. Polyploidy in arctic plants. Biological Journal of the Linnean Society. 2004;82:521–536. [Google Scholar]
- Cifuentes M, Grandont L, Moore G, Chèvre AM, Jenczewski E. Genetic regulation of meiosis in polyploidy species: new insights into an old question. New Phytologist. 2010;186:29–36. doi: 10.1111/j.1469-8137.2009.03084.x. [DOI] [PubMed] [Google Scholar]
- Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR. Long-term genome diploidization in allopolyploid section Repandae (Solanaceae) New Phytologist. 2005;168:241–252. doi: 10.1111/j.1469-8137.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–793. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gaeta RT, Pires JC. Homoeologous recombination in allopolyploids: the polyploidy ratchet. New Phytologist. 2010;186:18–28. doi: 10.1111/j.1469-8137.2009.03089.x. [DOI] [PubMed] [Google Scholar]
- Gajewski W. Cytotaxonomic investigations on Anemone L. I. Anemone janczewskii, a new amphidiploid species of hybrid origin. Acta Societatis Botanicorum Poloniae. 1946;17:129–194. [Google Scholar]
- Gerlach WL, Dyer TA. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Research. 1980;8:4851–4865. doi: 10.1093/nar/8.21.4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill BS. Nucleo-cytoplasmic interaction (NCI) hypothesis of genome evolution and speciation in polyploidy plants. In: Sasakuma T, Kinoshita T, editors. Nuclear and organellar genomes of wheat species. Proceedings of Dr. H. Kihara memorial international symposium on cytoplasmic engineering in wheat. Yokohama: Kihara Memorial Foundation; 1991. pp. 48–53. [Google Scholar]
- Guggisberg A, Baroux C, Grossniklaus U, Conti E. Genomic origin and organization of the allopolyploid Primula egaliksensis investigated by in situ hybridization. Annals of Botany. 2008;101:919–927. doi: 10.1093/aob/mcn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimburger M. Cytotaxonomic studies in the genus Anemone. Canadian Journal of Botany. 1959;37:587–612. [Google Scholar]
- Heimburger M. A karyotype study of Anemone drummondii and its hybrid with A. multifida. Canadian Journal of Botany. 1961;39:497–502. [Google Scholar]
- Heimburger M. Comparison of chromosome size in species of Anemone and their hybrids. Chromosoma. 1962;13:328–340. [Google Scholar]
- Hillis DM, Huelsenbeck JP. Signal, noise and reliability in molecular phylogenetic analyses. Journal of Heredity. 1992;83:189–195. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- Janczewski E. Les hybrides du genre Anemone III. Bull. intern. Academia Scientarum Cracoviensis. 1892;1892:228–230. [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam A, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–102. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Kay KM, Whittall JB, Hodges SA. A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evolutionary Biology. 2006;6(36) doi: 10.1186/1471-2148-6-36. http://dx.doi.org/10.1186/1471-2148-6-36 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LJ, Leitch AR, Clarkson JJ, Hunter RB, Knapp S, Chase MW. Intragenic recombination events and evidence for hybrid speciation in Nicotiana (Solanaceae) Molecular Biology and Evolution. 2010;27:781–799. doi: 10.1093/molbev/msp267. [DOI] [PubMed] [Google Scholar]
- Kotseruba V, Pistrick K, Blattner FR, et al. The evolution of the hexaploid grass Zingeria kochii (Mez) Tzvel. (2n = 12) was accompanied by complex hybridization and uniparental loss of ribosomal DNA. Molecular Phylogenetics and Evolution. 2010;56:146–155. doi: 10.1016/j.ympev.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Kovarik A, Pires JC, Leitch AR, et al. Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics. 2005;169:931–944. doi: 10.1534/genetics.104.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Książcyk T, Kovarik A, Eber F, et al. Immediate unidirectional epigenetic reprogramming of NORs occurs independently of rDNA rearrangements in synthetic and natural forms of a polyploidy species Brassica napus. Chromosoma. 2011;120:557–571. doi: 10.1007/s00412-011-0331-z. [DOI] [PubMed] [Google Scholar]
- Levan A, Fredga K, Sandberg A. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:210–220. [Google Scholar]
- Lim KY, Kovarík A, Matyásek R, Bezdek M, Lichstenstein CP, Leitch AR. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma. 2000;109:161–172. doi: 10.1007/s004120050424. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovarik A, Matyášek R, et al. Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytologist. 2007a;175:756–763. doi: 10.1111/j.1469-8137.2007.02121.x. [DOI] [PubMed] [Google Scholar]
- Lim KY, Matyasek R, Kovarik A, Leitch AR. Parental origin and genome evolution in the allopolyploid Iris versicolor. Annals of Botany. 2007b;100:219–224. doi: 10.1093/aob/mcm116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KY, Soltis DE, Soltis PS, et al. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae) PLoS ONE. 2008;3:e3353. doi: 10.1371/journal.pone.0003353. http://dx.doi.org/10.1371/journal.pone.0003353 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks GE, Schweizer D. Giemsa banding: karyotype differences in some species of Anemone and in Hepatica nobilis. Chromosoma. 1974;44:405–416. [Google Scholar]
- Meyer KM, Hoot SB, Arroyo MTK. Phylogenetic affinities of South American Anemone (Ranunculaceae) including the endemic segregate genera, Barneoudia and Oreithales. International Journal of Plant Sciences. 2010;171:323–331. [Google Scholar]
- Mlinarec J, Papeš D, Besendorfer V. Ribosomal, telomeric and heterochromatin sequences localization in the karyotype of Anemone hortensis. Botanical Journal of the Linnean Society. 2006;150:177–186. [Google Scholar]
- Mlinarec J, Chester M, Siljak-Yakovlev S, Papeš D, Besendorfer V. Molecular structure and chromosome distribution of three repetitive DNA families in Anemone hortensis L. (Ranunculaceae) Chromosome Research. 2009;17:331–343. doi: 10.1007/s10577-009-9025-2. [DOI] [PubMed] [Google Scholar]
- Mlinarec J, Šatović Z, Mihelj D, Malenica N, Besendorfer V. Cytogenetic and phylogenetic studies of diploid and polyploid members of tribe Anemoninae (Ranunculaceae) Plant Biology. 2012;14:525–536. doi: 10.1111/j.1438-8677.2011.00519.x. [DOI] [PubMed] [Google Scholar]
- Murray DF. Causes of arctic plant diversity: origin and evolution. In: Chapin FS, Körner C, editors. Arctic and alpine biodiversity: patterns, causes and ecosystem consequences. Heidelberg: Springer; 1995. pp. 21–32. [Google Scholar]
- Nicolas SD, Le Mignon G, Eber F, et al. Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics. 2007;175:487–503. doi: 10.1534/genetics.106.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas SD, Leflon M, Monod H, et al. Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. The Plant Cell. 2009;21:373–385. doi: 10.1105/tpc.108.062273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C, Alix K, Just J, et al. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytologist. 2010;186:37–45. doi: 10.1111/j.1469-8137.2009.03096.x. [DOI] [PubMed] [Google Scholar]
- Petit M, Guidat C, Daniel J, et al. Mobilization of retrotransposons in synthetic allotetraploid tobacco. New Phytologist. 2010;186:135–147. doi: 10.1111/j.1469-8137.2009.03140.x. [DOI] [PubMed] [Google Scholar]
- Puizina J, Sviben T, Krajačić-Sokol I, et al. Cytogenetic and molecular characterization of Abies alba genome and its relationship with other members of Pinaceae family. Plant Biology. 2008;10:256–267. doi: 10.1111/j.1438-8677.2007.00018.x. [DOI] [PubMed] [Google Scholar]
- Szadkowski E, Eber F, Huteau V, et al. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytologist. 2010;186:102–112. doi: 10.1111/j.1469-8137.2010.03182.x. [DOI] [PubMed] [Google Scholar]
- Szadkowski E, Eber F, Huteau F, et al. Polyploid formation pathways have an impact on genetic rearrangements in resynhesized Brassica napus. New Phytologist. 2011;191:884–894. doi: 10.1111/j.1469-8137.2011.03729.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods), Version 4. Sunderland, MA: Sinauer; 2003. [Google Scholar]
- Tamura M. Angiospermae: Ordnung Ranunculales, Fam. Ranunculaceae. In: Engler A., Prantl K., editors. Berlin: Duncker et Humboldt; 1995. pp. 1–555. Die Natürlichen Pflanzenfamilien. ed. 2, 17aIV, P. Hiepko (ed.) [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Hemleben V. Pattern and degree of methylation in ribosomal RNA genes of Cucurbita pepo L. Plant Molecular Biology. 1994;26:1167–1179. doi: 10.1007/BF00040697. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Schneeweiss GM, Stuessy TF, et al. Chromosomal stasis in diploids contrasts with genome restructuring in auto- and allopolyploid taxa of Hepatica (Ranunculaceae) New Phytologist. 2007;174:669–682. doi: 10.1111/j.1469-8137.2007.02019.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.