Abstract
The active form of Escherichia coli DNA polymerase V responsible for damage-induced mutagenesis is a multiprotein complex (UmuD′2C-RecA-ATP), called pol V Mut. Optimal activity of pol V Mut in vitro is observed on an SSB-coated single-stranded circular DNA template in the presence of the β/γ complex and a transactivated RecA nucleoprotein filament, RecA*. Remarkably, under these conditions, wild-type pol V Mut efficiently incorporates ribonucleotides into DNA. A Y11A substitution in the ‘steric gate’ of UmuC further reduces pol V sugar selectivity and converts pol V Mut into a primer-dependent RNA polymerase that is capable of synthesizing long RNAs with a processivity comparable to that of DNA synthesis. Despite such properties, Y11A only promotes low levels of spontaneous mutagenesis in vivo. While the Y11F substitution has a minimal effect on sugar selectivity, it results in an increase in spontaneous mutagenesis. In comparison, an F10L substitution increases sugar selectivity and the overall fidelity of pol V Mut. Molecular modeling analysis reveals that the branched side-chain of L10 impinges on the benzene ring of Y11 so as to constrict its movement and as a consequence, firmly closes the steric gate, which in wild-type enzyme fails to guard against ribonucleoside triphosphates incorporation with sufficient stringency.
INTRODUCTION
Among the multiple pathways that allow living organisms to overcome ubiquitous everyday genomic DNA damage, translesion DNA synthesis (TLS) plays an essential role. For more than three decades, it has been recognized that in bacterial cells TLS is largely dependent upon the products of the umuDC genes (1,2). However, the specific roles of the Umu proteins remained elusive, primarily because of the difficulty of purifying UmuC in soluble form (3). A major breakthrough in understanding the mechanisms of TLS came in the late 1990s when the Umu proteins were finally purified as a soluble heterotrimeric complex of UmuD′2C (4), that was subsequently shown to possess intrinsic polymerase activity capable of carrying out unassisted replication of damaged or distorted DNA (5).
As the fifth Escherichia coli polymerase reported in the literature, the UmuD′2C complex was named DNA polymerase V (pol V). The polymerase active site resides in the UmuC subunit of the complex (5,6) which shares sequence homology with other enzymes belonging to the Y-family of DNA polymerases (7). Despite that >10 years have passed since the discovery of intrinsic polymerase activity, further studies are needed to determine the precise biochemical mechanism of pol V-catalyzed TLS. It is currently accepted that for optimal activity on long single-stranded DNA (ssDNA) templates, pol V requires interactions with accessory factors that include RecA protein, β-sliding processivity clamp loaded on a circular template by the γ-complex and probably ssDNA binding protein (SSB) (8–10).
RecA stimulates pol V on several levels. It is well established that activation of pol V-dependent TLS in damaged cells requires RecA in a form of a nucleoprotein filament (RecA*) that is assembled on ssDNA. This pathway is initiated by RecA*-mediated autocleavage of LexA protein triggering the derepression of genes in the LexA-regulon and activation of the SOS response (11). As a result, umuDC gene expression is induced, which in conjunction with RecA*-mediated autocleavage of UmuD to UmuD′ (12,13) leads to the dramatic increase in the intracellular concentration of pol V (14). Multiple in vitro and in vivo studies have indicated that in addition to an indirect role in TLS, RecA is indispensable for direct stimulation of the catalytic activity of pol V (15–18). It was tacitly assumed that this stimulation also depends on the RecA filament assembled on the ssDNA immediately 3′ of the stalled replication complex. However, recent in vitro studies provide definitive evidence that activation of pol V for TLS requires an interaction with a single RecA protomer which is transferred along with an ATP molecule from the 3′ tip of a RecA* formed in trans (8). The resulting multiprotein complex composed of UmuD′2C-RecA-ATP in a 1:1:1 stoichiometry was termed pol V Mut [‘Mut’ is short for ‘Mutasome’ to acknowledge the mutagenic and multiprotein nature of TLS (19)]. Pol V Mut activity can be further stimulated by cofactors. Indeed, we have recently shown that pol V Mut is highly processive on an SSB-coated circular ssDNA template in the presence of the β/γ-complex (10).
Genetic evidence suggests that pol V is a highly error-prone polymerase (1,2). In agreement with the in vivo data, pol V copied an undamaged DNA in vitro with misincorporation frequency of ∼10−3–10−4 and was generally even less accurate when replicating damaged substrates (20). Since the overall catalytic activity and processivity of pol V Mut is significantly higher than that of pol V, we decided to examine its fidelity in vitro. However, our studies were complicated by the fact that pol V Mut in the presence of the accessory factors extended primers even in the absence of dNTPs. We hypothesized that extension occurs due to the apparent ability of pol V Mut to incorporate ATP into DNA (ATP that is intrinsically present in the reaction mixture for loading the β-clamp onto primer/template DNA). We show here that pol V Mut not only efficiently inserts ATP, but readily copies DNA templates in the presence of all four ribonucleoside triphosphates (rNTPs).
For DNA polymerases from the A-, B-, RT- and Y-family, a strict mechanism that protects cells from the aberrant incorporation of rNTPs into a nascent DNA strand during replication relies on a single amino acid, known as the ‘steric gate’, the bulky side chain of which sterically clashes with the ribose 2′-hydroxyl group of an incoming rNTP (21,22). The identity of the steric gate amino acid differs between the various polymerase families, but it is highly conserved within each family. In the case of the Y-family polymerases, it has been identified as a Tyr (Y), or Phe (F) residue (23). Based on primary amino acid sequence alignments the role of the steric gate in pol V was assigned to Y11 in UmuC (24).
In order to gain insights into sugar selectivity and overall fidelity of E. coli pol V, we have generated variants of UmuC with amino acid substitutions at, or adjacent to, its steric gate. The mutant pol V enzymes were purified and characterized for their fidelity and ability to incorporate ribonucleotides in vitro. The mutants were also cloned into vectors that allowed for their expression at physiological levels in E. coli and their effects on spontaneous polV-dependent ‘SOS’ mutagenesis monitored in vivo. We also performed structural modeling analyses to ascertain the molecular basis for the observed in vitro properties and in vivo phenotypes of the pol V ‘steric gate’ variants.
MATERIALS AND METHODS
Materials
Ultrapure NTPs and dNTPs were purchased from Affymetrix (Fremont, CA, USA), adenosine 5′ [γ-thio]triphosphate (ATPγS) was from Biolog Life Science Institute (Bremen, Germany) and β-d-arabinofuranosylcytosine was from TriLink Biotechnologies (San Diego, CA, USA). RecA and SSB proteins were supplied by New England Biolabs, (Ipswich, MA, USA) and Epicentre Biotechnologies, (Madison, WI, USA), respectively. Pol V, β-clamp and γ-complex were purified as previously described (10). All oligonucleotides were synthesized by Lofstrand Laboratories (Gaithersburg, MD, USA) and gel purified prior to use. M13mp18 ssDNA was from New England Biolabs (Ipswich, MA, USA). Undamaged pSO and pSOcpd, containing a unique CPD adduct, were constructed as previously described (25).
Bacterial strains and growth conditions
The E. coli K-12 strains used to study spontaneous mutagenesis were RW584 [full genotype: thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 glnV44 galK2 hisG4 rpsL31 xyl-5 mtl-1 argE3 thi-1 sulA211 ΔumuDC596::ermGT lexA51(Def) recA730] (26) and RW710 [full genotype: thr-1 araD139 Δ(gpt-proA)62 lacY1 tsx-33 glnV44 galK2 hisG4 rpsL31 xyl-5 mtl-1 argE3 thi-1 sulA211 ΔdinB61::ble ΔumuDC596::ermGT lexA51(Def) recA730 mutL211::Tn5] (27). Bacteria were grown in LB media containing zeocin (25 µg/ml), kanamycin (50 µg/ml) and spectinomycin (20 µg/ml) where appropriate. The E. coli B strain, RW644 [BL21(λDE3) ΔpolB::Spec ΔdinB61::ble ΔumuDC596::ermGT], (10) was used for the expression and purification of pol V variants.
Expression and purification of E. coli pol V variants
umuC mutant genes harboring the F10L, Y11A and Y11F mutations were synthesized and cloned into the pUC57 vector by Genscript (Piscataway, NJ, USA). The F10L, Y11A and Y11F mutations are each associated with a novel restriction site, AatII, HpaI and EagI, respectively, to facilitate their subsequent identification after subcloning into the UmuC expression vector, pHUC25 (10) to generate pJM948, pJM949 and pJM950, respectively. His-tagged pol V (and F10L, Y11A and Y11F variants) were purified from E. coli strain RW644 harboring plasmids pARAD2 (expressing UmuD′) and either pHUC25 or pJM948, pJM949, pJM950. Pol V was purified from 2 L cultures in three steps: Ni-NTA agarose affinity (QIAGEN), Superdex 200 HiLoad 26/60 size exclusion (GE Healthcare, Piscataway, NJ, USA) and Bio-Gel HTP Hydroxyapatite chromatography (Bio-Rad, Hercules, CA, USA) (10). The β-clamp and γ-complex were purified as described (10). The final concentration of the purified proteins was determined by Bradford assay and the purity of the eluted proteins was confirmed by separation on a 4–12% NuPAGE Bis–Tris gels followed by visualization with Coomassie blue staining (Supplementary Figure S1).
In vitro replication catalyzed by pol V variants
The sequences of primers used for the biochemical characterization of pol V mutants are listed in Supplementary Table S1. 5A17M, 5T17M, 5G17M and 5C17M primers were designed for hybridization to single stranded M13mp18 DNA to study the misincorporation specificity of pol V. 5′-32P labeled primers were annealed to the single-stranded circular plasmids at a 1.5:1 molar ratio by heating the required mixture in an annealing buffer [50 mM Tris–HCl (pH 8), 5 mM MgCl2, 50 µg/ml BSA, 1.42 mM 2-mercaptoethanol] for 10 min at 100°C followed by slow cooling to room temperature. RecA* (0.25 µM) was formed by incubating 4 µM RecA (New England Biolabs, Ipswich, MA, USA), 1 mM ATPγS and 0.25 µM 48-mer oligonucleotide in the 1× reaction buffer (20 mM Tris–HCl pH 7.5, 8 mM MgCl2, 8 mM DTT, 80 µg/ml BSA, 4% glycerol) at 37°C for 5 min. Purified pol V variants (80 nM) were first combined with RecA* to form pol V Mut complexes (8) and then added to the reaction mixture which had been pre-incubated for 3 min at 37°C. The reaction mixture contained 1 mM ATP, 50 µM dNTPs or rNTPs (unless otherwise indicated), 2 nM primed ssDNA templates (expressed as primer termini), 100 nM (as tetramer) SSB Epicentre Biotechnologies, (Madison, WI, USA), 50 nM (as a dimer) β clamp and 5 nM γ complex in the 1× reaction buffer. Reactions were incubated at 37°C for 0.5–20 min and terminated by adding 10 µl of 2× loading buffer (97% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). The products were heat-denatured and immediately resolved by denaturing PAGE (8 M urea, 15% acrylamide), followed by visualization and quantification using a Fuji image analyzer FLA-5100 and MultiGauge software. Primer elongation was calculated as percent of total primer termini. For processivity measurements, reaction mixtures contained DNA templates in sufficient excess over polymerase such as the probability of termination of synthesis at each template position was constant over a 0.5- to 16-min time course. Termination probabilities (expressed as a percentage) were calculated as the band intensity at the specific position divided by the intensity at that position plus all longer products (28).
Specificity of nucleotide incorporation
Reactions were performed as above except nucleotides with deoxy-, ribo- or arabinose sugar were added either individually, or in different combinations, as specified in the figure legends. Where indicated, reactions were split into two and treated with either 0.3 M KCl or 0.3 M KOH for 2 h at 55°C. Following addition of an equal volume of loading dye, samples were analyzed by PAGE as described earlier.
Spontaneous mutagenesis assays
The F10L, Y11A and Y11F UmuC alleles were sub-cloned from pJM948, pJM949 and pJM950, respectively, into the low-copy-number, spectinomycin resistant UmuD′C plasmid, pRW134 (29), to generate pJM964, pJM963 and pJM952, respectively. RW584 was freshly transformed with pGB2 (control vector) (30), or one of the low-copy-number plasmids containing wild-type (pRW134) or mutant pol V (pJM964, pJM963, pJM952) plasmids and grown overnight at 37°C on LB plates with 50 µg/ml spectinomycin. Three well-separated transformants were inoculated into 3 ml LB plus spectinomycin media and grown overnight with shaking at 37°C. The next day, cultures were harvested by centrifugation and resuspended in an equal volume of SM media (31). Aliquots (100 µl) of each culture were plated in triplicate on Davis and Mingioli minimal plates (32) containing 1 µg/ml l-Histidine and incubated at 37°C for 4 days. The data reported in the Figure 6 therefore, represent the average number of His+ mutants obtained from nine plates per plasmid-containing strain ± SEM. Statistical significance for the differences between the number of His+ revertants in the strains expressing wild-type and Y11F UmuC was estimated by comparing average data from different biological experiments using the paired t-test (SigmaPlot, Systat Software, Inc., Chicago, IL, USA).
Figure 6.
Qualitative analysis of spontaneous mutagenesis. Spontaneous pol V-dependent mutagenesis was measured in the E. coli strain, RW584 [recA730 lexA(Def) ΔumuDC] harboring low-copy-number vector control (pGB2) or a plasmid expressing wild-type pol V (pRW134), umuC F10L (pJM964), UmuC Y11A (pJM963) or UmuC Y11F (pJM952) by assaying reversion of the hisG4 (ochre) allele (leading to Histidine prototophy).
Spectra of spontaneous base-pair substitutions in the E. coli rpoB gene
Base-pair substitutions in the E. coli rpoB gene encoding the β-subunit of RNA polymerase are either lethal or result in rifampicin resistance. A single pair of oligonucleotide primers can be used for PCR amplification and a single primer for DNA sequencing because 88% of all rpoB mutations are localized in the central 202-bp region of the gene (33,34). Strain RW710 [relevant genotype: lexA(Def) recA730 ΔdinB ΔumuDC mutL] harboring pGB2, pRW134, pJM964, pJM963 or pJM952, was diluted from frozen stock cultures such that the initial inoculum contained ∼1000 viable cells. Cultures were grown in LB for 24 h at 37°C and appropriate dilutions spread on an LB agar plate containing 100 µg/ml rifampicin. Individual RifR colonies were picked from the plate using a toothpick and subjected to PCR in a 96-well microtiter plate. An <1-kb central region of the rpoB gene was amplified using the PCR primers RpoB1: 5′-CAC ACG GCA TCT GGT TGA TAC AG-3′ and RpoF1: 5′-TGG CGA AAT GGC GGA AAA C-3 by denaturation at 95°C for 3 min, followed by 30 cycles of 94°C for 30 s, 1 min at 59°C, 2 min at 72°C, followed by a final extension step at 72°C for 7 min (27,35). The ∼200-bp target region of rpoB in each individual PCR amplicon was determined by high-throughput DNA sequencing (Beckman Coulter Genomics, Danvers, MA, USA) using WOG923AP01 primer (5-CAG TTC CGC GTT GGC CTG-3). Only base-pair substitutions occurring between positions 1516 and 1717 of the rpoB gene were considered during data analysis. Nucleotide sequences obtained were aligned and analyzed using the ClustalW multiple sequence alignment program (Hinxton, UK).
Molecular modeling of UmuC
The E. coli UmuC protein sequence was aligned with all known structures of the Y-family polymerases using the server HHpred (toolkit.tuebingen.mpg.de/hhpred). Based on the alignment, human DNA polη structure [PDB accession code 3MR2, (36)] was chosen as a template and the UmuC sequence was introduced to replace corresponding residues in polη using the graphic program COOT (biop.ox.ac.uk/coot). Sidechain rotamers were optimized. Since residues surrounding the active site are fairly conserved, there was no need for energy minimization to avoid steric clashes.
RESULTS
Fidelity of pol V Mut-catalyzed DNA replication on an undamaged DNA template
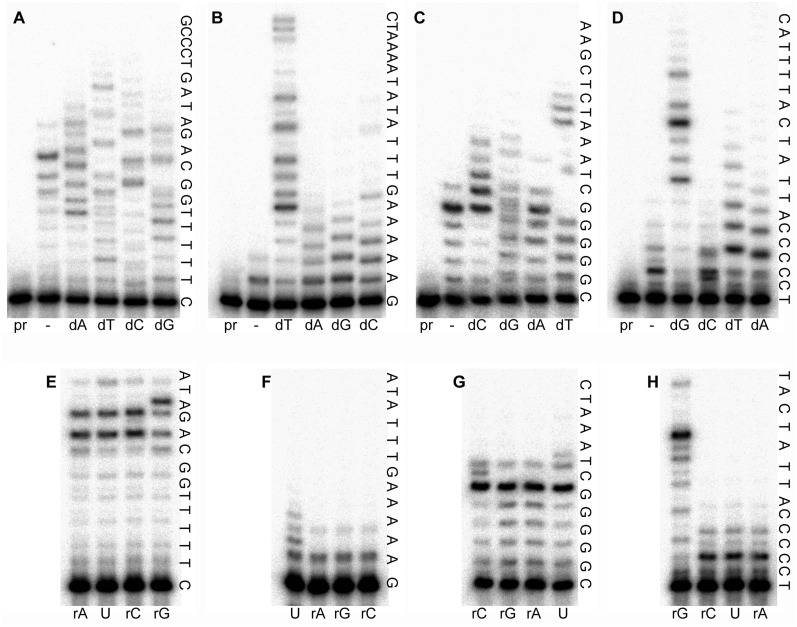
We have recently shown that pol V Mut is optimally active in vitro when reactions are carried out on SSB-coated circular DNA templates in the presence of the β/γ-complex (10). The original goal of the present study was to determine the fidelity of pol V Mut under these optimized reaction conditions. To accomplish this goal, we planned to utilize steady-state kinetic analysis (37). Although this assay has proven to be a very useful tool for fidelity characterization of different DNA polymerases, it quickly became apparent that such an approach would be unfeasible with pol V Mut. In our preliminary in vitro experiments on the undamaged template in the presence of each nucleotide added separately, we not only observed extremely high levels of misincorporation, but also detected intense bands above the primer position in the lanes with control reactions lacking dNTPs (similar to the one seen in the Figure 1A, lane indicated by dash).
Figure 1.
Gel images of primer extension reactions catalyzed by wild-type pol V in the presence of dNTPs (A–D) or rNTPs (E–H). Reactions in the presence of each individual nucleotide were carried out for 5 min. The identity of nucleotide incorporated is shown below the respective lanes. The extended sequence of templates with five consecutive Ts (A and E), As (B and F), Cs (C and G) or Gs (D and H), are indicated to the right of each gel panel. Determination of the specificity of nucleotide incorporation is complicated due to the efficient incorporation of ATP present in the reactions at 1 mM concentration. Lanes with reactions lacking polymerase are indicated as ‘pr’ (short for primer) and reactions with no additional nucleotide are indicated by dash (–).
As described earlier, pol V Mut catalyzes DNA replication in a multicomponent system; accordingly, the purity of each ingredient was our first concern [e.g. we have previously shown that commercially available RecA and SSB are often contaminated with polymerase and/or exonuclease activities (10)]. After thorough examination, we excluded the possibility that the protein preparations were contaminated with dNTPs (data not shown). Since ATP is an integral part of the reaction mixture required for proper assembly of the replication complex and the template we were using had T as a target base, our next logical assumption was that pol V Mut actually utilizes ATP as a substrate for primer extension. In order to test this hypothesis, we compared the specificity of dNTP incorporation using DNA templates with five consecutive Ts, As, Gs or Cs as targets for incorporation (Figure 1A–D).
The pattern of nucleotide (mis)incorporation did not contradict our hypothesis, but it did not provide an unambiguous answer either. For example, the origin of the intense bands corresponding to nucleotide incorporation opposite the template Gs (Figure 1C) in the absence of additional nucleotides (lane indicated by dash) remained unclear. Perhaps they had resulted from misinsertion of ATP opposite the Gs. However, the possibility of incorporation of other rNTPs or even dNTPs, present in trace amounts in the ATP sample, could not be excluded. Single ribonucleotide incorporation assays did not help resolve this question, since ATP was present in excess in all reactions and had a clear competitive advantage compared to other incorrect rNTPs (Figure 1E–H). Nevertheless, these data suggested that pol V Mut has very low sugar selectivity and all NTPs except uracil are utilized efficiently when the canonical Watson–Crick base pairing is preserved. While it is unclear which nucleotide, ATP or CTP, is responsible for the high level of incorporation opposite template G [Figure 1C and G, the product distribution pattern is similar for the lanes with reactions in the presence of ATP (- or rA) and CTP (rC)], it is obvious that uracil is a least favored substrate for pol V Mut (Figure 1F). This observation is not surprising since UTP insertion has generally proven especially difficult for a DNA polymerase to catalyze (38,39).
When reactions were performed in the presence of all four rNTPs, we observed unusually efficient and highly processive RNA synthesis (Figure 2B). However, the rate of primer extension was much slower than in the presence of dNTPs (Figure 2A); i.e. pol V Mut incorporated ∼1 dNTP every second, while the velocity of RNA synthesis was ∼10 rNTPs per min. Furthermore, the termination probability after each rNTP incorporation was in the range of 10–30% which is significantly higher than the 1–10% observed in the reactions with the dNTPs. Nevertheless, the overall primer extension was inhibited only by 10% when dNTPs were replaced by rNTPs. Furthermore, pol V Mut was able to produce elongated RNA products of remarkably long sizes within minutes of incubation. Such level of sugar promiscuity is rare, even for steric gate mutants, not to mention wild-type DNA polymerases. This unorthodox behavior of pol V prompted us to initiate a study to discover why pol V with an intact steric gate residue discriminates so poorly against rNTPs incorporation.
Figure 2.
Role of UmuC F10 and Y11 residues on sugar discrimination by pol V. Primer extension reactions catalyzed by wild-type pol V (A and B), F10L (C and D), Y11A (E and F) and Y11F (G and H) were compared in the presence of 100 µM dNTPs (A, C, E and G) or rNTPs (B, D, F and H). Reactions were performed for 20 s, 1, 2, 4, 8 or 16 min (lanes 1–6, respectively) under optimal conditions for pol V Mut. The numbers shown next to the nucleotide identity represent primer elongation as a percent of total primer termini.
Purification and biochemical characterization of pol V Mut variants with different rNTP discrimination capacities
To determine which amino acid in the catalytic subunit of pol V is responsible for reduced discrimination against rNTP incorporation, we constructed UmuC mutants with specific substitutions in the active site. The first logical candidate for amino acid alteration was the steric gate residue, Y11, which we replaced with alanine or the conservative phenylalanine that is often found at this position in other Y-family polymerases (40). Since there is no indication that sugar discrimination varies among members of Y-family polymerases depending on which residue, tyrosine or phenylalanine, occupies the steric gate position, we did not expect to find any significant effect of an Y11F substitution on the ability of pol V Mut to recognize rNTPs. In contrast, replacing tyrosine with alanine should theoretically ‘unlock’ the steric gate (23). We also constructed another mutant in which F10 adjacent to the steric gate was replaced by leucine. We chose this mutant based on the findings that amino acids flanking the steric gate could play an important role in maintaining the proper orientation of the incoming nucleotide relative to the side chain of the steric gate residue to achieve high sugar selectivity [(41) and references therein].
UmuC variants harboring F10L, Y11A and Y11F mutations were cloned into the UmuC expression vector pHUC25 (10) and introduced along with the UmuD′ expression plasmid pARAD2 into strain RW644 [BL21(λDE3) ΔpolB ΔdinB ΔumuDC] (10). Wild-type His-Pol V and F10L, Y11A and Y11F UmuC variants were purified by Ni2+-NTA affinity chromatography, followed by gel filtration and concentration on hydroxyapatite HTP resin (10). Two liters of E. coli culture yielded between 0.5–1.0 mg of pol V. Based on NuPAGE Bis–Tris Gel analysis of the final eluted fractions, the polymerases were >95% pure (Supplementary Figure S1).
Using these highly purified polymerases, we performed primer extension reactions in the presence of dNTPs or rNTPs (Figure 2). Wild-type pol V and all the mutant polymerases had similar overall catalytic parameters, i.e. processivity of DNA synthesis (measured by probability of reaction termination after 2-min incubation) and the percent of extended primers reached comparable levels for all polymerases when reactions were performed at equal enzyme/template ratios. However, several deviations from their general behavior were also detected for some characteristics of DNA synthesis. For example, the velocity of primer elongation by F10L (Figure 2C) was ∼2.5-fold lower compared to other pol V variants. For Y11A (Figure 2E) we detected a distinct pattern of size distribution of replication products, i.e. at short incubation times, there was a clear transient pause after incorporation of ∼13 nt followed by several less pronounced pause sites. At several positions, reaction products consisted of two bands with slightly different electrophoretic mobility. These doublets appeared mostly opposite certain template Ts suggesting that Y11A catalyzed a significant degree of misincorporation opposite these sites.
As anticipated, the Y11A substitution greatly increased the ability of pol V to insert rNTPs (Figure 2F). An estimate for the time-dependent product accumulation revealed that the Y11A mutant synthesizes RNA products at a 3-fold faster rate relative to the wild-type enzyme. The overall velocity of RNA synthesis by Y11A was only ∼1.3-fold less than that of DNA synthesis. Furthermore, the length distribution of reaction products and the maximum size of synthesized DNA and RNA were similar. In contrast, substitution of F10 with leucine yielded a mutant capable of efficiently blocking rNTP incorporation (Figure 2D). In fact, F10L was only able to extend the DNA primer by up to four rNTPs before synthesis stopped even when reactions were incubated as long as 16 min. These results suggest that Leu10 is a much better guard of genomic integrity against rNTPs incorporation than the designated steric gate, Y11. To our surprise, the Y11F mutation also modified sugar selectivity (Figure 2H); reactions were slower and maximum-sized products were shorter than with wild-type polV, suggesting that phenylalanine might sense the 2′-OH group better than tyrosine in UmuC.
Misincorporation assays (Figure 3 and Supplementary Figure S2) were unambiguous for F10L, i.e. they show that this enzyme is quite accurate, although it is still able to incorporate ATP opposite template T, C and G (Figure 3A–D and Supplementary Figure S2A–D). Unexpectedly, misincorporation of ATP opposite C and G (Supplementary Figure S2A–D) is actually more efficient than opposite the complementary T (Figure 3A–D). Misincorporation patterns for wild-type pol V and Y11F were very similar with slight, if any, increase in fidelity of dNTP incorporation for the Y11F mutant (c.f. Figures 1 and 3I–L; Supplementary Figure S2I–S2L). Nevertheless, both enzymes are highly error-prone and capable of incorporation of multiple wrong dNTPs outcompeting excessive amounts of ATP (as judged by the unique pattern of product distribution in various lanes).
Figure 3.
Specificity of nucleotide incorporation by mutant pol Vs. Reactions catalyzed by F10L (A–D), Y11A (E–H) or Y11F (I–L) in the presence 100 µM of each individual dNTP (A, B, E, F, I, J) or rNTP (C, D, G, H, K, L) were carried out for 5 min. All reactions contained 1 mM ATP. The identity of the nucleotide incorporated is shown below each lane and the extended sequence of templates with five consecutive Ts (A, C, E, G, I, K) or As (B, D, F, H, J, L) is indicated to the right of each gel panel. All lanes with reactions lacking polymerase are indicated as ‘pr’ and reactions with no additional nucleotide are indicated by dash (–).
The overall conclusion for pol V with an intact steric gate, is that the fidelity of matched rNTP incorporation appears to be comparable to mismatched dNTP incorporation with the exception observed for uracil, which is inserted opposite template A less efficiently. In contrast, results were obviously different for the Y11A polymerase (Figure 3E–H and Supplementary Figure S2E–S2H). While the interpretation is still complicated by high levels of ATP in all reactions, it is clear that all correct nucleotides (dNTPs as well as rNTPs) successfully competed with ATP present in 10-fold excess. However, when the wrong nucleotide was used, almost all primer extensions were ATP-dependent (i.e. the pattern of product distribution was similar in all lanes). While as many as six consecutive dNTP misinsertions could be found using different templates for the wild-type pol V [e.g. in Figure 1A, on template with target Ts, the products of dATP/ATP incorporations (lane 3) contain six successive mispairs with three Gs, one C and two As], only multiple ATP misincorporations are evident for Y11A. Therefore, there is no indication that pol V with a Y11A mutation in the UmuC subunit is any more error-prone for dNTP misincorporations (wrong base/correct sugar) than the wild-type enzyme even though its ability to incorporate rNTP (wrong sugar /correct base) is increased dramatically. Hence, the doublets observed on the gel with reactions in the presence of all four dNTPs (see above and Figure 2E) result from the competing incorporation of ATP opposite template T, rather than from dNTP misincorporation.
Substrate selectivity of UmuC variants
To characterize in greater depth the changes in fidelity and sugar discrimination of the pol V mutants, we performed more detailed analysis of nucleotide incorporations on templates with target Ts and As, followed by alkali cleavage of the resulting reaction products (Figure 4). When reactions were carried out using templates with five consecutive As adjacent to the 3′ primer end, ATP, dATP, UTP and dTTP were used either alone, or in different combinations (Figure 4A–D). For the templates with five target Ts, we used ATP and dATP along with CTP and dCTP since two Gs are the next available template bases (Figure 4E–H). The identity of the incorporated nucleotide was determined based on the differences in mobility of the reaction products and confirmed by their sensitivity to alkali digestion (since the DNA backbone is sensitive to alkaline hydrolysis at rNMPs).
Figure 4.
Discrimination against rNMP insertion by pol V mutants. Insertion of dA versus rA and T versus U by wild-type pol V (A), F10L (B), Y11A (C) or Y11F (D) were analyzed using a template containing five consecutive As. Insertion of dA/rA and dC/rC by wild-type pol V (E), F10L (F), Y11A (G) or Y11F (H) were analyzed using a template containing five consecutive Ts followed by two Gs. Reactions were performed in the presence of 100 µM dNTPs and/or 1 mM rNTPs for 10 min; half of each reaction mixture was subjected to alkaline hydrolysis (OH−) (lanes 2, 4, 6, 8 and 10 in each panel) under conditions that completely hydrolyze DNA chains at the positions of rNTP insertion. The identity of the nucleotide present in the reaction is shown below each track (I NTP, II NTP). The extended sequence of the templates with five consecutive As (A–D) or Ts (E–H) is indicated to the right of the gel panels. Reactions in lanes 1 and 2 contained only ATP; lanes 3, 4 ATP and dATP; lanes 5, 6 ATP with UTP (A–D) or with CTP (E–H); lanes 7, 8 ATP and dATP with UTP and dTTP (A–D) or with CTP and dCTP (E–H); lanes 9, 10 ATP and dATP with dTTP (A–D) or with dCTP (E–H). Bands corresponding to incorporation of UTP (in A and D) or dTTP (in C) are indicated by stars.
Consistent with our previous experiments, F10L remained highly accurate (it mainly incorporated the correct dNTP, although some misincorporation opposite G and C was also evident) and virtually all products formed by this enzyme were resistant to alkali cleavage (Figure 4B and F). On the other hand, the fidelity of wild-type pol V and Y11F was significantly lower and almost indistinguishable under these reaction conditions (c.f. Figure 4A, D, E and H). Both wild-type pol V and Y11F inefficiently misincorporated adenine ribo- and deoxyribonucleotides opposite template A (Figure 4A and D), and even in the presence of excess of rNTP, both polymerases preferred to incorporate dNTP rather than rNTP, as judged by the negligible alkali-sensitivity of reaction products (Figure 4A, D, E and H, lanes 7 and 8). It is evident, that wild-type pol V and Y11F incorporated ATP opposite template T (Figure 4E and H, lanes 1 and 2) much more efficiently than UTP opposite template A (Figure 4A and D, lanes 5 and 6), even though both preserve Watson–Crick base pairing. Furthermore, UTP and dTTP were inserted with equal efficiency opposite template A, even when dTTP was present in the reactions in trace amounts (as a contaminant of UTP; see Figure 4A and D, lanes 5 and 6).
Remarkably, sugar selectivity appears to reverse with the Y11A mutant (Figure 4C). Not only did the strong selectivity against UTP disappear, but UTP seems to be a preferred substrate for Y11A compared to dTTP. For example, when both UTP and dTTP were present in the reaction at equal concentrations, bands corresponding to inserted UMP were stronger than bands containing dTMP and most of the reaction products disappeared after alkali digestion (Figure 4C, lanes 7 and 8). Such selectivity is very unusual, since UTP has been proven to be an especially difficult substrate for many DNA polymerases (38,39). A preference for ribonucleotide incorporation was also observed in the case of ATP/dATP competition, i.e. Y11A preferentially incorporated ATP opposite template T (Figure 4G, lanes 3 and 4) and template A (Figure 4C, lanes 3 and 4). In terms of overall fidelity, these experiments unequivocally indicated that compared to the wild-type enzyme, Y11A is capable of much more efficient misincorporation of adenine and/or uracil opposite all DNA template bases. However, the comparable length of replication products when two or three dNTPs were omitted from the reactions (c.f. lanes 3, 7 and 9 in Figure 4A, C or E and G), argues that the overall fidelity of base selection (compared to sugar selection) is not appreciably lower for Y11A than for the wild-type polymerase.
To better understand the steric interactions between Y11A and an incoming rNTP, we compared reactions in the presence of dCTP and a steric isomer of CTP, cytosine-1-β-d-arabinofuranoside (Ara-C), which also has a 2′-hydroxyl group, only in a different conformation (Figure 5). Ara-C was incorporated as efficiently as dCTP by both wild-type pol V and Y11A, similarly outcompeting ATP and incorrect dNTPs incorporation opposite template G and equally interfering with further primer extension. No products beyond the +5 position are seen even in the presence of correct downstream nucleotides. Efficient incorporation of Ara-C is not unusual even for DNA polymerases with strict sugar selectivity, since a ribose with a 2′-OH group on the opposite side of the plane of the sugar (compared to rNTP) avoids the collision with the steric gate residue. However, extension of the resulting primer ends for these polymerases, as well as for wild-type pol V and Y11A, is largely inhibited despite the presence of a 3′-hydroxyl group on the sugar moiety of the terminal nucleotide. Therefore, even though the Y11A mutation greatly facilitates accommodation of a nucleotide with a 2′-OH group in the polymerase active site, it does not improve primer extension when the sugar pucker at its terminus is in a 2′-endo conformation. These experiments also indicated that dG and/or dC misincorporations occur more often when catalyzed by wild-type pol V than by Y11A [e.g. wild-type pol V in the presence of dGTP and dCTP synthesizes ∼5 nt longer product than Y11A (compare lanes 4 in Figure 5A and B), and the resulting 16-mer requires elongation of a primer with five consecutive C/A and C/T and/or G/A and G/T mispairs]. These data support our assumption that Y11A could actually be more accurate while incorporating dNTPs than the wild-type enzyme.
Figure 5.
The inhibitory effect of cytosine arabinoside incorporation on DNA synthesis by wild-type pol V (A) and the Y11A variant (B). Insertions of dCTP (lanes 2, 4, 6) versus ara-C (lanes 3, 5, 7) with, or without, dGTP alone (4, 5), or in combination with dATP and dTTP (6, 7) were analyzed using a substrate containing five consecutive Gs in the template. Reactions were carried out in the presence of 100 µM nucleotides for 5 min; all reactions contained 1 mM ATP. The identity of the nucleotide incorporated is shown below each track. The extended sequence of the template is indicated to the right of the gel. The reaction shown in lane 0 contained no polymerase and the reactions in lane 1 contained no other nucleotides except the 1 mM ATP. The well and primer locations are indicated by arrows at the top and bottom of the gel, respectively.
PolV-dependent spontaneous mutagenesis
To examine how differences in the biochemical properties of the pol V variants affect the phenotypes of strains expressing these polymerases, we determined the extent of pol V-dependent ‘SOS’ mutagenesis by assaying for reversion of the hisG4(ochre) allele in an E. coli ΔumuDC lexA(Def) recA730 strain (RW584) expressing UmuD′ and the various UmuC proteins at physiological levels. The lexA(Def) allele results in derepression of all LexA-regulated genes, including umuDC and recA, while the recA730 gene encodes for a constitutively activated RecA* protein and in this genetic background, spontaneous mutagenesis is largely dependent upon pol V, (17,42). As can be seen in Figure 6, there is minimal reversion of the hisG4 allele in the strain harboring the empty vector, pGB2, but considerable mutagenesis (∼770 His+ mutants per plate) in the presence of wild-type pol V (pRW134). It should be stressed that spontaneous mutagenesis does not reflect pol V-dependent translesion DNA synthesis of presumptive ‘cryptic’ DNA lesions (43), but rather the successful competition of pol V with the cell’s replicase, pol III, so as to gain access to the E. coli chromosome and replicate undamaged DNA with low-fidelity (26,43).
In contrast to wild-type pol V, the F10L variant only produced an average of ∼45 mutants per plate (Figure 6). Such an observation is entirely consistent with our in vitro studies showing that F10L substitution increases the fidelity of polV-catalyzed replication. Quite surprisingly, despite exhibiting low-fidelity DNA and RNA synthesis in vitro, Y11A-dependent spontaneous mutagenesis in vivo was about one-tenth of that promoted by wild-type pol V (∼75 versus 770 His+ revertants per plate). Interestingly, expression of the Y11F UmuC variant reproducibly resulted in ∼35% more His+ revertants than wild-type pol V (∼1030 versus ∼770 His+ mutants per plate) (Figure 6). This difference was statistically significant (P ≤ 0.002) and suggests that the Y11F substitution is, perhaps, not quite as neutral as we previously hypothesized.
Specificity of base-pair substitutions made by pol V variants
In order to establish base substitution fidelity of pol V variants in vivo, we determined the spectra of spontaneously arising mutations in strains expressing these polymerases. To do so, we detected mutations in an ∼200-bp region of the E. coli rpoB gene in rifampicin resistant derivatives of the E. coli strain, RW710 (Supplementary Table S2). The strain is isogenic with RW584, but also harbors two additional alleles; ΔdinB61::ble and mutL211::Tn5 (27). While spontaneous mutagenesis in the lexA(Def) recA730 strain is determined mainly by the mistakes made by pol V (Figure 6), a fraction of the mutations are also dependent upon dinB (pol IV) (27,44), presumably as a result of pol IV extending nascent DNA strands with terminal mismatches made by pol V. To avoid any potential complications resulting from the preferential extension of certain mispairs by pol IV, we utilized a ΔdinB strain for spectral analysis. The RW710 strain also lacks mismatch repair resulting from a Tn5 insertion in the mutL gene which allowed us to assay all replication errors, rather than only those that persist in the presence of active mismatch repair (45).
Sequence analysis of rifampicin-resistant mutants revealed that consistent with previous data, the spectrum of mutations with the vector control (pGB2) was dominated by transitions at particular mutagenic hot-spots (sites at which there are 10 or more mutations) (27) (Figure 7). These were AT → GC transitions located at positions 1532 and 1534 and CG → TA transitions at positions 1546 and 1691. Since the RW710 strain lacks pol IV and pol V, these are mutations generated by pols I, II or III (27). In the presence of wild-type pol V (pRW134), there was a general increase in transversions, most notably at positions 1547 and 1714 (AT → TA) and 1576 (CG → AT) (27). In contrast, the spectrum of rpoB mutations recovered from the F10L-expressing strain was very similar to the one observed in the absence of pol V, which is consistent with the higher fidelity of the F10L variant compared to wild-type pol V. The transversions observed at positions 1547 and 1576 for the wild-type enzyme were absent from the spectrum obtained in Y11A-expressing cells, instead there was one new CG → TA transition hot-spot at position 1535. In vivo, Y11F consistently produced higher levels of mutagenesis than the wild-type enzyme (Figure 6) even though in vitro it exhibited properties similar to the wild-type pol V. Interestingly, the Y11F- and wild-type pol V-dependent rpoB spectra differed significantly, with the spectrum in Y11F-expressing cells dominated by an AT → GC transition at position 1534 and a CG → TA transition at position 1691. Overall, our data demonstrate convincingly that the spectrum of pol V-dependent rpoB mutations observed in vivo is unique to each particular pol V allele.
Figure 7.
Spectrum of spontaneous rpoB mutations arising in a mutL recA730 lexA(Def) ΔdinB ΔumuDC strain expressing pol V variants. The types of base-pair substitutions observed in the rpoB gene in E. coli strain RW710 [recA730 lexA(Def) ΔdinB ΔumuDC mutL], that result in rifampicin resistance are color coded as shown in the figure. The arrows indicate mutagenic hot spots discussed in the article.
DISCUSSION
The fidelity of sugar discrimination by various DNA polymerases has attracted increasing attention in recent years (41,46). Multiple studies provide indisputable evidence that incorporation of rNTPs into DNA during replication in vivo is a frequent event that triggers a range of cellular responses. To minimize the negative effect of rNTPs incorporation into DNA, the active site of DNA polymerases is shaped so as to exclude the ribose 2′-OH group. The amino acid that is largely responsible for the discrimination against rNTPs is a steric gate that is highly conserved within DNA polymerase families. In addition to sugar selectivity, this residue has been implicated in other cellular pathways such as translesion DNA synthesis, incorporation of erroneous nucleotides, extension of a mismatched primers and fidelity of base substitution errors (41). It has also been reported that active site residues flanking the steric gate could modulate sugar selectivity.
We are interested in understanding the effects of such mutations on E. coli pol V. Wild-type pol V and the F10L, Y11A and Y11F variants exhibited similar primer extension efficiencies. This is in contrast to a steric gate mutant of human pol η (F18A), in which an increased capacity to incorporate rNTPs into DNA coincided with a substantial reduction in the ability to incorporate dNTPs (47). Both, wild-type pol V and the Y11F mutant were able to efficiently synthesize long stretches of RNA. This is particularly interesting since steric gate polymerase mutants selected for the superior capacity to incorporate rNTPs are rarely able to synthesize RNAs longer than several nucleotides (41). Our data suggest that the additional constraints on RNA primer extension that normally exist in most DNA polymerases are relaxed in pol V. Indeed, molecular modeling of the UmuC active site suggests that UmuC is different from other Y-family polymerases in the region surrounding the steric gate. For example, a loop structure immediately before Y74 in UmuC, is not conserved, and in UmuC is two residues longer than most Y-family polymerases and five residues shorter than human polη (Supplementary Figure S3). The different number of residues in this region likely changes the backbone structure around Y74, which may result in more flexibility of Y11 in UmuC and allow for ribonucleotide incorporation by wild-type pol V (Figure 8A and B).
Figure 8.
Structural models of UmuC and variants. Models of wild-type UmuC or steric gate mutants inserting dATP or ATP opposite a template T. These models are based on the structure of human DNA pol η (PDB 3MR3), and amino acid substitutions were made following the alignment by HHpred (toolkit.tubebingen.mpg.de/hhpred). Residues surrounding the incoming nucleotide are modeled and labeled according to the UmuC sequence. (A) Wild-type UmuC showing that Y11 coordinates a conserved water molecule (red sphere) that interacts with the incoming nucleotide, dATP (shown in yellow). (B) Wild-type pol V can accommodate ATP (shown in blue) by rotation of the benzene ring of Y11 (alternate positions shown in pink and grey). (C) Substitution of Y11 with Ala leaves a void in the active site of UmuC that can readily accommodate ATP. (D) Modeling of Phe10 (shown in grey), and Leu10 (shown in blue), reveals that the side chain of Leu presses on the benzene ring of Y11, so as to close the distance between the C2 position of the incoming deoxyribonucleotide (dATP, shown in yellow), and concomitantly precludes ribonucleotide incorporation.
An amino acid with a smaller side chain (alanine) replacing the Y11 steric gate leaves a void around the incoming nucleotide, which not only relaxes selection of the sugar (as a modeled ribose can be readily accommodated), but also relaxes the minor groove alignment for correct Watson–Crick base pair (Figure 8C). It was therefore not surprising that the Y11A substitution not only reduced the selectivity against single rNTP incorporation, but also effectively converted the resulting mutant into a bona fide RNA polymerase. Clearly, pol V Y11A synthesizes RNA nearly as efficiently as DNA (Figure 2). What was unforeseen was that pol V in which F10 was replaced with leucine had much greater sugar selectivity than wild-type polV with the conserved steric gate, Y11. As a consequence, RNA synthesis was inefficient with F10L, even after prolonged incubation times and the mutant polymerase was extremely limited in its ability to incorporate multiple rNTPs sequentially. Our observations can, however, be explained by the fact that the branched side-chain of L10 presses on the benzene ring of Y11 (indicated by the arrows in Figure 8D), which, in turn, requires Y11 to rotate slightly. As a result, the side-chain of Y11 is closer to the C2′ position of the incoming nucleotide than in the native protein, thereby restricting any possible movement of the steric gate residue. The tightened constrains for ribonucleotide accommodation therefore leads to firm closure of the steric gate.
The involvement of a steric gate in shaping base substitution fidelity of a DNA polymerase is not unequivocal. For example, the F12 residue of the archaeal DinB polymerase, Dbh, plays a minor role in fidelity (22,48), while the steric gate of human pol κ (Y112) modulates its accuracy by regulating the mismatched termini extension and incorporation of erroneous nucleotides (47,49). While our study did not delineate with certainty whether the steric gate of pol V has any significant impact on misincorporation of dNTPs, we clearly established that misincorporated dNTPs cannot outcompete ATP misincorporation catalyzed by Y11A (Figure 3 and Supplementary Figure S2). This is in contrast to polymerases with an intact steric gate (Y11 and F11) which incorporated nucleotides that change sugar identity and nucleotides that distort Watson–Crick base pairing with comparable frequency (Figures 1 and 3; Supplementary Figure S2). The only exception was reduced incorporation of uracil, the most challenging substrate for wild-type and Y11F pol V.
While reduced mutagenesis in strains expressing UmuC F10L (Figure 6) undoubtedly results from an increase in fidelity (Figure 3 and Supplementary Figure S2), a similar scenario cannot explain the poor mutability of the strains expressing the UmuC Y11A variant. The Y11A mutant is considerably more error-prone than F10L pol V (Figure 3 and Supplementary Figure S2). Furthermore, compared to the wild-type enzyme, Y11A does not appear to be significantly less prone to make base substitutions but it readily misincorporates ribonucleotides with high efficiency (Figures 1 and 3; Supplementary Figure S2). Perhaps therein lies the answer for the poor mutability? It is possible that excessive rNMPs in the E. coli genome might have detrimental biological consequences due to the compromised chemical stability of DNA, or through the modulation of nucleic acid–protein interactions that could obstruct DNA processing. However, it seems more plausible to assume that the errant ribonucleotides incorporated in vivo might trigger downstream pathways directed at rNMP removal, which would simultaneously repair deoxyribonucleotide base mismatches. Further studies are needed to test this hypothesis.
Since Phe is often found at the position of the steric gate in pol V orthologs and Tyr and Phe amino acids are structurally similar [the only difference being that Tyr has an additional –OH on the para (4th) position of its benzene ring], we tacitly assumed that the Y11F UmuC substitution would have no effect on fidelity of pol V. On the contrary, however, the substitution actually resulted in a consistent ∼35% increase in pol V-dependent spontaneous mutagenesis in vivo (Figure 6). We also observed differences in the types of base substitutions found in the rpoB spectra in strains expressing the wild-type or Y11A UmuC (Figure 7). Most notably, the AT → TA and CG → AT transversions observed with wild-type pol V were absent in the Y11F spectrum. Instead, the Y11F-dependent spectrum was dominated by two AT → GC and CG → TA hot-spots. Both these transitions are likely to have arisen through the formation of a G:T mispair. Molecular modeling of the UmuC structure (Figure 8A) suggests that Y11 coordinates a conserved water molecule that interacts with the replicating base pair in the minor groove. A similar interaction with a water molecule was found in Dpo4, as well as in pol η structures (50). However, in the Y11F mutant (Figure 9), the absence of the hydroxyl group destabilizes the water molecule and favors the accommodation of a G:T mispair, which helps explain why we observed an increase in the overall level of Y11F-dependent mutagenesis in vivo and the unique Y11F mutagenic rpoB spectra in particular.
Figure 9.
Model of UmuC Y11F. UmuC Y11 normally coordinates a conserved water molecule that interacts with the incoming nucleotide. However, in the UmuC Y11F mutant, the absence of the hydroxyl group destabilizes the water molecule and in its absence allows for the misincorporation of dGTP (shown in blue) and the formation of a G:T mispair (dotted lines). For comparison, the correct base (dATP) with the water molecule is shown in light yellow.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–3.
FUNDING
National Institute of Child Health and Human Development/National Institutes of Health Intramural Research Program (to R.W.); the National Institute of Diabetes and Digestive and Kidney Diseases/National Institutes of Health Intramural Research Program (to W.Y.); the Special Coordination Funds for Promoting Science and Technology of the Japanese Ministry of Education, Culture, Sports, Science and Technology (to K.K.); and National Institutes of Health [GM21422, ESO12259] (to M.F.G.). Funding for open access charge: NIH Intramural Research Program.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol. Gen. Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 2.Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol. Gen. Genet. 1978;165:87–93. doi: 10.1007/BF00270380. [DOI] [PubMed] [Google Scholar]
- 3.Woodgate R, Rajagopalan M, Lu C, Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD′. Proc. Natl Acad. Sci. USA. 1989;86:7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruck I, Woodgate R, McEntee K, Goodman MF. Purification of a soluble UmuD′C complex from Escherichia coli: cooperative binding of UmuD′C to single-stranded DNA. J. Biol. Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 5.Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. UmuD′2C is an error-prone DNA polymerase, Escherichia coli, DNA pol V. Proc. Natl Acad. Sci. USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and Is specialized for translesion replication. J. Biol. Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 7.Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF. The active form of DNA polymerase V is UmuD′2C-RecA-ATP. Nature. 2009;460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii S, Fuchs RP. Biochemical basis for the essential genetic requirements of RecA and the β-clamp in Pol V activation. Proc. Natl Acad. Sci. USA. 2009;106:14825–14830. doi: 10.1073/pnas.0905855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karata K, Vaisman A, Goodman MF, Woodgate R. Simple and efficient purification of E. coli DNA polymerase V: cofactor requirements for optimal activity and processivity in vitro. DNA Repair. 2012;11:431–440. doi: 10.1016/j.dnarep.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington,D.C: American Society for Microbiology; 1995. [Google Scholar]
- 12.Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc. Natl Acad. Sci. USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burckhardt SE, Woodgate R, Scheuermann RH, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification and cleavage by RecA. Proc. Natl Acad. Sci. USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommer S, Boudsocq F, Devoret R, Bailone A. Specific RecA amino acid changes affect RecA-UmuD′C interaction. Mol. Microbiol. 1998;28:281–291. doi: 10.1046/j.1365-2958.1998.00803.x. [DOI] [PubMed] [Google Scholar]
- 15.Nohmi T, Battista JR, Dodson LA, Walker GC. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc. Natl Acad. Sci. USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutreix M, Moreau PL, Bailone A, Galibert F, Battista JR, Walker GC, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J. Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J. Bacteriol. 1990;172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pham P, Bertram JG, O’Donnell M, Woodgate R, Goodman MF. A model for SOS-lesion targeted mutations in E. coli involving pol V, RecA, SSB and β sliding clamp. Nature. 2001;409:366–370. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- 19.Echols H, Goodman MF. Mutation induced by DNA damage: a many protein affair. Mutat. Res. 1990;236:301–311. doi: 10.1016/0921-8777(90)90013-u. [DOI] [PubMed] [Google Scholar]
- 20.Tang M, Pham P, Shen X, Taylor J-S, O’Donnell M, Woodgate R, Goodman M. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 21.Astatke M, Ng K, Grindley ND, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl Acad. Sci. USA. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLucia AM, Chaudhuri S, Potapova O, Grindley ND, Joyce CM. The properties of steric gate mutants reveal different constraints within the active sites of Y-family and A-family DNA polymerases. J. Biol. Chem. 2006;281:27286–27291. doi: 10.1074/jbc.M604393200. [DOI] [PubMed] [Google Scholar]
- 23.DeLucia AM, Grindley ND, Joyce CM. An error-prone family Y DNA polymerase (DinB homolog from Sulfolobus solfataricus) uses a ‘steric gate’ residue for discrimination against ribonucleotides. Nucleic Acids Res. 2003;31:4129–4137. doi: 10.1093/nar/gkg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shurtleff BW, Ollivierre JN, Tehrani M, Walker GC, Beuning PJ. Steric gate variants of UmuC confer UV hypersensitivity on Escherichia coli. J. Bacteriol. 2009;191:4815–4823. doi: 10.1128/JB.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karata K, Vidal AE, Woodgate R. Construction of a circular single-stranded DNA template containing a defined lesion. DNA Repair. 2009;8:852–856. doi: 10.1016/j.dnarep.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandewiele D, Fernández de Henestrosa AR, Timms AR, Bridges BA, Woodgate R. Sequence analysis and phenotypes of five temperature sensitive mutator alleles of dnaE, encoding modified a-catalytic subunits of Escherichia coli DNA polymerase III holoenzyme. Mutat. Res. 2002;499:85–95. doi: 10.1016/s0027-5107(01)00268-8. [DOI] [PubMed] [Google Scholar]
- 27.Curti E, McDonald JP, Mead S, Woodgate R. DNA polymerase switching: effects on spontaneous mutagenesis in Escherichia coli. Mol. Microbiol. 2009;71:315–331. doi: 10.1111/j.1365-2958.2008.06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase | and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 29.Szekeres ESJ, Woodgate R, Lawrence CW. Substitution of mucAB or rumAB for umuDC alters the relative frequencies of the two classes of mutations induced by a site-specific T-T cyclobutane dimer and the efficiency of translesion DNA synthesis. J. Bacteriol. 1996;178:2559–2563. doi: 10.1128/jb.178.9.2559-2563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor,N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Davis BD, Mingioli ES. Mutants of Escherichia coli requiring methionine or vitamin B12. J. Bacteriol. 1950;60:17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, Diep A, Hu K, Iverson A, Yang H, et al. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair. 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 34.Wolff E, Kim M, Hu K, Yang H, Miller JH. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J. Bacteriol. 2004;186:2900–2905. doi: 10.1128/JB.186.9.2900-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mead S, Vaisman A, Valjavec-Gratian M, Karata K, Vandewiele D, Woodgate R. Characterization of polVR391: a Y-family polymerase encoded by rumA′B from the IncJ conjugative transposon, R391. Mol. Microbiol. 2007;63:797–810. doi: 10.1111/j.1365-2958.2006.05561.x. [DOI] [PubMed] [Google Scholar]
- 36.Biertümpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase η. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 38.Patel PH, Loeb LA. Multiple amino acid substitutions allow DNA polymerases to synthesize RNA. J. Biol. Chem. 2000;275:40266–40272. doi: 10.1074/jbc.M005757200. [DOI] [PubMed] [Google Scholar]
- 39.Xia G, Chen L, Sera T, Fa M, Schultz PG, Romesberg FE. Directed evolution of novel polymerase activities: mutation of a DNA polymerase into an efficient RNA polymerase. Proc. Natl Acad. Sci. USA. 2002;99:6597–6602. doi: 10.1073/pnas.102577799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boudsocq F, Ling H, Yang W, Woodgate R. Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass. DNA Repair. 2002;1:343–358. doi: 10.1016/s1568-7864(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 41.Brown JA, Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciesla Z. Plasmid pKM101-mediated mutagenesis in Escherichia coli is inducible. Mol. Gen. Genet. 1982;186:298–300. doi: 10.1007/BF00331866. [DOI] [PubMed] [Google Scholar]
- 43.Fijalkowska IJ, Dunn RL, Schaaper RM. Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J. Bacteriol. 1997;179:7435–7445. doi: 10.1128/jb.179.23.7435-7445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuban W, Banach-Orlowska M, Schaaper RM, Jonczyk P, Fijalkowska IJ. Role of DNA polymerase IV in Escherichia coli SOS mutator activity. J. Bacteriol. 2006;188:7977–7980. doi: 10.1128/JB.01088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaaper RM, Dunn RL. Spectra of spontaneous mutations in Escherichia coli strains defective in mismatch correction: the nature of in vivo DNA replication errors. Proc. Natl Acad. Sci. USA. 1987;84:6220–6224. doi: 10.1073/pnas.84.17.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katafuchi A, Sassa A, Niimi N, Gruz P, Fujimoto H, Masutani C, Hanaoka F, Ohta T, Nohmi T. Critical amino acids in human DNA polymerases η and κ involved in erroneous incorporation of oxidized nucleotides. Nucleic Acids Res. 2010;38:859–867. doi: 10.1093/nar/gkp1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu M, Gruz P, Kamiya H, Kim SR, Pisani FM, Masutani C, Kanke Y, Harashima H, Hanaoka F, Nohmi T. Erroneous incorporation of oxidized DNA precursors by Y-family DNA polymerases. EMBO Rep. 2003;4:269–273. doi: 10.1038/sj.embor.embor765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niimi N, Sassa A, Katafuchi A, Gruz P, Fujimoto H, Bonala RR, Johnson F, Ohta T, Nohmi T. The steric gate amino acid tyrosine 112 is required for efficient mismatched-primer extension by human DNA polymerase κ. Biochemistry. 2009;48:4239–4246. doi: 10.1021/bi900153t. [DOI] [PubMed] [Google Scholar]
- 50.Pata JD. Structural diversity of the Y-family DNA polymerases. Biochim. Biophys. Acta. 2010;1804:1124–1135. doi: 10.1016/j.bbapap.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.