A radiogenomics strategy to accelerate the identification of prognostically important imaging biomarkers is presented, and preliminary results were demonstrated in a small cohort of patients with non-small cell lung cancer for whom CT and PET images and gene expression microarray data were available but for whom survival data were not available.
Abstract
Purpose:
To identify prognostic imaging biomarkers in non–small cell lung cancer (NSCLC) by means of a radiogenomics strategy that integrates gene expression and medical images in patients for whom survival outcomes are not available by leveraging survival data in public gene expression data sets.
Materials and Methods:
A radiogenomics strategy for associating image features with clusters of coexpressed genes (metagenes) was defined. First, a radiogenomics correlation map is created for a pairwise association between image features and metagenes. Next, predictive models of metagenes are built in terms of image features by using sparse linear regression. Similarly, predictive models of image features are built in terms of metagenes. Finally, the prognostic significance of the predicted image features are evaluated in a public gene expression data set with survival outcomes. This radiogenomics strategy was applied to a cohort of 26 patients with NSCLC for whom gene expression and 180 image features from computed tomography (CT) and positron emission tomography (PET)/CT were available.
Results:
There were 243 statistically significant pairwise correlations between image features and metagenes of NSCLC. Metagenes were predicted in terms of image features with an accuracy of 59%–83%. One hundred fourteen of 180 CT image features and the PET standardized uptake value were predicted in terms of metagenes with an accuracy of 65%–86%. When the predicted image features were mapped to a public gene expression data set with survival outcomes, tumor size, edge shape, and sharpness ranked highest for prognostic significance.
Conclusion:
This radiogenomics strategy for identifying imaging biomarkers may enable a more rapid evaluation of novel imaging modalities, thereby accelerating their translation to personalized medicine.
© RSNA, 2012
Supplemental material: http://radiology.rsna.org/lookup/suppl/doi:10.1148/radiol.12111607/-/DC1
Introduction
Personalized medicine aims to tailor medical care to the individual by characterizing molecular heterogeneity. Advances in high-throughput molecular technologies promise to produce biomarkers that will drive the future of patient-specific medical care (1–3). However, a limitation of these approaches is the need to acquire tissue through invasive biopsy. In such biopsies, samples are often obtained from only a portion of a generally heterogeneous lesion and cannot completely represent the lesion’s anatomic, functional, and physiologic properties, particularly its size, location, and morphology. Yet many of these lesion-specific features are acquired in routine imaging examinations and are known to be highly informative in diagnosis, clinical staging, and treatment planning. Despite the importance of these image features, only a handful of studies (4–7) have generated a radiogenomics map that integrates the genomic and image data.
Our work extends the use of radiogenomics mapping to address the growing demand for prognostic image-based biomarkers. Rapid advances in imaging technologies permit better anatomic resolution and provide noninvasive measurements of functional and physiologic tissue- and lesion-specific properties. Yet the ability to determine the clinical significance of newly identified image features relies on clinical studies that can be costly and require long-term follow-up periods. We propose a novel radiogenomics strategy that may identify new imaging biomarkers when long-term clinical outcomes are not immediately available. Our approach relies on first acquiring paired gene expression data and medical images at diagnosis from a study cohort, then leveraging the public gene expression data that contain clinical outcomes from a closely matched population (Fig 1a). A critical step in this approach involves predicting the image features in the study cohort in terms of gene signatures. We evaluate the prognostic significance of these gene signatures in public data sets for which gene expression and survival data are available. Predicting image features from gene signatures may not only enable immediate translational potential but may also suggest potential molecular mechanisms that may give rise to imaging phenotypes.
Figure 1a:
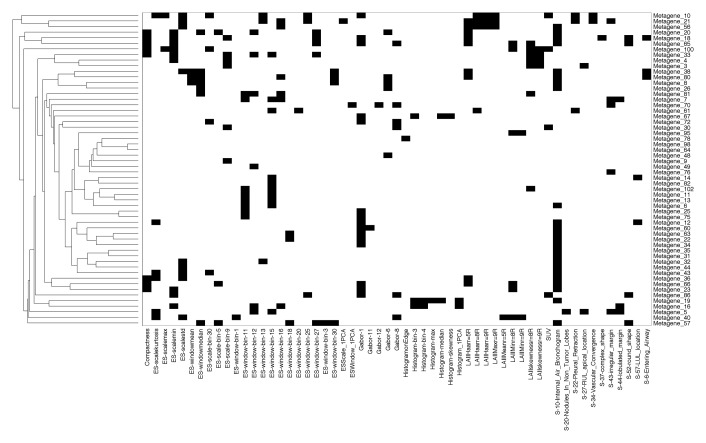
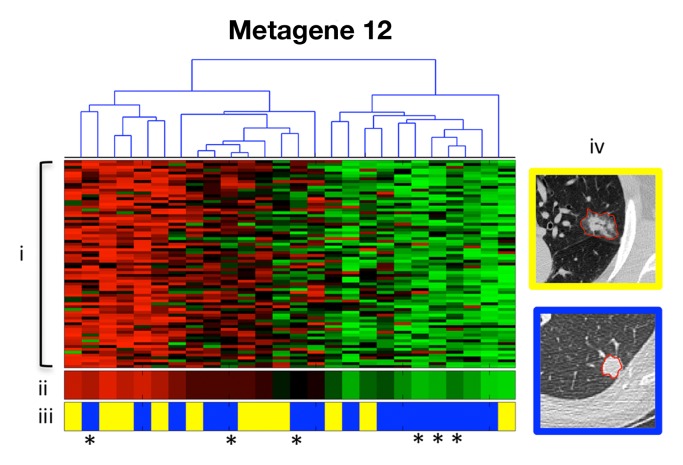
Creation of radiogenomics map for non-small cell lung cancer (NSCLC). (a) Strategy for creation and use of the radiogenomics map in NSCLC. Step 1 integrates the computed tomographic (CT) and positron emission tomographic (PET)/CT image and the gene microarray data from our study cohort. Step 2 maps the metagenes to publicly available microarray data with survival. Step 3 links image features expressed in terms of metagenes to public gene expression data. The dashes in this link highlight its indirectness, because by leveraging public gene expression data, we are able to associate the image features in the study cohort with survival, even without survival data in the study cohort. (b) Hierarchical clustering of radiogenomics correlations map with metagenes (in rows) and image features (in columns). Black squares = 243 significant associations between an image feature and a metagene with q < 5%. (c) Association between metagene 12 and the image feature for the internal air bronchogram; (i) gene expression of genes in metagene 12; (ii) metagene 12 expression; (iii) presence of an internal air bronchogram, where * = squamous lung carcinoma cases; and (iv) sample CT images of a lesion with (top) an internal air bronchogram present versus (bottom) a lesion with an internal air bronchogram absent. For gene expression, red = overexpression and green = underexpression; for image features, blue = absence of the feature and yellow = presence of the feature.
As a proof-of-concept, we applied our radiogenomics strategy to a human study cohort of patients with NSCLC in whom we retrospectively acquired medical images (CT and PET/CT images) and gene expression microarray data. We focused on NSCLC because it is the leading cause of cancer death, with an overall 5-year survival rate of 16% that has not changed appreciably over the past 15 years (8). Also, imaging has an important role in the management of NSCLC and will likely have an increased clinical role, on the basis of the results of the National Lung Screening Trial (9,10). Moreover, gene expression of NSCLC is abundantly available in public databases with clinical outcomes (11,12). To identify prognostic imaging biomarkers, we propose a radiogenomics strategy that integrates gene expression and medical images of patients for whom survival outcomes are not available by leveraging survival data in public gene expression data sets.
Materials and Methods
Image and Microarray Data Collection
With institutional review board approval, we studied data in 26 patients with NSCLC who underwent preoperative CT and fluorine 18 fluorodeoxyglucose (FDG) PET/CT between April 7, 2008, and May 21, 2010, and who had archived frozen tissue available for gene expression analysis (Table E1 [online]). We gathered preoperative thin-section CT and FDG PET/CT images, and a representative cross section of the excised lesion was used for microarray analysis. Image and gene expression data were gathered as described in Appendix E1 (online). Briefly, CT and FDG PET/CT images were deidentified, and image features were extracted manually by a radiologist using a controlled vocabulary and computationally by using predefined analytical software tools to characterize the lesion’s properties. This resulted in 153 computational image features, 26 semantic image features, and a PET standardized uptake value (SUV) for each patient’s tumor. Figure E1 (online) shows a single representative CT cross section of each of the 26 nodules included in this study, Table E2 (online) lists all of the semantic annotations applied to each tumor, and Table E8 (online) provides a description of the computational features.
From the same patients, we retrieved frozen tissue and extracted the RNA, which was analyzed with gene expression microarrays (Illumina HT-12). The gene expression data were first clustered on the basis of coexpression, and 56 clusters were selected for subsequent analysis because their cluster homogeneity was conserved in external data (Gene Expression Omnibus accession number, GSE8894) (13). Each of the clusters was represented by its metagene, which was defined as the first principal component of the cluster (Fig E2 [online]).
Creating the Radiogenomics Correlation Map
Initially, we established a radiogenomics correlation map for pairwise associations between metagenes with image features, using Significance Analysis of Microarrays (SAM) in R (version 1.28), with the false discovery rate (FDR) for multiple hypothesis testing correction (14). We used the two-class-unpaired and the continuous-response types of SAM for the binary- and continuous-valued image features, respectively. The SAM “Δ threshold” was set at 0.3, and 1000 permutations were used to estimate the FDR. A q value filter of 0.05 or less was used to identify statistically significant associations between metagenes and image features.
Creating the Radiogenomics Predictive Models
We built a predictive model of the metagenes in terms of the image features, using generalized linear regression with lasso regularization (glmnet package in R, version 1.5.1) (15). The regularization parameter was set such that at least 80% of the deviance is captured by the model. Similarly, we predicted each image feature in terms of the metagenes. Depending on the type of image feature, the response variable was set as binomial, multinomial, or Gaussian. The resulting predictive models of image features expressed in terms of metagenes can be regarded as surrogates for image features, and we define them as “predicted image features” (Fig 2a). We used leave-one-out cross validation to assess the model’s performance. The performance metric of the predicted semantic image features was the AUC. The performance metric for the predicted computational image features, which were continuously valued, was termed “accuracy” and was defined as 1 minus the error, where the error was defined as the average absolute error divided by the numeric range of the feature. Predictions with at least 65% AUC or 65% accuracy were selected for subsequent analysis.
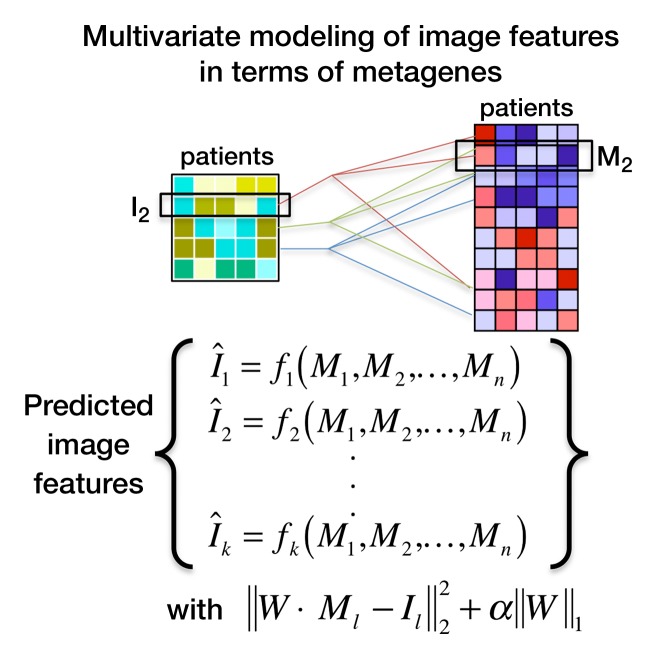
Figure 2a:
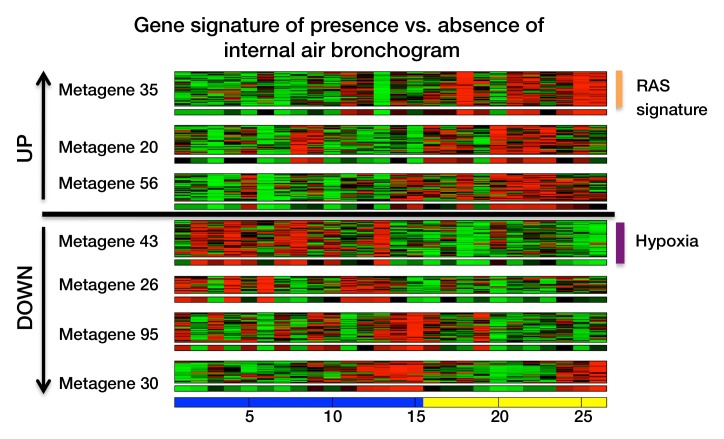
Multivariate modeling of image features in terms of metagenes. (a) Strategy for multivariate modeling of image features in terms of metagenes. Each image feature is modeled as a linear combination of metagenes, using L1 regularization to induce sparsity in the number of metagenes that are selected. I1 = the first image feature of k image features in total; Mj = jth metagene; fi = linear regression for the ith image feature; α = regularization parameter; Wj (not shown) = weight for each metagene M1, M2, to Mn; and W = matrix with all weights. (b) Semantic features predicted by metagenes with an area under the receiver operating characteristic curve (AUC) of 65% or greater, based on leave-one-out cross-validation analysis. (c) Multivariate metagene prediction model for the presence versus absence of internal air bronchogram at CT. The top seven metagenes, representing 95% of the weight of the multivariate model, are shown; the top three metagenes are upregulated when an air bronchogram is present, and the bottom four metagenes are downregulated. The downregulated metagenes are enriched in hypoxia-related pathways; the upregulated metagenes contain a Ras signature and genes upregulated by Ras. (d) Receiver operating characteristic curve for the predicted presence versus absence of internal air bronchogram at CT, when expressed in terms of metagenes. (e) Multivariate model for internal air bronchogram corresponding to c. For gene expression, red = overexpression and green = underexpression; for image features, blue = absence of the feature and yellow = presence of the feature. CI = confidence interval.
Figure 2e:
Multivariate modeling of image features in terms of metagenes. (a) Strategy for multivariate modeling of image features in terms of metagenes. Each image feature is modeled as a linear combination of metagenes, using L1 regularization to induce sparsity in the number of metagenes that are selected. I1 = the first image feature of k image features in total; Mj = jth metagene; fi = linear regression for the ith image feature; α = regularization parameter; Wj (not shown) = weight for each metagene M1, M2, to Mn; and W = matrix with all weights. (b) Semantic features predicted by metagenes with an area under the receiver operating characteristic curve (AUC) of 65% or greater, based on leave-one-out cross-validation analysis. (c) Multivariate metagene prediction model for the presence versus absence of internal air bronchogram at CT. The top seven metagenes, representing 95% of the weight of the multivariate model, are shown; the top three metagenes are upregulated when an air bronchogram is present, and the bottom four metagenes are downregulated. The downregulated metagenes are enriched in hypoxia-related pathways; the upregulated metagenes contain a Ras signature and genes upregulated by Ras. (d) Receiver operating characteristic curve for the predicted presence versus absence of internal air bronchogram at CT, when expressed in terms of metagenes. (e) Multivariate model for internal air bronchogram corresponding to c. For gene expression, red = overexpression and green = underexpression; for image features, blue = absence of the feature and yellow = presence of the feature. CI = confidence interval.
Leveraging Public Gene Expression Data Sets for Identifying Image Biomarkers
Despite the lack of survival outcomes in our study cohort, we identified candidate prognostic imaging biomarkers by mapping the predicted image features to public availability of gene expression data sets with clinical outcomes across hundreds of patients (Fig 1a). In particular, we used the NSCLC gene expression data set by Lee et al (13) because it was a relevantly large study (n = 138), it contains clinical outcomes, and it has has a histologic composition of NSCLC that is similar to that in our study cohort. Because prognostic signatures for adenocarcinoma and squamous carcinoma differ, we limited our survival analysis to cases of adenocarcinoma (n = 63), because they constituted a larger fraction of our study cohort. First, we mapped each predicted image feature to the Lee et al data to assess its prognostic significance separately. Next, we applied Cox proportional hazards modeling and Kaplan-Meier survival analysis to investigate the prognostic significance of predicted image features (survival R package, version 2.35–8). Kaplan-Meier curves were evaluated by splitting the predictor at its median to identify a good versus poor prognostic group. We used Cox proportional hazards modeling to determine whether the predicted image features added independent information in the presence of the clinical covariates, namely: age, sex, smoking, nodal stage, and tumor size. Finally, we built a multivariate survival model based on the predicted image features by using generalized linear regression models with lasso regularization (glmnet package in R, version 1.5.1) and evaluated its performance with 10-fold cross validation. We included the clinical covariates to determine whether the predicted image features provided independent prognostic value.
Results
Radiogenomics Correlation Map of NSCLC
Figure 1b shows a NSCLC radiogenomics correlation map of 243 statistically significant pairwise associations between metagenes with CT image features and PET SUV (Table E3 [online]). Several provocative associations were identified. For example, the presence or absence of the internal air bronchogram feature at CT was associated with metagene 12 (Fig 1c). The corresponding gene cluster contains genes that are specifically overexpressed in NSCLC, including KRAS (16,17). This particular association suggests that the presence of an internal air bronchogram is related to the overexpression of KRAS and its targets. Interestingly, none of the six patients with squamous carcinoma in our study cohort had internal air bronchograms, and none showed KRAS overexpression, which is consistent with the reported observations that activating KRAS mutations are exclusively associated with adenocarcinoma (18) and that Ras pathway activation is characteristic of adenocarcinoma (19) (Fig 1c). Such analyses of the radiogenomics map enable the synthesis of hypotheses that relate image phenotypes with genotype.
Figure 1b:
Creation of radiogenomics map for non-small cell lung cancer (NSCLC). (a) Strategy for creation and use of the radiogenomics map in NSCLC. Step 1 integrates the computed tomographic (CT) and positron emission tomographic (PET)/CT image and the gene microarray data from our study cohort. Step 2 maps the metagenes to publicly available microarray data with survival. Step 3 links image features expressed in terms of metagenes to public gene expression data. The dashes in this link highlight its indirectness, because by leveraging public gene expression data, we are able to associate the image features in the study cohort with survival, even without survival data in the study cohort. (b) Hierarchical clustering of radiogenomics correlations map with metagenes (in rows) and image features (in columns). Black squares = 243 significant associations between an image feature and a metagene with q < 5%. (c) Association between metagene 12 and the image feature for the internal air bronchogram; (i) gene expression of genes in metagene 12; (ii) metagene 12 expression; (iii) presence of an internal air bronchogram, where * = squamous lung carcinoma cases; and (iv) sample CT images of a lesion with (top) an internal air bronchogram present versus (bottom) a lesion with an internal air bronchogram absent. For gene expression, red = overexpression and green = underexpression; for image features, blue = absence of the feature and yellow = presence of the feature.
Figure 1c:
Creation of radiogenomics map for non-small cell lung cancer (NSCLC). (a) Strategy for creation and use of the radiogenomics map in NSCLC. Step 1 integrates the computed tomographic (CT) and positron emission tomographic (PET)/CT image and the gene microarray data from our study cohort. Step 2 maps the metagenes to publicly available microarray data with survival. Step 3 links image features expressed in terms of metagenes to public gene expression data. The dashes in this link highlight its indirectness, because by leveraging public gene expression data, we are able to associate the image features in the study cohort with survival, even without survival data in the study cohort. (b) Hierarchical clustering of radiogenomics correlations map with metagenes (in rows) and image features (in columns). Black squares = 243 significant associations between an image feature and a metagene with q < 5%. (c) Association between metagene 12 and the image feature for the internal air bronchogram; (i) gene expression of genes in metagene 12; (ii) metagene 12 expression; (iii) presence of an internal air bronchogram, where * = squamous lung carcinoma cases; and (iv) sample CT images of a lesion with (top) an internal air bronchogram present versus (bottom) a lesion with an internal air bronchogram absent. For gene expression, red = overexpression and green = underexpression; for image features, blue = absence of the feature and yellow = presence of the feature.
Predictive Models of Metagenes in Terms of Image Features
We found that image features predicted all 56 metagenes with a mean accuracy of 72% (range, 59%–83%; Table E4 [online]). In particular, metagene 43, which is enriched with genes in the hypoxia pathway (20,21), had the presence or absence of internal air bronchograms at CT as its top-ranking predictive semantic feature. When we tried to predict the 56 metagenes by using only the top 25 image features selected in an additional leave-one-out cross validation, we found that all metagenes were predicted with an average accuracy of 71% (range, 56%–85%). In other words, the metagenes could be predicted from 25 image features with reasonable accuracy. We also investigated whether either semantic or computational image features alone were sufficient to predict the metagenes with similar accuracy. Interestingly, the semantic features alone predicted 21 of the 56 metagenes (mean accuracy, 74%; range, 61%–83%); however, the computational features predicted all 56 metagenes (mean accuracy, 71%; range, 56%–83%). This finding suggests that, in our study cohort, semantic annotations were not needed for predicting the metagenes if the computational image features were available (Table E4 [online]).
Predictive Models of Image Features in Terms of Metagenes
We found that 115 of 153 image features were predicted from the metagenes with an accuracy of 65% or greater. These image features consisted of PET SUV, 10 semantic features, and 104 computational features (Fig 2b and Table E5 [online]). The top 10 predicted computational image feature models had an average accuracy of 85% (84%–86%) and were predominately associated with lesion size, edge shape, and edge sharpness.
Figure 2b:
Multivariate modeling of image features in terms of metagenes. (a) Strategy for multivariate modeling of image features in terms of metagenes. Each image feature is modeled as a linear combination of metagenes, using L1 regularization to induce sparsity in the number of metagenes that are selected. I1 = the first image feature of k image features in total; Mj = jth metagene; fi = linear regression for the ith image feature; α = regularization parameter; Wj (not shown) = weight for each metagene M1, M2, to Mn; and W = matrix with all weights. (b) Semantic features predicted by metagenes with an area under the receiver operating characteristic curve (AUC) of 65% or greater, based on leave-one-out cross-validation analysis. (c) Multivariate metagene prediction model for the presence versus absence of internal air bronchogram at CT. The top seven metagenes, representing 95% of the weight of the multivariate model, are shown; the top three metagenes are upregulated when an air bronchogram is present, and the bottom four metagenes are downregulated. The downregulated metagenes are enriched in hypoxia-related pathways; the upregulated metagenes contain a Ras signature and genes upregulated by Ras. (d) Receiver operating characteristic curve for the predicted presence versus absence of internal air bronchogram at CT, when expressed in terms of metagenes. (e) Multivariate model for internal air bronchogram corresponding to c. For gene expression, red = overexpression and green = underexpression; for image features, blue = absence of the feature and yellow = presence of the feature. CI = confidence interval.
The most accurately predicted semantic feature was the presence or absence of the internal air bronchogram at CT (AUC = 86%, Fig 2c). This feature was predicted by using 10 metagenes, seven of which captured 95% of the weight in the linear regression (Fig 2d). The associated gene clusters that were upregulated in the presence of an internal air bronchogram were enriched with Ras targets and confirmed our earlier univariate association of this feature (19,22); the downregulated gene clusters were enriched in hypoxia-related pathways (20,21). This finding is consistent with the observation that inactivation of HIF2alpha is related to upregulation of KRAS-driven lung tumors (23) and suggests that the presence of internal air bronchograms may be a biomarker of high Ras and low hypoxia pathway activity.
Figure 2c:
Multivariate modeling of image features in terms of metagenes. (a) Strategy for multivariate modeling of image features in terms of metagenes. Each image feature is modeled as a linear combination of metagenes, using L1 regularization to induce sparsity in the number of metagenes that are selected. I1 = the first image feature of k image features in total; Mj = jth metagene; fi = linear regression for the ith image feature; α = regularization parameter; Wj (not shown) = weight for each metagene M1, M2, to Mn; and W = matrix with all weights. (b) Semantic features predicted by metagenes with an area under the receiver operating characteristic curve (AUC) of 65% or greater, based on leave-one-out cross-validation analysis. (c) Multivariate metagene prediction model for the presence versus absence of internal air bronchogram at CT. The top seven metagenes, representing 95% of the weight of the multivariate model, are shown; the top three metagenes are upregulated when an air bronchogram is present, and the bottom four metagenes are downregulated. The downregulated metagenes are enriched in hypoxia-related pathways; the upregulated metagenes contain a Ras signature and genes upregulated by Ras. (d) Receiver operating characteristic curve for the predicted presence versus absence of internal air bronchogram at CT, when expressed in terms of metagenes. (e) Multivariate model for internal air bronchogram corresponding to c. For gene expression, red = overexpression and green = underexpression; for image features, blue = absence of the feature and yellow = presence of the feature. CI = confidence interval.
Figure 2d:

Multivariate modeling of image features in terms of metagenes. (a) Strategy for multivariate modeling of image features in terms of metagenes. Each image feature is modeled as a linear combination of metagenes, using L1 regularization to induce sparsity in the number of metagenes that are selected. I1 = the first image feature of k image features in total; Mj = jth metagene; fi = linear regression for the ith image feature; α = regularization parameter; Wj (not shown) = weight for each metagene M1, M2, to Mn; and W = matrix with all weights. (b) Semantic features predicted by metagenes with an area under the receiver operating characteristic curve (AUC) of 65% or greater, based on leave-one-out cross-validation analysis. (c) Multivariate metagene prediction model for the presence versus absence of internal air bronchogram at CT. The top seven metagenes, representing 95% of the weight of the multivariate model, are shown; the top three metagenes are upregulated when an air bronchogram is present, and the bottom four metagenes are downregulated. The downregulated metagenes are enriched in hypoxia-related pathways; the upregulated metagenes contain a Ras signature and genes upregulated by Ras. (d) Receiver operating characteristic curve for the predicted presence versus absence of internal air bronchogram at CT, when expressed in terms of metagenes. (e) Multivariate model for internal air bronchogram corresponding to c. For gene expression, red = overexpression and green = underexpression; for image features, blue = absence of the feature and yellow = presence of the feature. CI = confidence interval.
To validate our approach for predicting image features with a gene signature, we focused on the accuracy of predicting image features that describe the tumor’s dimensions, because the actual tumor size was available in the Lee et al (13) data set. In particular, we focused on the gene signatures that predict the three computed CT image features: minor axis, major axis, and lesion size. We found that all three predicted image features were statistically significantly correlated with actual tumor size (Fig 3a). Moreover, this significance was also correlated with the accuracy of each predictive model (Fig 3b). Interestingly, the upregulated genes for large predicted tumor sizes were associated with extracellular matrix remodeling (P < .0001) (24) and the epithelial-to-mesenchymal transition (25–27).
Figure 3a:
Validation of the predicted image features related to computed lesion size at CT. (a) Graph shows the correlation of the predicted image feature “lesion size” and the actual lesion size in the Lee et al (13) data (P < .0001). (b) Graph shows comparison of the accuracy of the predicted image features lesion size, minor axis, and major axis with their correlation with actual tumor size in the Lee et al data.
Figure 3b:
Validation of the predicted image features related to computed lesion size at CT. (a) Graph shows the correlation of the predicted image feature “lesion size” and the actual lesion size in the Lee et al (13) data (P < .0001). (b) Graph shows comparison of the accuracy of the predicted image features lesion size, minor axis, and major axis with their correlation with actual tumor size in the Lee et al data.
Identification of Image Biomarkers by Leveraging Public Gene Expression Data
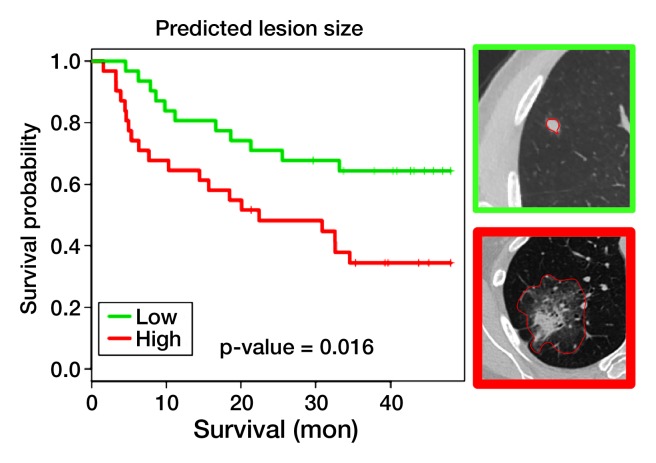
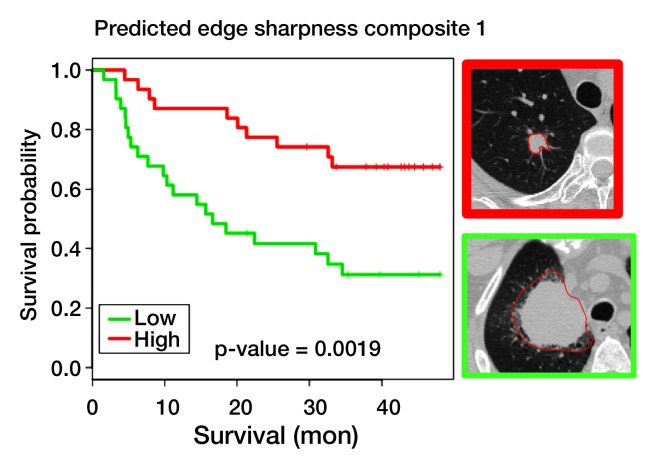
Univariate survival analysis of predicted image features.—We found 26 and 22 predicted image features that were significantly associated with recurrence-free survival (RFS) (log-rank P and Cox P < .05 [Fig 4, Table) and overall survival (Table E6 [online]), respectively, at 4 years from diagnosis, in a univariate survival analysis of the Lee et al (13) adenocarcinoma data. Among the 26 predicted image features associated with RFS were all three image features that characterize tumor dimensions on CT (ie, minor axis, major axis, and lesion size [Fig 4a]), a finding consistent with the literature (28). In addition, several semantic image features were prognostically significant, including the presence of satellite nodules in the primary tumor lobe, lobulated margins, and pleural attachment (Fig 4b and Table). We also found that edge sharpness was correlated with RFS (hazard ratio, 0.35; 95% CI: 0.13, 0.91 [Fig 4c]); in particular, blurry versus sharp edges were associated with poor versus good survival. In addition, the presence of an internal air bronchogram, which we previously associated with KRAS, was associated with poor RFS (hazard ratio, 3.1; 95% CI: 1.5, 6.6 [Fig 4d]). This is consistent with the reported association between upregulation of KRAS and poor survival (29,30). However, this appears to contrast with the studies that correlate the presence of an internal air bronchogram, in stage 1 tumors with pleural retraction, with good prognosis (31). Yet a recent study found a nonsignificant correlation between the presence of an air bronchogram and RFS, suggesting that larger studies that can adjust for confounding factors are needed (32). Consistent with our observations, Travis et al (33) reported a positive correlation between the presence of KRAS mutations and the presence of an internal air bronchogram. Overall, many image features remained prognostically significant (Table) in a multivariate analysis that included clinical covariates such as age, nodal stage, and tumor size (28).
Figure 4a:
Univariate survival analysis for four predicted image features evaluated in the Lee et al (13) data for RFS. Predicted image features are expressed in terms of a multivariate gene expression signature. (a) Survival curves for predicted lesion size at CT, with top right image showing that a small CT lesion is associated with good prognosis (green curve) and bottom right image showing that a large CT lesion is associated with poor prognosis (red curve). (b) Survival curves for predicted presence versus absence of pleural attachment at CT. (c) Survival curves based on predicted edge sharpness composite 1, with top right image showing that high edge sharpness is associated with good prognosis (red curve) and bottom right image showing that low edge sharpness is associated with poor prognosis (green curve). (d) Survival curves based on predicted presence versus absence of internal air bronchogram.
Predicted Image Features That Are Significantly Related to RFS in the Data Set of Lee et al

Note.—The univariate results refer to evaluating a predicted image feature without other clinical covariates. The multivariate results show the results of the univariate analysis after correcting for clinical covariates available in the Lee et al (13) data set. N stage refers to nodal stage.
Single image feature plus clinical covariates.
Semantic feature.
Figure 4b:
Univariate survival analysis for four predicted image features evaluated in the Lee et al (13) data for RFS. Predicted image features are expressed in terms of a multivariate gene expression signature. (a) Survival curves for predicted lesion size at CT, with top right image showing that a small CT lesion is associated with good prognosis (green curve) and bottom right image showing that a large CT lesion is associated with poor prognosis (red curve). (b) Survival curves for predicted presence versus absence of pleural attachment at CT. (c) Survival curves based on predicted edge sharpness composite 1, with top right image showing that high edge sharpness is associated with good prognosis (red curve) and bottom right image showing that low edge sharpness is associated with poor prognosis (green curve). (d) Survival curves based on predicted presence versus absence of internal air bronchogram.
Figure 4c:
Univariate survival analysis for four predicted image features evaluated in the Lee et al (13) data for RFS. Predicted image features are expressed in terms of a multivariate gene expression signature. (a) Survival curves for predicted lesion size at CT, with top right image showing that a small CT lesion is associated with good prognosis (green curve) and bottom right image showing that a large CT lesion is associated with poor prognosis (red curve). (b) Survival curves for predicted presence versus absence of pleural attachment at CT. (c) Survival curves based on predicted edge sharpness composite 1, with top right image showing that high edge sharpness is associated with good prognosis (red curve) and bottom right image showing that low edge sharpness is associated with poor prognosis (green curve). (d) Survival curves based on predicted presence versus absence of internal air bronchogram.
Figure 4d:
Univariate survival analysis for four predicted image features evaluated in the Lee et al (13) data for RFS. Predicted image features are expressed in terms of a multivariate gene expression signature. (a) Survival curves for predicted lesion size at CT, with top right image showing that a small CT lesion is associated with good prognosis (green curve) and bottom right image showing that a large CT lesion is associated with poor prognosis (red curve). (b) Survival curves for predicted presence versus absence of pleural attachment at CT. (c) Survival curves based on predicted edge sharpness composite 1, with top right image showing that high edge sharpness is associated with good prognosis (red curve) and bottom right image showing that low edge sharpness is associated with poor prognosis (green curve). (d) Survival curves based on predicted presence versus absence of internal air bronchogram.
Multivariate survival analysis of predicted image features.—We computed a multivariate model of the predicted image features that significantly correlated with 4-year RFS (log-rank P = .00166 [Fig 5a) in the Lee et al (13) adenocarcinoma data. The top-ranked image features were edge sharpness, length of the major axis of the lesion, and lesion texture (Table E7 [online]). The multivariate model was also predictive of RFS when clinical covariates were included (Cox P = .0017; hazard ratio, 3.88; 95% CI: 1.7, 9.1). Because predicted tumor size was in this model, we repeated the analysis without all predicted tumor size features, and found that the model was still predictive of RFS (log-rank P = .0257 [Fig 5b]). Even when the clinical variables were included, a combination of predicted image features was independently significant (Cox P = .020; hazard ratio, = 1.79; 95% CI: 1.1, 2.9). Top-ranking predicted image features included computational features that describe the lesion texture and edge sharpness and a semantic image feature that describes the presence or absence of entering airways (Table E7 [online]). When we repeated this analysis using only semantic features, the resulting model was also significantly correlated with RFS (log-rank P = .0251 [Fig 5c]), and the top-ranking semantic features included the presence or absence of entering airways, lobulated margins, and pleural retraction (Table E7 [online]).
Figure 5:
Multivariate survival analysis for predicted image features evaluated in the Lee et al data set (13) for RFS. Graphs show Kaplan-Meier survival curves for selected multivariate models on the Lee et al external data set. Left: Multivariate model using all predicted image features. Middle: Multivariate model using all image features except the three size features: lesion size, minor axis, and major axis. Right: Multivariate model using only semantic features.
Discussion
While there has been much recent activity in developing quantitative imaging biomarkers of disease, linking these biomarkers to clinical outcomes such as progression-free and overall survival and response to treatment is problematic because of the length of time required to obtain these outcomes in study cohorts. Here, we demonstrate a radiogenomics strategy for rapidly identifying prognostically significant image biomarkers that requires only the paired acquisition of image and gene expression data and the existence of a large public gene expression data set where survival outcomes are available. We demonstrated that once the image features can be predicted from metagenes, the clinical significance of the predicted image features can be explored by mining existing public gene expression microarray databases that contain clinical outcomes. As a proof-of-concept, our radiogenomics strategy identified image features that are known to be prognostically significant without relying on paired observations of image features and survival outcomes.
We applied our radiogenomics strategy in a cohort of 26 patients with NSCLC for whom we had medical images (CT and PET/CT images) and gene expression microarray data. Initially, we identified several statistically significant correlations between gene expression profiles of NSCLC and imaging phenotypes at CT and PET/CT. Similar to Segal et al (4), who studied the radiogenomics of hepatocellular carcinoma, we demonstrated that the metagenes of NSCLC can be predicted from a linear combination of a small set of image features with a reasonable accuracy to warrant further investigation in a larger study that includes an independent validation data set. Moreover, we demonstrated that many CT image features and PET SUV can be predicted from a linear combination of the metagenes with similar accuracy. For example, we provided a gene signature for image-based tumor size that implicates processes in extracellular modeling and the epithelial-to-mesenchymal transition. This particular result suggests that from cross-sectional observations of tumors, we may be able to identify molecular characteristics of a single tumor as if it were observed longitudinally with increasing size. Finally, we showed that a linear combination of predicted image features was prognostically significant in a publicly available gene expression data set of NSCLC outcomes, even when clinical covariates that are known to be prognostically significant were considered. This last analysis may prove to be valuable because many emerging and evolving imaging technologies change during the time required to establish a clinical end point.
A strength in our radiogenomics strategy is the use of a large set of computer-derived image features in addition to semantic features. We extracted computational features from CT that describe tumor shape, edge shape, and edge variability. Interestingly, we showed that the computational image features were necessary and sufficient for accurate modeling of metagenes and predicting survival outcomes. This finding alone suggests that the imaging community may greatly benefit from the creation of a large-scale publicly available database for associating computationally derived image features with clinical outcomes (11). Moreover, the creation of a lexicon and ontology of reproducible semantic and computed image features further permits images to be minable like genomic data (34). Over the longer term, a minable image database, together with genomics data and clinical outcomes, would enable an analysis of the added value of image and genomic features in clinical decision making.
The limitations of our radiogenomics analysis relate to the data used as proof-of-concept. We analyzed data in a small cohort of patients with NSCLC, which did not fully capture the variability of the disease at imaging and gene expression profiling, nor the variability due to histologic subtype. Also owing to the absence of a high-quality thin-section CT validation data set, we used cross-validation techniques to estimate the correlation with survival. In addition, we did not consider variability in image acquisition, reconstruction, and interpretation. The CT-derived computational image features may be sensitive to the methods of image formation, including CT reconstruction kernel, section thickness, and pixel size. Similarly, PET SUVs may show similar variations as a function of scanner design and reconstruction parameters. We limited the feature extraction of the PET image to the most important clinical value—namely, maximum SUV—yet it is certainly possible to extract a multitude of image features from PET. Finally, a single thoracic radiologist identified a region of interest in a representative cross section for computational feature extraction and selected the descriptive semantic features from a controlled vocabulary. Multiple radiologists would capture variability in these interpretations. Variability in selected regions of interest may result in variations in the computational features and their associations with the image features. A larger study and an external validation cohort will enable investigation of these limiting factors underlying our analysis.
In summary, by mapping image features to gene expression data, we were able to leverage public gene expression microarray data to assess prognosis and, where available, therapeutic response, as a function of image features. Even though we focused on CT and PET images in patients with NSCLC, our methods are sufficiently general that they can be applied to other imaging modalities and diseases.
Advances in Knowledge.
• Fifty-six high-quality metagenes captured from non–small cell lung cancer (NSCLC) gene expression microarrays can be predicted by CT image features with a mean accuracy of 72% (range, 59%–83%).
• Ten semantic, 104 computationally derived CT image features, and the PET/CT standardized uptake value of NSCLC can be predicted by using the 56 high-quality metagenes, with an accuracy or area under the receiver operating characteristic curve of between 65% and 86%.
• A prognostic signature of image biomarkers, composed of imaging features that are expressed in terms of their predictive gene expression signature, was identified by leveraging publicly available microarray data with survival outcomes.
Implication for Patient Care.
• Prognostically significant patient-specific molecular signatures may be predicted noninvasively from image features, further advancing the role of imaging in personalized medicine.
Disclosures of Potential Conflicts of Interest: O.G. No potential conflicts of interest to disclose. J.X. No potential conflicts of interest to disclose. C.D.H. No potential conflicts of interest to disclose. A.N.L. No potential conflicts of interest to disclose. Y.X. No potential conflicts of interest to disclose. A.Q. Financial activities related to the present article: none to disclose. Financial activities not related to the present article: has served as an expert witness for Hassard Bonnington. Other relationships: none to disclose. D.L.R. No potential conflicts of interest to disclose. S.N. No potential conflicts of interest to disclose. S.K.P. Financial activities related to the present article: institution received grant from GE Healthcare and GE Medical Systems. Financial activities not related to the present article: none to disclose. Other relationships: none to disclose.
Supplementary Material
Acknowledgments
We are grateful to Andrew Gentles, PhD, and Professors Robert Tibshirani, PhD, and Maximilian Diehn, MD, PhD, for helpful discussions. We are also grateful to Professor Jhingook Kim and his colleagues for providing additional clinical annotation for the Lee et al data set.
Received August 5, 2011; revision requested September 30; revision received November 26; accepted December 29; final version accepted February 21, 2012.
See also the editorial by Jaffe et al in this issue
Supported by Information Sciences in Imaging at Stanford, the Center for Cancer Systems Biology at Stanford, and GE Healthcare. O.G. is a fellow of the Fund for Scientific Research Flanders (FWO-Vlaanderen); an Honorary Fulbright Scholar of the Commission for Educational Exchange between the United States of America, Belgium, and Luxembourg; and a Henri Benedictus Fellow of the King Baudouin Foundation and the Belgian American Educational Foundation.
Funding: This research was supported by the National Cancer Institute (grants R01 CA160251, U54 CA149145, and U01 CA142555).
Abbreviations:
- AUC
- area under the receiver operating characteristic curve
- CI
- confidence interval
- FDG
- fluorine 18 fluorodeoxyglucose
- NSCLC
- non–small cell lung cancer
- RFS
- recurrence-free survival
- SUV
- standardized uptake value
References
- 1.Sotiriou C. Molecular biology in oncology and its influence on clinical practice: gene expression profiling [abstr]. Ann Oncol 2009;20(Suppl 4):v10 [Google Scholar]
- 2.Pao W, Kris MG, Iafrate AJ, et al. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res 2009;15(17):5317–5322 [DOI] [PubMed] [Google Scholar]
- 3.Gevaert O, De Moor B. Prediction of cancer outcome using DNA microarray technology: past, present and future. Expert Opin Med Diagn 2009;3(2):157–165 [DOI] [PubMed] [Google Scholar]
- 4.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007;25(6):675–680 [DOI] [PubMed] [Google Scholar]
- 5.Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 2008;105(13):5213–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol 2007;18(7):821–831 [DOI] [PubMed] [Google Scholar]
- 7.Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol 2009;70(2):232–241 [DOI] [PubMed] [Google Scholar]
- 8.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010;60(5):277–300 [DOI] [PubMed] [Google Scholar]
- 9.National Lung Screening Trial Research Team , Aberle DR, Berg CD, et al. The National Lung Screening Trial: overview and study design. Radiology 2011;258(1):243–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365(5):395–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 2009;37(Database issue):D885–D890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkinson H, Sarkans U, Kolesnikov N, et al. ArrayExpress update—an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res 2011;39(Database issue):D1002–D1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee ES, Son DS, Kim SH, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res 2008;14(22):7397–7404 [DOI] [PubMed] [Google Scholar]
- 14.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 2001;98(9):5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33(1):1–22 [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue A, Nukiwa T. Gene mutations in lung cancer: promising predictive factors for the success of molecular therapy. PLoS Med 2005;2(1):e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc 2009;6(2):201–205 [DOI] [PubMed] [Google Scholar]
- 18.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res 2002;62(23):6997–7000 [PubMed] [Google Scholar]
- 19.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439(7074):353–357 [DOI] [PubMed] [Google Scholar]
- 20.Manalo DJ, Rowan A, Lavoie T, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 2005;105(2):659–669 [DOI] [PubMed] [Google Scholar]
- 21.Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J Biol Chem 2006;281(22):15215–15226 [DOI] [PubMed] [Google Scholar]
- 22.Sweet-Cordero A, Mukherjee S, Subramanian A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet 2005;37(1):48–55 [DOI] [PubMed] [Google Scholar]
- 23.Mazumdar J, Hickey MM, Pant DK, et al. HIF-2alpha deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci U S A 2010;107(32):14182–14187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergamaschi A, Tagliabue E, Sørlie T, et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J Pathol 2008;214(3):357–367 [DOI] [PubMed] [Google Scholar]
- 25.Charafe-Jauffret E, Ginestier C, Monville F, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 2006;25(15):2273–2284 [DOI] [PubMed] [Google Scholar]
- 26.Seo DC, Sung JM, Cho HJ, et al. Gene expression profiling of cancer stem cell in human lung adenocarcinoma A549 cells. Mol Cancer 2007;6:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xi LQ, Lyons-Weiler J, Coello MC, et al. Prediction of lymph node metastasis by analysis of gene expression profiles in primary lung adenocarcinomas. Clin Cancer Res 2005;11(11):4128–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Port JL, Kent MS, Korst RJ, Libby D, Pasmantier M, Altorki NK. Tumor size predicts survival within stage IA non-small cell lung cancer. Chest 2003;124(5):1828–1833 [DOI] [PubMed] [Google Scholar]
- 29.Massarelli E, Varella-Garcia M, Tang X, et al. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res 2007;13(10):2890–2896 [DOI] [PubMed] [Google Scholar]
- 30.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer 2005;92(1):131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshino I, Nakanishi R, Kodate M, et al. Pleural retraction and intra-tumoral air-bronchogram as prognostic factors for stage I pulmonary adenocarcinoma following complete resection. Int Surg 2000;85(2):105–112 [PubMed] [Google Scholar]
- 32.Ikehara M, Saito H, Kondo T, et al. Comparison of thin-section CT and pathological findings in small solid-density type pulmonary adenocarcinoma: prognostic factors from CT findings. Eur J Radiol 2012;81(1):189–194 [DOI] [PubMed] [Google Scholar]
- 33.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6(2):244–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubin DL, Rodriguez C, Shah P, Beaulieu C. iPad: Semantic annotation and markup of radiological images. AMIA Annu Symp Proc 2008:626–630 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.