Abstract
Environmental agents induce intragenic alterations in the FRA3B/FHIT chromosome fragile site, resulting in fragile FHIT allele loss early in cancer development. Fhit knockout mice are predisposed to tumor development and Fhit gene therapy reduces tumor burden. Repair-deficient cancers are likely to be Fhit-deficient and Fhit-deficient cells show enhanced resistance to ultraviolet C, mitomycin C, camptothecin and oxidative stress-induced cell killing. Loss of Fhit leads to alterations in the DNA damage response checkpoint and contributes to DNA instability. Hsp60/Hsp10 are Fhit interactors, suggesting a direct role for Fhit in stress responses. Fhit also interacts with and stabilizes ferrodoxin reductase (Fdxr), a mitochondrial flavoprotein that transfers electrons from NADPH to cytochrome P450, suggesting a role for Fhit in the modulation of reactive oxygen species production and of genomic damage.
Keywords: carcinogens, DNA instability, ferredoxin reductase, FHIT, HSP, reactive oxygen species, tumor suppressor gene
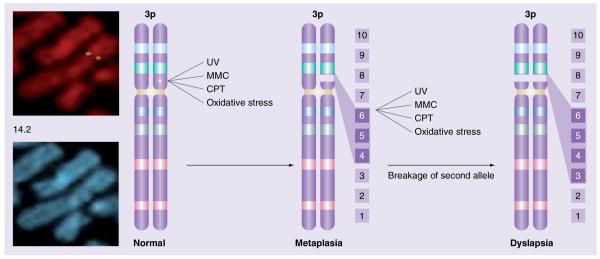
Malignant transformation is a multistep process involving numerous genetic changes, which include loss of tumor-suppressor gene function, oncogene activation and alterations of modifier genes [1,2]. These genetic changes affect cellular processes such as survival, proliferation and genomic stability. Precancerous cells experience selective pressure to escape from the cell cycle block induced by checkpoint responses to DNA damage, and DNA damage checkpoint genes are frequently mutated to overcome the block and allow neoplastic progression [3,4]. The short arm of human chromosome 3 is a common site of chromosomal alterations in human malignant disease. In four major regions of 3p (3p25, 3p21.3, 3p14.2 and 3p12), allelic losses have been reported in cancers of the kidney, lung and breast, among others [5,6]. The 3p14.2 region is particularly interesting to cancer researchers because it includes other genetic landmarks, including the most active common fragile site (the FRA3B locus) [7], a familial kidney cancer-associated breakpoint t(3;8)(p14.2;q24) [8], and a papilloma virus integration site [9]. The familial translocation indicated that a gene important for cancer initiation or progression might be located at 3p14.2. In 1996, an intensive search of this genomic region resulted in the identification of the fragile histidine triad (FHIT) gene [10,11]. Subsequent studies established that FHIT is a target of chromosomal rearrangements at 3p14.2 [12]. Loss of Fhit expression is observed in premalignant lesions of lung, esophagus, cervix and other organs, suggesting that loss of Fhit expression, due to the susceptibility of FHIT/FRA3B to carcinogen damage, plays a role in initial stages of multistep carcinogenesis (Figure 1) [13,14]. Since the FHIT gene is prone to breakage and deletion in precancer or early carcinogenesis, and precancerous lesions and cancers show clonal expansion of cells with specific FHIT gene alterations, it was proposed that FHIT gene alteration and loss of Fhit function provides a selective advantage for this clonal growth [14]. The abnormal checkpoint responses and genome instability of Fhit-deficient cells could clearly contribute to selective growth of precancerous cells with damaged FHIT alleles. For example, carcinogens cause damage at FRA3B, leading to breakage of a FHIT allele with loss of exons 4–6 [13]. Further carcinogen exposure can lead to damage at the second FHIT allele with loss of other exons, 3–5 for example (illustrated in Figure 1). Loss of the second FHIT allele can lead to total loss of Fhit protein expression, as observed in many dysplastic lesions (Figure 1). In this review we summarize new studies describing FHIT alterations in human cancers, from analysis of Fhitdeficient mice to identification of important Fhit biological functions.
Figure 1. Chromosome 3, showing the gap or break at 3p14 (right).
Hybridization of fluorescent genomic fragments of the 5′-end of the FHIT gene (green, left) shows the position of one end of FHIT flanking the fragile region. The carcinogens in cigarette smoke and other carcinogens also cause damage at FRA3B, leading to breakage of one FHIT allele with loss of exons 4–6, for example. Further carcinogen exposure can lead to damage at the second FHIT allele with loss of exons 3–5. Loss of one FHIT allele is frequently detected in the non-neoplastic epithelium of current and former smokers, and might lead to areas of metaplasia with reduced Fhit protein expression. Loss of the second FHIT allele can lead to total loss of Fhit protein expression (lower right), as observed in many dysplastic lesions. CPT: Cisplatin; MMC: Mitomycin C; UV: Ultraviolet.
FHIT alterations occur in most cancers
The presence of the FHIT gene in the most active common chromosome fragile region has been proposed as an example of a tumor suppressor gene altered by chromosome translocations and deletions rather than by point mutation; several reports had suggested that the FHIT gene was altered in cancer simply because it was in a fragile region and not because it had contributed to the clonal expansion [15,16]. If this were the case, it would be difficult to explain why the FHIT genomic alteration, within all cells of a specific cancer-derived cell line, was identical to the nucleotide. Many cancer cell lines and primary cancers exhibiting hemi- or homo-zygous deletions with end points within the FHIT gene and reduced or absent Fhit expression have been reported [13]. Furthermore, many studies have reported altered FHIT loci and protein expression in precancerous lesions, suggesting that FHIT alterations are an early event in carcinogenesis [17]. In esophageal cancer, Mori et al. reported that most of the in situ lesions, 50% of severe and moderate dysplasias and 33% of mild dysplasias were Fhit negative [18]; in the study of Kitamura et al., reduced Fhit expression was observed in 68% of in situ and 43.5% of esophageal dysplastic lesions [19]. Hao et al. found reduced Fhit expression only in a small fraction of adenomatous colon lesions, but reduced Fhit expression was associated with a greater degree of dysplasia [20]. In cervical cancer, Connolly et al. observed reduced or absent Fhit staining in 71% of invasive cancers and in 52% of highgrade intraepithelial lesions (HSILs) with invasive cancer [21]. In approximately 85% of bronchial dysphasia there was loss of Fhit expression [22]. In our study of ductal carcinoma in situ (DCIS), reduced Fhit expression was observed in 70% of pure DCIS and 52% of DCIS adjacent-to-invasive tumor cases. In total, 20% of pure DCIS cases exhibited individual glands of adjacent normal tissue with absence of expression [23]. These clinical findings supported the proposal that FHIT inactivation occurs in the early steps of carcinogenesis in many organs. Several studies have shown that FHIT alterations are common in environmental carcinogen-related cancers, such as those of lung and esophagus. An association between smoking and loss of Fhit expression or FHIT deletion was shown in lung and esophageal cancers. In fact, alterations in Fhit expression in lung carcinomas were more frequent and occurred earlier than p53 mutations or deregulation of the epidermal growth factor receptor (EGFR) [22]. Interestingly, Fhit inactivation was almost twice as frequent in tumors of smokers (75%) than nonsmokers (39%) [22,24]. These studies suggested that loss of Fhit is an early event in the development of lung cancer, and that predisposing genetic changes have occurred even in normal-appearing epithelium in cases heavily exposed to environmental carcinogens. Recent studies point to detection of FHIT deletions in purified bronchial epithelial cells from sputum as a way to improve early detection of lung cancer with 58% sensitivity [25]. Exposure to asbestos and to γ-irradiation during the Chernobyl accident caused an increase in FHIT inactivation in lung cancer and preneoplastic bronchial lesions [26,27]. Smoking history and alcohol abuse associated with higher frequency of loss of Fhit expression was also reported in esophageal cancer [18]. If FHIT is one of the first targets of carcinogens, the ability of the host to repair this initial damage or to eliminate cells carrying damage to the FHIT locus may prevent clonal expansion. In support of this idea, loss of FHIT function is observed more frequently in cancers developing in individuals with constitutional alterations to genes involved in DNA repair, such as the BRCA1 and 2 and mismatch repair genes [28–30].
The Fhit-deficient mouse: a model to study the role of Fhit in carcinogen-induced tumors
The mouse Fhit ortholog also encompasses a common fragile site, Fra14A2 on murine chromosome 14, and sustains homozygous deletions in murine cancer cell lines [12]; therefore, Fhit-knockout mice have served as models for the study of Fhit function. To establish an animal model and to explore the role of Fhit in tumorigenesis, our laboratory developed a mouse strain carrying one or two inactivated Fhit alleles. Fhit+/− and Fhit−/− mice, although healthy and fertile, showed increased susceptibility to spontaneous and carcinogen-induced tumors [31,32]. Epidemiological studies have linked exposure to nitrosamines to a high incidence of esophageal cancer [33]; to better understand the role of Fhit in this neoplasia, our laboratory performed a carcinogenesis study with N-nitrosomethyl-benzylamine (NMBA), an environmental nitrosamine [34]. All Fhit+/− mice developed several fore stomach tumors, and some developed sebaceous gland tumors after six doses of intragastric NMBA, while only 25% of wild-type mice developed tumors. The tumors were a mixture of benign, in situ and invasive lesions. The NMBA-induced tumor spectrum in Fhit+/− mice was similar to a human syndrome called Muir–Torre syndrome, a variant of the hereditary nonpolyposis colorectal cancer (HNPCC or Lynch) syndrome, which is caused by inactivation of a mismatch repair gene, usually MSH2. Homozygous deletions of FHIT were observed in half of the cancer cell lines with a mutant mismatch repair gene; among nine Msh2-negative human colon cancer cells, eight were negative for Fhit, and Fhit loss was reported in 90% of BRCA1- and BRCA2-associated cancers [29,35,36]. These correlations suggest that fragile genes are especially vulnerable to damage-induced alterations in cells with ‘caretaker’ gene deficiencies.
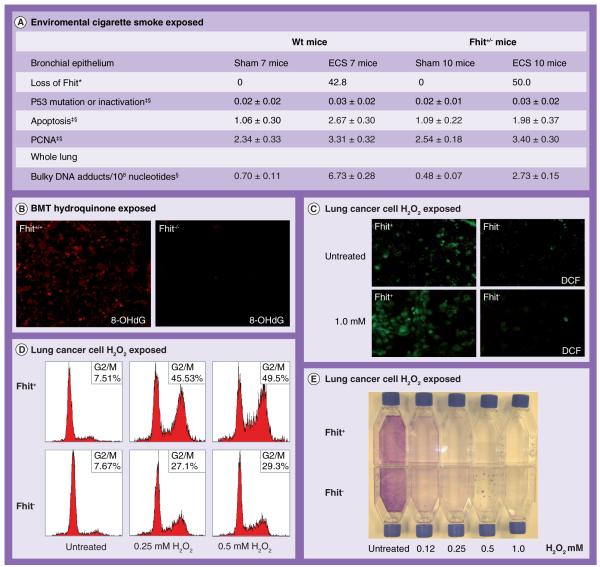
As discussed previously, several studies have observed more frequent FHIT gene deletions in tumors of smokers than tumors of nonsmokers [14]. Recently D’Agostini et al., to assess the role of Fhit under controlled experimental conditions, exposed mice to environmental cigarette smoke (ECS) and evaluated Fhit RNA or protein in the respiratory tract of rodents (Figure 2A) [37]. Confirming previous studies conducted in humans, they found that after 14 days of exposure to ECS, there was loss of Fhit protein in bronchial/bronchiolar epithelium of half of the mice, both wild-type or Fhit+/−. Interestingly, they also found that the oral administration of N-acetylcysteine (NAC), a well-known antioxidant, attenuated the ECS-related loss of Fhit. Another possible role of Fhit in response to carcinogen was demonstrated in the study of Balansky et al., who found that after treatment with benzo[a]pyrene (B[a]P), a prototypic genotoxic, carcinogenic polycyclic aromatic hydrocarbon (PAH), preneoplastic lesions of the uterus were more frequent in Fhit+/− mice [38]. They also found that B6/129 F1 mice underwent spontaneous alopecia areata and hair bulb cell apoptosis, which was greatly accelerated either by Fhit heterozygosity or by B[a]P treatment, suggesting that Fhit plays a role in the pathogenesis of alopecia areata. Intriguingly, the oral administration of NAC inhibited occurrence of this inflammatory skin disease. Thus, this thiol compound with anti-inflammatory properties, which can inhibit apoptosis consequent to DNA damage and redox imbalances, also inhibits the stimuli that cause loss of Fhit protein due to cigarette smoke in the bronchial epithelium of rats, and prevents spontaneous alopecia areata and hair bulb cell apoptosis in Fhit heterozygous mice. Recently, Ishii et al. have shown that in vivo-transplanted, hydroquinone-exposed, Fhit-deficient mouse bone marrow cells escaped the bone marrow suppression exhibited by wild-type bone marrow [39]. After hydroquinone exposure, occurrence of the oxidized base 8-hydroxyguanosine, a marker of DNA damage, was also reduced in Fhit-deficient bone marrow, as was production of intracellular reactive oxygen species (ROS) (Figure 2B). Also, in this experimental model, treatment with NAC relieved hydroquinone-induced suppression of colony formation by wild-type hematopoietic cells, suggesting that decreased oxidative damage to Fhit-deficient cells, relative to wild-type hematopoietic cells, accounts for the survival advantage of Fhit-xsdeficient bone marrow. Zanesi et al. reported that 4-methylnitrosamino-1–3-pyridyl-1-butanone induced lung tumors (adenomas and carcinomas) in 100% of Fhit−/−Vhl+/− mice and adenomas in 40% of Fhit−/− mice by 20 months of age [40]. Thus, double deficiency in murine homologues of 3p suppressor genes, including haploinsufficiency of Vhl, predisposes to spontaneous and induced lung cancers, demonstrating that Fhit-deficient mice will be useful, in combination with other 3p tumor suppressors, in recapitulating a pattern of lung cancer development similar to the human pattern; such double- or triple-deficient mice will be excellent lung cancer prevention and therapy models. This summary of effects in several mouse models illustrates the strong correlation between FHIT-allele loss and early steps in the neoplastic process. It is especially interesting that in varied experiments conducted in different laboratories, an anti-inflammatory and ROS scavenging agent such as NAC could inhibit Fhit loss or prevent its negative effect.
Figure 2. Fhit in carcinogen-induced cancer and stress response.
(A) Table summarizing alterations induced in B6–129(F1) mice, either wildtype or Fhit+/−, by whole-body exposure to ECS for 15 days [36]. *Percentage of mice showing extensive loss of Fhit within each experimental group; ‡Percentage of mice with the reported alteration; §Means ± SE among all mice within each experimental group. (B) Detection of 8-OHdG representing the oxidative DNA damage in hydroquinone-exposed transplanted bone marrow cells; Fko and Wt bone marrow cells were exposed to hydroquinone and transplanted to recipient mice [38]. (C) Increased green fluorescent DCF signal in H1299 Fhit-expressing cells (D1) under stress condition [43]. (D) FACS ana lysis of D1 and E1 cell-cycle kinetics at 48 h after oxidative stress treatment; cells were treated with increasing concentrations of H2O2 (0.25, 0.5 mM) for 4 h. (E) Colony-formation assay of H1299/D1 (Fhit+) and H1299/E1 (Fhit−) cells after 5 h treatment with H2O2 at indicated concentrations [43].
BMT: Bone marrow transplant; DCF: Dichlorofluorescein diacetate; ECS: Environmental cigarette smoke; PCNA: Proliferating cell nuclear antigen protein.
Fhit function in the stress response
Despite strong evidence of Fhit tumor suppressor function, our knowledge of specific Fhit signalling pathways and mechanisms involved in its suppressor activity is limited. It is well-known that overexpression of Fhit results in apoptosis in Fhit-deficient cancer cells [41], and recent studies have demonstrated a role for Fhit in responses to genotoxic damage induced by ultraviolet C (UVC) light, mitomycin C, camptothecin and ionizing radiation; approximately tenfold more colonies of Fhit-deficient cells survived exposure to high UVC doses [42,43]. After mitomycin C treatment approximately sixfold, and after UVC treatment 3.5-fold more Fhit-positive human cancer cells than Fhit-negative cells had died, and UVC-surviving Fhit−/– cells showed more than fivefold increased mutation frequency. Furthermore, a recent study from our laboratory has shown that p53-negative lung cancer cells expressing Fhit are more sensitive to H2O2 treatment compared with Fhit-negative cells undergoing apoptosis or G2/M arrest when Fhit was present (Figure 2) [44]. After oxidative stress, Fhit-positive cells also produced higher ROS (Figure 2C, D & E) and 8-hydroxyguanosine levels. Fhit-deficent cancer cells show a mild response to oxidative stress, producing less ROS, crucial mediators of chemotherapy-induced cell death, confirming that Fhit deficiency could negatively influence treatment outcome.
It has also been shown that introduction of exogenous Fhit into cells in vitro leads to modulation of expression of the checkpoint proteins Hus1 and Chk1 at mid-S checkpoint, modulation that led to induction of apoptosis in esophageal cancer cells, but not in noncancerous primary cultures [45]. The results suggested that the DNA-damage-susceptible FRA3B/FHIT chromosome fragile region encodes a protein that is necessary for protecting cells from accumulation of DNA damage through its role in modulation of checkpoint proteins; and the inactivation of Fhit contributes to the accumulation of abnormal checkpoint phenotypes in cancer development [45,46]. The study showed that when Fhit was down-modulated in 293 kidney cells, the level of Hus1 and Rad1 proteins was reduced, and experiments further suggested that Fhit protein stabilizes Hus1 protein, preventing its degradation by the proteasome pathway.
Together, these studies are consistent with the conclusion that Fhit, as a modifier of stress responses, can affect stabilization of proteins involved in activation of checkpoints, cell-cycle block or apoptosis.
Fhit protein structure
In contrast to the gene, the ubiquitously expressed FHIT mRNA is only 1.1 kb long and encodes a 146 amino acid chain that forms a protein of 16.8 kD. Fhit is a member of the histidine triad (HIT) nucleotide-binding protein superfamily, encoding a diadenosine polyphosphate (ApnA) hydrolase that cleaves substrates such as diadeno sine triphosphate (Ap3A) and diadenosine tetraphosphate (Ap4A) to AMP plus the other nucleo tide [13]. Structural studies have shown that Fhit is a dimer that binds two Ap3A substrates, presenting a highly phosphorylated surface, with five phosphate groups and two adenosine moieties [47]. The conserved HIT motif (His-X-His-X-His-X-X, where X is a hydro phobic residue) of Fhit is located near its C-terminal end and the hydrolytic activity of Fhit is lost when histidine 96 (H96) is replaced with asparagine (Fhit-H96N), showing that the H96 central histidine residue of the triad is essential for Ap3A hydrolase activity [13]; however, the FhitH96N protein suppresses tumorigenicity about as well as wild-type Fhit. This finding led Garrison and colleagues to suggest that the Fhit enzyme–substrate complex might send the tumor-suppression signal [49]. Fhit-induced apoptosis in cancer cells was correlated with the apparent substrate-binding activity (Km) but not the substrate hydrolytic activity (kcat). Recently, it was observed that the sequence DSIY114EEL of Fhit, which fits the consensus for targets of phosphorylation by Src tyrosine kinase family members, could be phosphorylated in vitro and in vivo [48]. Garrison et al. determined the steadystate Km and kcat values for the Ap3A hydrolase activity of recombinant nonphospho-Fhit, monophospho-Fhit and diphospho-Fhit, and found that the Km and kcat values for monophospho-Fhit and diphospho-Fhit are lower than for nonphospho-Fhit [49]. Recent studies have shown that a Fhit mutant that carries a phenylalanine instead of a tyrosine at position 114 (Y114F), and thus unable to be phosphorylated on tyrosine 114 by Src, does not induce apoptosis in cancer cells and prevents Fhit degradation [50,51]. Furthermore, Bianchi et al. have shown that during the signaling of activated tyrosine kinase receptors after EGF treatment, phosphorylation of Fhit leads to its degradation; the subsequent reduction in Fhit protein level allows transmission of the mitogenic signal [51]. This would suggest a key role for Fhit in the balance of proliferation/survival/apoptosis signals, and indicates that Fhit phosphorylation at Y114 may be a key feature of Fhit molecular function.
Identification of Fhit effectors to define Fhit function
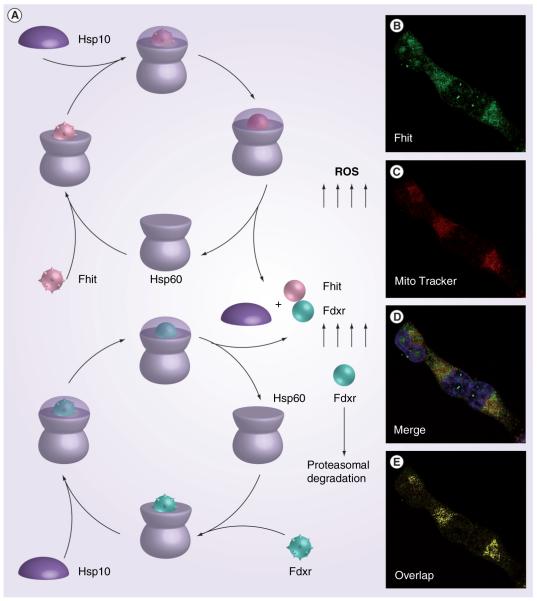
One way to define specific protein signal pathways is through identification of interacting proteins that could be effectors of function. Earlier searches for Fhit-interacting proteins pointed to several candidate proteins, none of which we could confirm as interactors [Huebner K et al., Ohio State University, Columbus, OH, USA. Unpublished Data] by co-immunoprecipitaton experiments, including Ubc9, α-tubulin, Mdm2 and β-catenin [52–55]. To readdress the question of Fhit protein interactors, and find the molecular basis for the important role of Fhit in cancer prevention, we used adenovirus transduced Fhit-His6 protein for Fhit complex purification after cross-linking, and Fhit-bound proteins, Hsp60, Hsp10, ferredoxin reductase (Fdxr), malate dehydrogenase (Mdh), electrontransfer flavoprotein (Etfb) and mitochondrial aldehyde dehydrogenase 2 (Adh 2) were identified (Table 1) [44]. The mitochondrial localization of these proteins led to our determination that Fhit localizes to mitochondria, as well as cytosol. We found that Fhit is important for Fdxr stability and that Fhit–Fdxr interaction leads to increased ROS generation through electron leakage from the shuttle system NADPH-cytochrome P450 via ferrodoxin. Using HCT116 cells with one or three copies of the FDXR gene, we found that cells with only one copy were less susceptible to Fhit-induced apoptosis and that the level of Fdxr protein was stabilized in the presence of Fhit protein. Furthermore, the finding that Fhit interacts with Hsp ‘stress proteins’ [56], in particular the chaperone machinery Hsp60/10, suggests that the Hsp complex may be important for Fhit stability, correct folding and mitochondrial addressing; it is even possible that Fhit itself is part of important stress machinery, able to protect vital proteins such as Fdxr from degradation under stress conditions (see model, Figure 3). Intriguingly, Hsp60 interacts directly with Fdxr [Pichiorri F et al. Ohio State University, Columbus, OH, USA. Unpublished Data] and we speculate that Hsp60/10 may also mediate the correct folding and mitochondrial import of Fdxr and Mdh [52] and protect them from stress denaturation (see model, Figure 3). The finding that Fhit interacts with Fdxr and thereby increases Fdxr stability suggests that Fhit may be part of specific molecular machinery to protect important proteins from degradation and affecting the cellular response to the damaging effects of ROS.
Table 1.
Candidate Fhit protein partners isolated from Trapasso et al. [44].
| Protein | Accession no. |
Mr (kDa) |
Function/category | Subcellular localization |
|---|---|---|---|---|
| Hsp60 | NP_002147 | 60 | 60 kDa heat shock protein | Cytosol/mitochondria |
| Malate dehydrogenase | NP_005909 | 33 | Catalyzes the reversible oxidation of malate to oxaloacetate |
Mitochondrial matrix |
| Electron transfer flavoprotein | NP_001976 | 28 | Specific electron acceptor for mitochondrial dehydrogenases |
Mitochondrial matrix |
| Hsp10 | AAC96332 | 10 | 10 kDa heat shock protein | Cytosol/mitochondria |
| Mitochondrial aldehyde dehydrogenase 2 |
NP_000681 | 55 | Second enzyme of the major oxidative pathway of alcohol metabolism |
Mitochondrial matrix |
| Ferredoxin reductase | P22570 | 54 | First electron transfer protein in all the mitochondrial p450 systems |
Mitochondrial matrix |
Figure 3. Model for the role of Fhit in protection of Fdxr from proteaosomal degradation.
(A) Non-native Fhit (rough pink ball) and Fdxr (rough green ball) proteins bind to the trans ring of a Hsp60–Hsp10 complex. End-to-end exchange of Hsp10 results in the encapsulation of the protein substrate in the cis cavity. Release of Hsp10 and substrate proteins, Fhit and Fdxr, in the folded conformation (pink and green smooth balls, respectively) for mitochondrial addressing; we hypothesized that absence of Fhit leads to enhanced proteasomal degradation of Fdxr [43]. The immunofluorescence microscopy (B–E) was performed with antiFhit serum on H1299 Fhit-positive cells; Fhit staining was detected using fluorescein isothiocyanate (green) conjugated antirabbit immunoglobulin (IgG); MitoTracker Red staining, which identifies mitochondria, shows partial colocalization with Fhit. (E) The yellow color shows the colocalizations points.
Conclusion
The FHIT gene was discovered in 1996, and its protein product, Fhit, was shown in numerous studies by laboratories worldwide, to be a tumor suppressor that was reduced in expression or lost in the majority of cancers. Progress in understanding the Fhit signal pathways was less rapid. Very recently, several pathways affected by Fhit loss, as summarized in this review, have been identified: a stress response pathway; a DNA damage response checkpoint pathway; and a role in the production of reactive oxygen species on exposure to oxidative stress. Participation of Fhit in these pathways suggests reasons why Fhit-deficient cells show increased resistance to certain cytotoxic therapeutic drugs, and suggest that in normal cells Fhit is involved in the protection of cells from preneoplastic changes.
Future perspective
Delineation of direct downstream effectors of the Fhit-suppressor pathway will lead to:
▪ Intensification of mechanistic studies of Fhit function that may influence future preventive and therapeutic strategies to activate the Fhit pathway or to specifically target Fhit-deficient cancers;
▪ Clarification of mechanisms by which this gene product protects cells from carcinogens and modulates sensitivity to external insults.
The finding that ROS generation precedes Fhitmediated apoptosis in lung cancer cells is in satisfying accord with previous reports showing:
▪ That exposure to carcinogens in cigarette smoke was associated with FHIT gene loss;
▪ The importance of Fhit loss as a negative prognostic factor in various clinical settings (for example, assessment of Fhit status in preneoplastic or neoplastic conditions may be predictive of responses to antioxidant treatments).
Finally, the fact that Fhit interacts with Hsp chaperones and this interaction appears to be important for its stability and correct localization in mitochondria, where it can initiate apoptosis through affecting stability of mitochondria respiratory chain proteins, suggests that drugs targeted to such chaperones might have efficacy in preneoplastic, neoplastic or other conditions associated with Fhit loss.
Executive summary.
FHIT alterations occur in most cancers
▪ The FHIT gene is a tumor-suppressor gene altered by chromosome translocations and deletions.
▪ FHIT alterations are an early event in carcinogenesis.
▪ FHIT alterations are common in environmental carcinogen-related cancers, such as those of the lung and esophagus.
▪ Loss of FHIT function is observed more frequently in cancers with alterations to genes involved in DNA repair.
The Fhit-deficient mouse: a model to study the role of Fhit in carcinogen-induced tumors
▪ The mouse Fhit ortholog also encompasses a common fragile site, Fra14A2 on murine chromosome 14.
▪ Fhit−/− mice, although healthy and fertile, showed increased susceptibility to spontaneous and carcinogen-induced tumors.
▪ The N-nitrosomethyl-benzylamine (NMBA)-induced tumor spectrum in Fhit+/− mice was similar to a human syndrome known as Muir–Torre syndrome.
▪ Rodents exposed to environmental cigarette smoke showed a loss of Fhit protein in bronchial/bronchiolar epithelium.
▪ Fhit plays a role in the pathogenesis of alopecia areata.
▪ 4-methylnitrosamino-1–3-pyridyl-1-butanone induced lung tumors in 100% of Fhit−/−Vhl+/− mice and adenomas in 40% of Fhit−/− mice.
▪ In different experiments conducted in different laboratories, the anti-inflammatory agent N-acetylcysteine (NAC) could inhibit Fhit loss or prevent its negative effect.
Fhit function in the stress response
▪ The knowledge of specific Fhit signaling pathways and mechanisms involved in its suppressor activity is limited.
▪ Overexpression of Fhit results in apoptosis in Fhit-deficient cancer cells.
▪ Fhit-deficient cells are less sensitive to genotoxic damage induced by ultraviolet C light, mitomycin C, camptothecin and ionizing radiation.
▪ Lung cancer cells expressing Fhit are more sensitive to H2O2 treatment.
▪ After oxidative stress, Fhit-positive cells also produced higher reactive oxygen species (ROS) and 8-hydroxyguanosine levels.
▪ Expression of exogenous Fhit in cells in vitro leads to modulation of expression of checkpoint proteins Hus1 and Chk1 at the mid-S checkpoint.
Fhit structure
▪ FHIT mRNA is only 1.1 kb long and codes for an 146 amino acid chain that forms a protein of 16.8 kD.
▪ Fhit is a member of the histidine triad (HIT) nucleotide-binding protein superfamily, encoding a diadenosine polyphosphate (ApnA) hydrolase.
▪ The Fhit mutant H96N has shown that the H96 residue is essential for Ap3A hydrolase activity, but suppresses the tumorigenicity just as well as wild-type FHIT.
▪ The Fhit mutant Y114F, unable to be phosphorylated on tyrosine 114 by Src, does not induce apoptosis in cancer cells and prevents Fhit degradation.
Identification of Fhit effectors to define Fhit function
▪ Using adenovirus transduced Fhit-His6, Fhit-linked proteins, Hsp60, Hsp10, ferredoxin reductase (Fdxr), malate dehydrogenase (Mdh), electron-transfer flavoprotein (Etfb) and mitochondrial aldehyde dehydrogenase 2 (Adh 2) were identified.
▪ Fhit localizes to mitochondria, as well as cytosol.
▪ Fhit is important for Fdxr stability and Fhit–Fdxr interaction.
▪ Fhit interacts with the chaperone machinery Hsp60/10, suggesting that the Hsp complex may be important for Fhit stability.
▪ The finding that Fhit interacts with Fdxr may be part of specific molecular machinery to protect important proteins from degradation, and also affects the cellular response to the damaging effects of ROS.
Footnotes
Financial & competing interests disclosure The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Flavia Pichiorri, Ohio State University Comprehensive Cancer Center, Department of Molecular Virology, Molecular Virology and Medical Genetics. 460 W 12th Avenue, 43210 Columbus, OH, USA.
Tiziana Palumbo, Ohio State University Comprehensive Cancer Center, Department of Molecular Virology, Molecular Virology and Medical Genetics. 460 W 12th Avenue, 43210 Columbus, OH, USA.
Sung-Suk Suh, Ohio State University Comprehensive Cancer Center, Department of Molecular Virology, Molecular Virology and Medical Genetics. 460 W 12th Avenue, 43210 Columbus, OH, USA.
Hiroshi Okamura, Ohio State University Comprehensive Cancer Center, Department of Molecular Virology, Molecular Virology and Medical Genetics. 460 W 12th Avenue, 43210 Columbus, OH, USA.
Francesco Trapasso, Ohio State University Comprehensive Cancer Center, Department of Molecular Virology, Molecular Virology and Medical Genetics. 460 W 12th Avenue, 43210 Columbus, OH, USA.
Hideshi Ishii, Department of Gastroenterological Surgery, Osaka University Graduate School of Medicine, Suita, Osaka, 565-087, Japan.
Kay Huebner, Ohio State University Comprehensive Cancer Center, Department of Molecular Virology, Molecular Virology and Medical Genetics. 460 W 12th Avenue, 43210 Columbus, OH, USA.
Carlo M Croce, Director Human Cancer Genetics, Ohio State University, 460 W 12th Avenue, 43210 Columbus, OH, USA Tel.: +1 614 292 4930 Fax: +1 614 292 3558 carlo.croce@osumc.edu.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Croce CM. Role of chromosome translocations in human neoplasia. Cell. 1987;49(2):155–156. doi: 10.1016/0092-8674(87)90552-6. ▪▪ One of the most important reviews explaining the role of chromosome translocation and how they are related to cancer initiation.
- 2.Soloman E, Borrow J, Goddart A. Chromosome aberrations and cancer. Science. 1991;254(5035):1153–1160. doi: 10.1126/science.1957167. [DOI] [PubMed] [Google Scholar]
- 3.Bartkova J, Horejsí Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. ▪▪ Reports an elegant study describing how cancer development is associated with DNA replication stress, which leads to DNA double-strand breaks, including breaks at FRA3B, checkpoint activation and selective pressure for p53 mutations.
- 4.Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):829–830. doi: 10.1038/nature03485. ▪▪ Reports an elegant study describing how cancer development is associated with DNA replication stress, which leads to DNA double-strand breaks, including breaks at FRA3B, checkpoint activation and selective pressure for p53 mutations.
- 5.Naylor SL, Johnson BE, Minna JD, et al. Loss of heterozygosity of chromosome 3p markers in small cell lung cancer. Nature. 1987;329(6138):451–454. doi: 10.1038/329451a0. ▪ One of the first studies supporting the hypothesis that loss of alleles of chromosome 3p contributes to tumorigenesis in small-cell lung cancer.
- 6.Hibi K, Takahashi T, Yamakawa K, et al. Three distinct regions involved in 3p deletions in human lung cancer. Oncogene. 1992;7(3):445–449. [PubMed] [Google Scholar]
- 7.Glover TW, Stein CK. Chromosome breakage and recombination at fragile sites. Am. J. Hum. Genet. 1988;43(3):265–273. [PMC free article] [PubMed] [Google Scholar]
- 8.Glover TW, Coyle-Morris JF, Frederick PL, et al. Translocation t(3;8) (p14.2;q24.1) in renal cell carcinoma affects expression of the common fragile site at 3p14 (FRA3B) in lymphocytes. Cancer Genet. Cytogenet. 1988;31(1):69–73. doi: 10.1016/0165-4608(88)90013-1. [DOI] [PubMed] [Google Scholar]
- 9.Rassool FV, McKeithan TW, Neilly ME, et al. Preferential integration of marker DNA into the chromosomal fragile site at 3p14.2: a novel approach to cloning fragile sites. Proc. Natl Acad. Sci. USA. 1991;88(15):6657–6661. doi: 10.1073/pnas.88.15.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohta M, Inoue H, Cotticelli MG, et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84(4):587–597. doi: 10.1016/s0092-8674(00)81034-x. ▪▪ Very important paper on the identification and cloning of the human FHIT gene.
- 11.Zimonjic D, Druck T, Ohta M, et al. Positions of chromosome 3p14.2 fragile sites (FRA3B) within the FHIT gene. Cancer Res. 1997;57(6):1166–1170. [PubMed] [Google Scholar]
- 12.Huebner K, Croce CM. Cancer and the FRA3B/FHIT fragile locus: it’s a hit. Br. J. Cancer. 2003;88(10):1501–1506. doi: 10.1038/sj.bjc.6600937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huebner K, Croce CM. FRA3B and other common fragile sites: the weakest links. Nat. Rev. Cancer. 2001;1(3):214–221. doi: 10.1038/35106058. ▪▪ Nice review summarizing evidence that the product of the FHIT gene is partially or entirely lost in most human cancers, indicating that it has a tumor-suppressor function.
- 14.Pekarsky Y, Zanesi N, Palamarchuk A, Huebner K, Croce CM. FHIT: from gene discovery to cancer treatment and prevention. Lancet Oncol. 2002;3(12):748–754. doi: 10.1016/s1470-2045(02)00931-2. [DOI] [PubMed] [Google Scholar]
- 15.Iliopoulos D, Guler G, Han SY, et al. Roles of FHIT and WWOX fragile genes in cancer. Cancer Lett. 2006;232(1):27–36. doi: 10.1016/j.canlet.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 16.Le Beau MM, Drabkin H, Glover TW, et al. An FHIT tumor suppressor gene? Genes. Chromosomes Cancer. 1998;21(13):281–289. doi: 10.1002/(sici)1098-2264(199804)21:4<281::aid-gcc1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 17.Ishii H, Ozawa K, Furukawa Y. Alteration of the fragile histidine triad gene early in carcinogenesis: an update. J. Exp. Ther. Oncol. 2003;3(6):291–296. doi: 10.1111/j.1533-869x.2003.01101.x. [DOI] [PubMed] [Google Scholar]
- 18.Kuroki T, Tajima Y, Furui J, Kanematsu T. Common fragile genes and digestive tract cancers. Surg. Today. 2006;36(1):1–5. doi: 10.1007/s00595-005-3094-4. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura A, Yashima K, Okamoto E, et al. Reduced Fhit expression occurs in the early stage of esophageal tumorigenesis: no correlation with p53 expression and apoptosis. Oncology. 2001;61(3):205–211. doi: 10.1159/000055376. [DOI] [PubMed] [Google Scholar]
- 20.Hao XP, Willis JE, Pretlow TG, et al. Loss of fragile histidine triad expression in colorectal carcinomas and premalignant lesions. Cancer Res. 2000;60(1):18–21. [PubMed] [Google Scholar]
- 21.Connolly DC, Greenspan DL, Wu R, et al. Loss of Fhit expression in invasive cervical carcinomas and intraepithelial lesions associated with invasive disease. Clin. Cancer Res. 2000;6(9):3505–3510. [PubMed] [Google Scholar]
- 22.Sozzi G, Pastorino U, Moiraghi L, et al. Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res. 1998;58(22):5032–5037. ▪ Well-conducted study describing the loss of FHIT in preinvasive bronchial lesions; suggests a potential use of this gene in the early detection of lung cancer and in chemopreventive studies as an intermediate biomarker.
- 23.Guler G, Uner A, Güler N, et al. Concordant loss of fragile gene expression early in breast cancer development. Pathol. Int. 2005;55(8):471–478. doi: 10.1111/j.1440-1827.2005.01855.x. [DOI] [PubMed] [Google Scholar]
- 24.Mao L, Lee JS, Kurie JM, et al. Clonal genetic alterations in the lungs of current and former smokers. J. Natl. Cancer Inst. 1997;89(12):857–862. doi: 10.1093/jnci/89.12.857. [DOI] [PubMed] [Google Scholar]
- 25.Qiu Q, Todd NW, Li R, et al. Magnetic enrichment of bronchial epithelial cells from sputum for lung cancer diagnosis. Cancer. 2008;114(4):275–283. doi: 10.1002/cncr.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pylkkanen L, Wolff H, Stjernvall T, et al. Reduced Fhit protein expression and loss of heterozygosity at FHIT gene in tumours from smoking and asbestos-exposed lung cancer patients. Int. J. Oncol. 2002;20(2):285–290. [PubMed] [Google Scholar]
- 27.Chizhikov V, Chikina S, Gasparian A, et al. Molecular follow-up of preneoplastic lesions in bronchial epithelium of former Chernobyl clean-up workers. Oncogene. 2002;21(15):2398–2405. doi: 10.1038/sj.onc.1205310. [DOI] [PubMed] [Google Scholar]
- 28.Ingvarsson S, Agnarsson BA, Sigbjornsdottir BI, et al. Reduced Fhit expression in sporadic and BRCA2-linked breast carcinomas. Cancer Res. 1999;59(11):2682–2689. [PubMed] [Google Scholar]
- 29.Mori M, Mimori K, Masuda T, et al. Absence of Msh2 protein expression is associated with alteration in the FHIT locus and Fhit protein expression in colorectal carcinoma. Cancer Res. 2001;61(20):7379–7382. [PubMed] [Google Scholar]
- 30.Turner BC, Ottey M, Zimonjic DB, et al. The fragile histidine triad/common chromosome fragile site 3B locus and repair-deficient cancers. Cancer Res. 2002;62(14):4054–4060. [PubMed] [Google Scholar]
- 31.Zanesi N, Fidanza V, Fong LY, et al. The tumor spectrum in FHIT-deficient mice. Proc. Natl. Acad. Sci. USA. 2001;98(18):10250–10255. doi: 10.1073/pnas.191345898. ▪ First study describing spontaneous and induced tumor spectra observed in mice with one or both Fhit alleles inactivated; suggested that FHIT is a haplo-insufficient tumor suppressor, so that loss of only one FHIT allele would predispose to cancer.
- 32.Zanesi N, Pekarsky Y, Croce CM. A mouse model of the fragile gene FHIT: from carcinogenesis to gene therapy and cancer prevention. Mutat. Res. 2005;591(1–2):103–109. doi: 10.1016/j.mrfmmm.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Magee PN. The experimental basis for the role of nitroso compounds in human cancer. Cancer Surv. 1989;8(2):207–239. [PubMed] [Google Scholar]
- 34.Fong LY, Fidanza V, Zanesi N, et al. Muir-Torre-like syndrome in Fhit-deficient mice. Proc. Natl. Acad. Sci. USA. 2000;97(9):4742–4747. doi: 10.1073/pnas.080063497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingvarsson S, Agnarsson BA, Sigbjornsdottir BI, et al. Reduced Fhit expression in sporadic and BRCA2-linked breast carcinomas. Cancer Res. 1999;59(11):2682–2689. [PubMed] [Google Scholar]
- 36.Turner BC, Ottey M, Zimonjic DB, et al. The fragile histidine triad/common chromosome fragile site 3B locus and repair-deficient cancers. Cancer Res. 2002;62(14):4054–4060. [PubMed] [Google Scholar]
- 37.De Flora S, D’Agostini F, Balansky R, et al. High susceptibility of neonatal mice to molecular, biochemical and cytogenetic alterations induced by environmental cigarette smoke and light. Mutat. Res. 2008;659(1–2):137–146. doi: 10.1016/j.mrrev.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Balansky R, D’Agostini F, Ganchev G, et al. Influence of FHIT on benzo[a] pyrene-induced tumors and alopecia in mice: chemoprevention by budesonide and N-acetylcysteine. Proc. Natl Acad. Sci. USA. 2006;103(20):7823–7828. doi: 10.1073/pnas.0601412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ishii H, Mimori K, Ishikawa K, et al. Fhit-deficient hematopoietic stem cells survive hydroquinone exposure carrying precancerous changes. Cancer Res. 2008;68(10):3662–3670. doi: 10.1158/0008-5472.CAN-07-5687. [DOI] [PubMed] [Google Scholar]
- 40.Zanesi N, Mancini R, Sevignani C, et al. Lung cancer susceptibility in Fhit-deficient mice is increased by Vhl haploinsufficiency. Cancer Res. 2005;65(15):6576–6582. doi: 10.1158/0008-5472.CAN-05-1128. [DOI] [PubMed] [Google Scholar]
- 41.Ishii H, Dumon KR, Vecchione A, et al. Potential cancer therapy with the fragile histidine triad gene: review of the preclinical studies. J. Am. Med. Assoc. 2001;286(19):2441–2449. doi: 10.1001/jama.286.19.2441. [DOI] [PubMed] [Google Scholar]
- 42.Ottey M, Han SY, Druck T, et al. Fhit-deficient normal and cancer cells are mitomycin C and UVC resistant. Br. J. Cancer. 2004;91(9):1669–1677. doi: 10.1038/sj.bjc.6602058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu B, Han SY, Wang X, et al. Involvement of the Fhit gene in the ionizing radiation-activated ATR/CHK1 pathway. J. Cell Physiol. 2005;202(2):518–523. doi: 10.1002/jcp.20139. [DOI] [PubMed] [Google Scholar]
- 44.Trapasso F, Pichiorri F, Gaspari M, et al. Fhit interaction with ferredoxin reductase triggers generation of reactive oxygen species and apoptosis of cancer cells. J. Biol. Chem. 2008;283(20):13736–13744. doi: 10.1074/jbc.M709062200. ▪Describes Fhit molecular interactions and sublocalization.
- 45.Pichiorri F, Ishii H, Okumura H, Trapasso F, Wang Y, Huebner K. Molecular parameters of genome instability: Roles of fragile genes at common fragile sites. J. Cell Biochem. 2008;104(5):1525–1533. doi: 10.1002/jcb.21560. [DOI] [PubMed] [Google Scholar]
- 46.Ishii H, Wang Y, Huebner K. A Fhit-ing role in the DNA damage checkpoint response. Cell Cycle. 2007;6(9):1044–1048. doi: 10.4161/cc.6.9.4213. [DOI] [PubMed] [Google Scholar]
- 47.Campiglio M, Bianchi F, Andriani F, et al. Diadenosines as FHIT-ness instructors. J. Cell Physiol. 2006;208(2):274–281. doi: 10.1002/jcp.20633. [DOI] [PubMed] [Google Scholar]
- 48.Pekarsky Y, Garrison PN, Palamarchuk A, et al. Fhit is a physiological target of the protein kinase Src. Proc. Natl Acad. Sci. USA. 2004;101(11):3775–3779. doi: 10.1073/pnas.0400481101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garrison PN, Robinson AK, Pekarsky Y, Croce CM, Barnes LD. Phosphorylation of the human Fhit tumor suppressor on tyrosine 114 in Escherichia coli and unexpected steady state kinetics of the phosphorylated forms. Biochemistry. 2005;44(16):6286–6292. doi: 10.1021/bi047670s. [DOI] [PubMed] [Google Scholar]
- 50.Semba S, Trapasso F, Fabbri M, et al. Fhit modulation of the Akt–survivin pathway in lung cancer cells: Fhit-tyrosine 114 (Y114) is essential. Fhit modulation of the Akt–survivin pathway in lung cancer cells: Fhit-tyrosine 114 (Y114) is essential. Oncogene. 2006;25(20):2860–2872. doi: 10.1038/sj.onc.1209323. [DOI] [PubMed] [Google Scholar]
- 51.Bianchi F, Magnifico A, Olgiati C, et al. FHIT-proteasome degradation caused by mitogenic stimulation of the EGF receptor family in cancer cells. Proc. Natl Acad. Sci. USA. 2006;103(50):18981–18986. doi: 10.1073/pnas.0605821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi Y, Zou M, Farid NR, Paterson MC. Association of FHIT (fragile histidine triad), a candidate tumour suppressor gene, with the ubiquitin-conjugating enzyme hUBC9. Biochem. J. 2000;352(2):443–448. [PMC free article] [PubMed] [Google Scholar]
- 53.Chaudhuri AR, Khan IA, Prasad V, Robinson AK, Ludueña RF, Barnes LD. The tumor suppressor protein Fhit. A novel interaction with tubulin. J. Biol. Chem. 1999;274(34):24378–24382. doi: 10.1074/jbc.274.34.24378. [DOI] [PubMed] [Google Scholar]
- 54.Nishizaki M, Sasaki J, Fang B, et al. Synergistic tumor suppression by coexpression of FHIT and p53 coincides with FHIT-mediated MDM2 inactivation and p53 stabilization in human non-small cell lung cancer cells. Cancer. Res. 2004;64(16):5745–5752. doi: 10.1158/0008-5472.CAN-04-0195. [DOI] [PubMed] [Google Scholar]
- 55.Weiske J, Albring KF, Huber O. The tumor suppressor Fhit acts as a repressor of β-catenin transcriptional activity. Proc. Natl Acad. Sci. USA. 2007;104(51):20344–20349. doi: 10.1073/pnas.0703664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levy-Rimler G, Bell RE, Ben-Tal N, Azem A. Type I chaperonins: not all are created equal. FEBS Lett. 2002;529(1):1–5. doi: 10.1016/s0014-5793(02)03178-2. [DOI] [PubMed] [Google Scholar]
- 57.Chen J, Walter S, Horwich AL, Smith DL. Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nat. Struct. Biol. 2001;8(8):721–728. doi: 10.1038/90443. [DOI] [PubMed] [Google Scholar]