Abstract
The bacterial homologue of C4orf14, YqeH, has been linked to assembly of the small ribosomal subunit. Here, recombinant C4orf14 isolated from human cells, co-purified with the small, 28S subunit of the mitochondrial ribosome and the endogenous protein co-fractionated with the 28S subunit in sucrose gradients. Gene silencing of C4orf14 specifically affected components of the small subunit, leading to decreased protein synthesis in the organelle. The GTPase of C4orf14 was critical to its interaction with the 28S subunit, as was GTP. Therefore, we propose that C4orf14, with bound GTP, binds to components of the 28S subunit facilitating its assembly, and GTP hydrolysis acts as the release mechanism. C4orf14 was also found to be associated with human mitochondrial nucleoids, and C4orf14 gene silencing caused mitochondrial DNA depletion. In vitro C4orf14 is capable of binding to DNA. The association of C4orf14 with mitochondrial translation factors and the mitochondrial nucleoid suggests that the 28S subunit is assembled at the mitochondrial nucleoid, enabling the direct transfer of messenger RNA from the nucleoid to the ribosome in the organelle.
INTRODUCTION
The eukaryotic cell maintains small circles of DNA in mitochondria in order to produce a handful of proteins. Although few in number, the 13 proteins encoded in human mitochondrial DNA (mtDNA) nevertheless make critical contributions to aerobic adenosine triphosphate (ATP) production. An accurate estimate of the number of gene products that are required to maintain and express mtDNA still eludes us, but it is expected to exceed 200. Based on other systems (1), 200 proteins could well be involved in mitochondrial translation, as mitochondria harbour dedicated ribosomes (2). Mitochondrial ribosomes are more closely related to their prokaryotic antecedents, than their immediate physical neighbours in the cytosol, as indicated by their sensitivity to a range of antibiotics that target bacterial ribosomes (3). In addition to ribosomal proteins, mitochondria have inherited translation initiation, elongation and termination factors from bacteria based on sequence homology. Mitochondrial protein synthesis is also of medical importance as defects in this process account for an increasing number of cases of mitochondrial disease (4). Several studies have defined the components of the mitochondrial ribosome and allied proteins (5–7) yet our understanding of mitochondrial ribosomal biogenesis is far from complete.
mtDNA is organized in nucleoprotein complexes, or nucleoids. The identification of proteins in enriched preparations of mtDNA from mammalian cells and tissues has provided an extensive list of candidate nucleoid proteins (8–11), although little is known of the functions of many of them, at least in respect of mtDNA metabolism. Most progress has been made in yeasts, where several seemingly unlikely candidates, such as HSP60, α-ketoglutarate dehydrogenase and ilv5, an enzyme involved in amino acid biosynthesis, have been shown to contribute to mtDNA maintenance (12–15). Thus, mtDNA organization and maintenance appear to be quite different to nuclear DNA. mtDNA is also unlike nuclear DNA in that there is no physical barrier to prevent concurrent transcription and translation in mitochondria, and there is some evidence that mitochondrial transcription and translation are linked (16–18). If this is so, then one might expect numerous translation factors to co-purify with mtDNA. Here, we report that chromosome 4 open reading frame 14, C4orf14 (or NOA1) is linked to mitochondrial nucleoids and to the apparatus of mitochondrial translation, specifically the small mitochondrial ribosomal (28S) subunit. It was identified in one preparation of TFAM (mitochondrial transcription factor A) affinity-purified nucleoids (19), and its prokaryotic homologue YqeH is a guanosine triphosphate (GTP) binding protein of Era/Obg family involved in the biogenesis of the bacterial small ribosomal subunit (20–21). For this reason, and because C4orf14 has also been implicated in DNA replication in prokaryotes (23), we selected it for further study. Recently, C4orf14 (NOA1) was reported to be required for normal mitochondrial translation and respiratory functions (24). It contains both a functional and highly conserved circularly permuted GTPase domain (25), and a predicted TRAP domain, suggesting that it could bind to RNA (25,26). Here, we show that C4orf14 binds to the small (28S) subunit of the mitochondrial ribosome and to other mitochondrial translation factors, via a GTP-dependent mechanism. C4orf14 is also a DNA-binding protein, and so it potentially links the processes of protein synthesis and DNA maintenance in the mitochondrion.
MATERIALS AND METHODS
Cell culture
Human osteosarcoma (HOS 143B cells) and human embryonic kidney cells (HEK293T) were grown in Dulbecco’s Modified Eagle’s Medium and 10% fetal bovine serum. In the case of transgenic HEK293T cells, the serum was tetracycline-free, and the medium included 15 µg/ml of blasticidin and 100 µg/ml of Zeocin or 100 µg/ml of hygromycin and 15 µg/ml of blasticidin for control and C4orf14 expressing cells, respectively.
Affinity purification of C4orf14 and TFAM from HEK293T cells
Human complementary DNAs (cDNAs) of TFAM or C4orf14, with a carboxy-terminal Strep II followed by a FLAG tag, were introduced into HEK293T cells (Invitrogen), to establish inducible, transgenic cell lines. Transgene expression was induced with 2–20 ng/ml doxycycline for 24–48 h. Mitochondria from HEK293T cells were isolated by a method modified from (11), and the tagged protein affinity purified as detailed in (19), except that the second affinity purification step was not applied.
Mass spectrometry
Protein samples were fractionated by SDS–PAGE, stained with Coomassie blue dye, excised and subjected to in-gel tryptic digestion and then analysed in a MALDI-TOF-TOF mass spectrometer (model 4800; ABI-Sciex) using matrix α-cyano-4-hydroxycinnamic acid. Acquired peptide mass data were compared with sequence databases using MASCOT, and proteins were identified from a significant peptide mass fingerprint (P < 0.05) or from peptide fragmentation data (P < 0.05). Most proteins were identified by both criteria, see Supplementary Table S1. Stable isotope labelling of proteins in cell culture (SILAC) experiments were performed as for ATAD3B (ATPase family AAA domain-containing protein 3) (19).
GDP treatment of strepactin columns with bound C4orf14
The strepactin column with bound recombinant C4orf14 from HEK293T-purified mitochondria was washed five times with 20 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) pH 7.6, 5 mM ethylenediaminetetraacetic acid, 150 mM NaCl, 2 mM dithitothreitol (DTT), 0.2 mM phenylmethanesulfonylfluoride (PMSF), 1v/50v Roche protease inhibitor solution, 0.05% n-dodecyl-β-d-maltoside (wash buffer) and then incubated with wash buffer supplemented with 20 mM guanosine diphosphate (GDP) pH 7.6, for 30 min at 4°C, and washed a further five times with wash buffer containing GDP. All five washes with GDP were pooled and dialysed in 100 × sample volume wash buffer overnight at 4°C. After dialysis, the ‘GDP-eluted’ material was incubated with or without 20 mM GTP and then re-loaded on two new strepactin columns; thereafter the standard washes and desthiobiotin elution procedure were applied.
Cell transfection
For RNAi experiments, 143B cells were transfected with lipofectamine 2000 (Invitrogen) and 5 or 10 nM double-stranded (ds) RNA. After 48 or 72 h, cells were lysed and total RNA or DNA was extracted. The two pairs of dsRNAs (Qiagen) targeting C4orf14 were 5′-TTGGGTTTCAGTAACACCTAA-3′, and 5′-TTGGGTGCTATAGGCCGCATA-3′, respectively, known as C4orf14-3 and C4orf14-6. Qiagen: allstar neg. control siRNA, cat: 1027281 was used as a control dsRNA. Plasmid DNA transfection was similar except that 1.5 µg plasmid DNA replaced dsRNA, and antibiotic selection was applied after 24 or 48 h.
Estimation of transcript and mtDNA copy number
Mitochondrial mRNA abundance was estimated relative to the mRNA for glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Total RNA was extracted from cells with TRIzol reagent (Invitrogen) and cDNA was generated with an Omniscript reverse transcription kit with random hexamer primers and oligo dT (Qiagen) according to the manufacturer’s instructions. mtDNA copy number and the relative abundance of mRNA was estimated by quantitative PCR, as described before (27). Primers with the following sequences were employed: GAPDH, forward 5′-GAAGGTGAAGGTCGGAGTCAAC-3′, reverse 5′-CAGAGTTAAAAGCAGCCC TGGT-3′, probe 5′-TTTGGTCCGTATTGGGCG-3′; APP, forward 5′-TTTTTGTGTGCTCTCCCAGGTCT-3′, re-verse 5′-TGGTCACTGGT TGGTTGGC-3′, APP probe 5′-CCCTGAACTGCAGATCACCAATGCGGTAG-3′; COXII, forward 5′-CGTCTGAACTATCCTGCCCG-3′, reverse 5′-TGGTAAGGGAGGG ATCGTTG-3′, probe 5′-CGCCCTCCCATCCCTACGCATC-3′; Cyt b forward 5′-GCCTGCCTGATCCTCCAAAT-3′, reverse 5′-AAGGTAGCGGATGATTCAGCC-3′, probe 5′-CACCAGACGCCTCAACCGCCTT-3′; C4orf14, forward 5′-TCCTGCAGGGAAATCAGTC-3′, reverse primer 5′-GTTCTTTTCCACCCAT TGGA-3′, probe 5′-ATCAGAAGCATGCAGGTCATACGT-3′. Probes contained a 5′ Familial Mediterranean fever fluorophore and a 3′ TAMRA quencher (Sigma Genosys). Cycle conditions were the default setting on the ABI sequence detection system 7700.
DNA analysis
DNA extraction and Southern blotting were performed as described previously (9). mtDNA copy number was estimated by comparing the signal from a radiolabelled probe to the major non-coding region, nucleotides 16343-151 of human mtDNA with that of a nuclear 18S rRNA probe.
Immunoblotting
Immunodetection utilized antibodies to goat anti-human POLG1 1 : 500 (Santa Cruz), rabbit anti-Cyt (cytochrome) c 1 : 200 (Abcam), goat anti-VDAC (1 : 1000, Santa Cruz), mouse anti-TUFM 1 : 1000 (Abnova), mouse monoclonal anti-FLAG M2 1 : 1000 (Sigma), rat anti-HA peroxidase 1 : 1000 (Roche), rabbit anti-POLMRT 1 : 1000 (Abcam), rabbit anti-TOM20 1 : 20 000 (Santa Cruz), rabbit anti-MRPS29 and MRPL11 1 : 1000 (Cell Signaling), goat anti-MRPS6, anti-MRPS10, anti-MRPL15 1 : 200 (Santa Cruz), rabbit anti-Prohibitin 1 : 500 (BioLegend), rabbit anti-DHX30 1 : 200 (Abcam), mouse anti-MRPS27 1 : 500 (Abcam), mouse anti-MRPS2 1 : 500 (Abcam), rabbit anti-LRP130 1 : 500 (Santa Cruz technology), mouse anti-SF2P321 1 : 1000 (BD Bioscience), rabbit anti-PTCD3 1 : 500 (Abcam), rabbit anti-HSPD1 1 : 5000 (Abcam), rabbit anti-UQCRC (complex III): 1 : 3000 (Sigma), mouse anti-SDH (SDHB, complex II), 1 : 500 (Mitoscience), rabbit anti-cytochrome oxidase II 1 : 200 (Abcam), rabbit anti-ERAL1 (E. coli Ras-like protein-Like 1) (1 : 2000) (ProteinTech), C4orf14 1 : 500 (Abcam). Rabbit anti-SSBP1 (1 : 500) and rabbit anti-TFAM (1 : 20 000) were kind gifts of Drs M. Zeviani and R. Wiesner, respectively. Rabbit anti-ATAD3 (1 : 50 000), anti-NIPSNAP1 (1 : 1000) anti-CRIF1 (1 : 2000), anti-ATPase β-subunit 1 : 2000, anti-POLG2 (1 : 200) and anti-NDUFS1 (1 : 10 000) were raised against recombinant proteins produced in-house. Secondary antibodies were anti-rabbit or anti-mouse HRP 1 : 1000 (Promega).
Mitochondrial translation
The translation of proteins encoded in mtDNA was assayed by 35S-methionine labelling for 30 min, in the presence of emetine, after which total cellular protein was fractionated by SDS–PAGE, as described before (28). The proteins were fixed for 30 min in 7% acetic acid, 20% methanol, dried and exposed to phosphorimager plates. Radiolabelled proteins were quantified with a Typhoon™ phosphorimager (GE Healthcare).
Confocal microscopy
FLAG-tagged C4orf14 was detected in transgenic HEK293T cells with IgG mouse monoclonal anti-FLAG antibody M2 (1 : 1000, Sigma) and goat anti-mouse-Alex-Fluro®488 (1 : 1000, Invitrogen); TFAM was detected with rabbit polyclonal anti-TFAM antibody (1 : 400) and goat anti-rabbit Alex-Fluro®647 (1 : 1000, Invitrogen). Antibodies were diluted in phosphate buffered saline in 5% serum. Slides were mounted using ProLong Gold antifade with DAPI (4′,6-diamidino-2-phenylindole, Invitrogen). Cells were imaged with a Zeiss LSM 510 Meta confocal microscope used in conjunction with LSM 510 software. Images were acquired with a Zeiss 63×/1.40 oil immersion objective, set at zoom 2.4. MitoTracker orange signal was pseudocoloured red to improve contrast. Images were edited in Photoshop Elements (Adobe).
Isokinetic sucrose-gradient analysis of mitochondrial ribosomes
Mitochondria were isolated from control HEK293T cells and cells induced to express recombinant C4orf14 with 20 ng/ml doxycycline for 24 h. After, isolation of mitochondria from the interface of a 1/1.5 M sucrose-gradient, 10 mg of mitochondria were lysed with 1% n-dodecyl-β-d-maltoside, 20 mM HEPES–KOH pH 7.6, 100 mM KCl, 1/25 v/v inhibitor, 0.4 mM PMSF, 2 mM DTT, 20 mM MgAc2, centrifuged at 1000gmax, 10 min and the 0.5 ml supernatant was loaded onto a 10–30% (v/v) linear sucrose gradient in buffer (20 mM HEPES–KOH pH 7.6, 100 mM NaCl, 1/50 v/v inhibitor, 0.2 mM PMSF, 2 mM DTT, 20 mM MgAc2) containing 0.05% n-dodecyl-β-d-maltoside and 50 µg/ml chloramphenicol, and centrifuged for 16 h at 100 000gmax, 4°C. Fractions (0.5 ml) were collected, and the distribution of ribosomal subunits through the gradient was assessed by the detection of MRPS29 and MRPL11 and other proteins by immunoblotting.
Iodixanol gradient analysis of mitochondrial lysates
Iodixanol gradient fractionation was as previously described (10). Briefly, sucrose gradient purified HEK293T cell mitochondria were lysed with 0.4% n-dodecyl-β-d-maltoside, and a 1000gmax supernatant loaded on a 20–42.5% iodixanol gradient and centrifuged at 100 000gmax for 14 h. Fractions of 0.5 ml were collected from the base of the tube, and nucleic acids and proteins from the various fractions were resolved by 1% agarose and 4–12% polyacrylamide gel electrophoresis, respectively.
Protein expression and purification
The C4orf14 cDNA was purchased from Cambridge Bioscience (clone # 3504209), and a DNA fragment corresponding to residues 63–698 of the protein was amplified and cloned into pMAL-c2 (NEB), and transfected into shuffle T7 cells. Expression in exponentially growing cells was induced with 200 μM isopropyl 1-thio-β-d-galactopyranoside. After 18 h at 30°C, cells were collected and stored at −80°C. Amylose column affinity purification was performed according to the manufacturer’s instructions (NEB), and fractions of interest were pooled, concentrated and applied to a size exclusion chromatography column (Superose 6, GE Health care) in 50 mM Tris–HCl, pH 7.5, 200 mM NaCl, 10% glycerol, and 5 mM dithiothreitol, to purify further the proteins. Protein concentrations were evaluated using DC protein assay reagent (Bio-Rad). The protein’s identity was confirmed by mass spectrometry (MS) analysis. Rat TFAM (NM_031326.1) was C-terminally FLAG tagged and purified from Escherichia coli carrying the pLysS-Rare plasmid (Novagen). The soluble fraction of E. coli cell lysate was mixed with anti-FLAG M2 affinity gel (Sigma, A2220) and the recombinant TFAM eluted by competition with FLAG peptide (Sigma, F3290), according to the manufacturer’s instructions.
DNA and RNA binding assays
For DNA–protein gel retardation analysis, reaction mixtures of 10 µl, containing 50 mM Tris (pH 7.4), 0.2 mg/ml bovine serum albumin, 2 mM dithiothreitol, 32P-dCTP labelled DNA (∼170-bp of pUC19), and 1 µl protein were incubated at 37°C for 20 min. After the incubation, 2 µl of loading buffer (50% glycerol, 0.1% bromophenol, 0.1% xylene-cyanol) was added and after a further 20 min on ice, 8 µl of each sample was loaded onto a 1 × tris-borate-EDTA (TBE), 4% polyacrylamide gel (PAG). The gel was electrophoresed at 100 V for 2 h at 4°C, dried and exposed to a phosphorimager screen overnight. For RNA–protein gel retardation analysis, Trizol™-extracted and DNase-treated RNA from purified mitochondria of HEK293T cells was end-labelled with γ32P-ATP and mixed with different amounts of maltose-binding protein (MBP), or C4orf14-MBP, in reaction volumes of 10 μl, at 37°C for 20 min. The products of the reaction were separated on 1% TBE agarose gels by electrophoresis at 120 V for 3 h at 4°C. C4orf14-MBP binding to mtRNA was tested by mixing 60 µg protein with 1.6 µg mtRNA. After immobilizing C4orf14-MBP on 100 µl amylose beads (New England Biolab) and washing five times with 1 ml 150 mM NaCl, the captured RNA was released by boiling, end-labelled and used to probe membranes containing the equal amounts of amplified mtDNA. The PCR products were derived from the following primers: COX2, forward 5′-TATTCCTAGAACCAGGCGACCTGC-3′, reverse 5′-TTTCGTTCATTTTGGTTCTCAGGGTTTG-3′; ND1, forward 5′-CAT GGCCAACCTCCTACTCCTCATTG-3′, reverse 5′-GGCAGGAGTAATCAGAGG TGTTCTTG-3′; 12S rRNA, forward 5′-GCATCCCCGTTCCAGTGAGTTC-3′, reverse 5′-GTTAAATGTCCTTTGAAGTATACTTG-3′; 16S rRNA, forward 5′-AAATAGTGGGAAGATTTATAGGTAG-3′, reverse 5′-AGGATTGCGCTGTTATC CCTAG-3′.
RESULTS
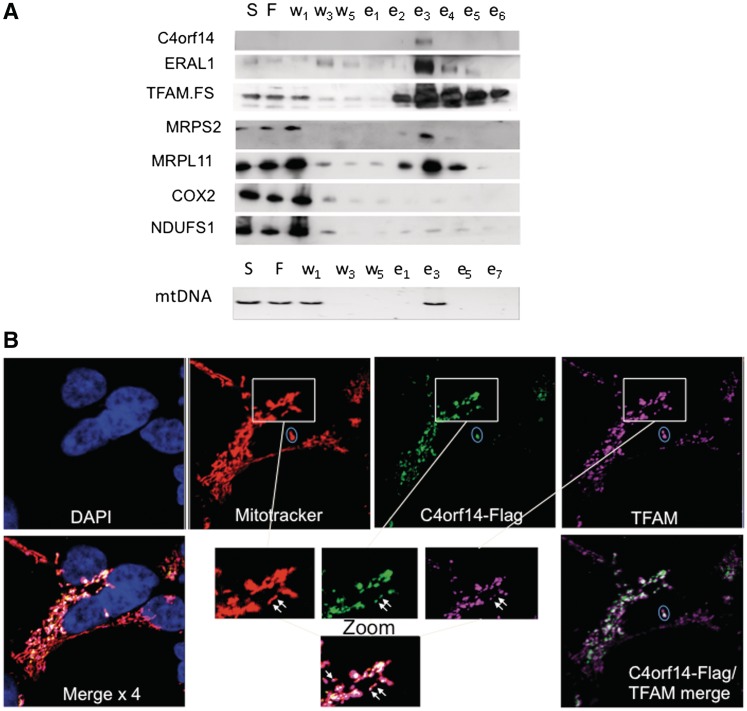
C4orf14 (NOA1) was identified in one preparation of TFAM affinity-purified nucleoids by mass spectrometry, together with many proteins involved in mitochondrial translation (19). Its prokaryotic homologue, YqeH, has been implicated in the biogenesis of the bacterial small ribosomal subunit, and in DNA replication (20,21,23), and so we speculated that C4orf14 might play dual roles in protein synthesis and DNA metabolism in mitochondria. First, we confirmed that C4orf14 was enriched in preparations of affinity-purified recombinant TFAM isolated from human embryonic kidney (HEK293T) cells, via immunodetection (Figure 1A). Moreover, mitochondrial ribosomal proteins and ancillary factors were also captured with recombinant TFAM, whereas highly abundant components of respiratory chain complexes were depleted almost to extinction during the purification process (Figure 1A).
Figure 1.
C4orf14 co-purifies with FLAG-StrepII-tagged TFAM and C4orf14.FLAG.StrepII is targeted to mitochondria in HEK293T cells. (A) Affinity purified TFAM.FLAG.StrepII protein was isolated from mitochondria fractions of HEK293T cells. Proteins from various stages of the purification procedure were analysed by immunoblotting, after separation via 4–12% SDS–PAGE. S, supernatant; F, flow-through; w, washes; e, eluted fractions. PvuII digested mtDNA was detected by Southern hybridization. (B) Immunocytochemistry of C4orf14. FLAG.StrepII expressing HEK293T cells with an anti-FLAG antibody (green) 24 h after transgene induction. Additionally, TFAM was labelled with an antibody (false-colour violet), mitochondria were stained with MitoTracker (false-colour red) and the nucleus was stained with DAPI (blue). Bottom right: merged image of C4orf14 and TFAM. The 4 × merged image (bottom left) is C4orf14, TFAM, DAPI and MitoTracker. White arrows in the enlarged images (bottom centre) indicate foci where TFAM and C4orf14 coincide within the mitochondrial network. The ellipses each enclose two TFAM labeled foci, one of which coincides with C4orf14.
C4orf14 physically and functionally interacts with the small mitochondrial ribosomal subunit
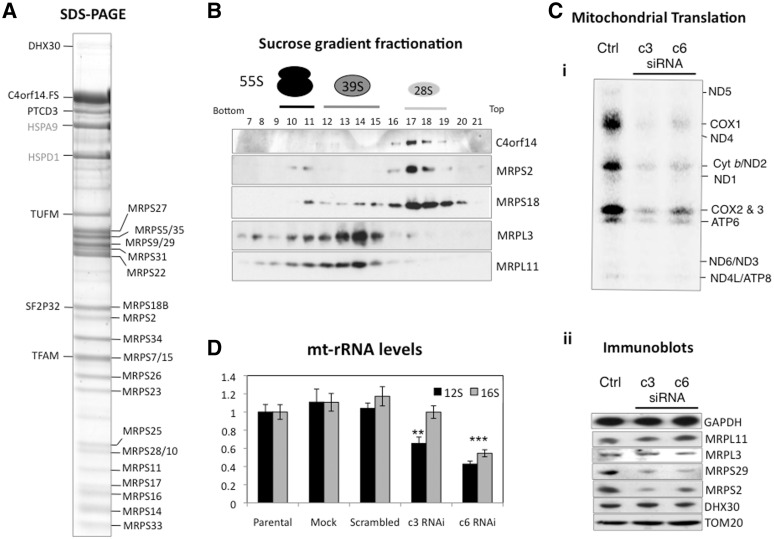
In order to investigate C4orf14 in further detail, a human cDNA of C4orf14 with FLAG and StrepII tags was cloned and expressed in HEK293T Flp-In T-Rex cells. Exclusive targeting to mitochondria of C4orf14.FLAG.StrepII protein was demonstrated by immunocytochemistry and its distribution in mitochondria was found to be similar, albeit not identical, to that of TFAM (Figure 1B). In an effort to identify the binding partners of C4orf14, three TAP experiments were performed using C4orf14 as the bait, and the bound proteins were identified by MALD-TOF-TOF MS. A substantial majority of the proteins identified in each preparation were protein components of the 28S mitochondrial ribosomal subunit [Figure 2A and Supplementary Table S1 (2,29,30)]. In marked contrast, MS identified no component of the large 39S subunit. The absence of components of the 39S subunit co-purifying with C4orf14 strongly suggests C4orf14 interacts specifically with the small mitochondrial ribosomal subunit (directly, or indirectly).
Figure 2.
Association of C4orf14 with the mitochondrial small ribosomal subunit. (A) Affinity purified C4orf14.FLAG.StrepII protein was isolated from mitochondria fractions of transgenic HEK293T cells and the concentrated eluted fractions resolved by SDS–PAGE. Proteins identified by MS are indicated on the left and right of the gel. They included 22 polypeptides of the 28S subunit (MRPS; see Supplementary Table S1); (B) sucrose-gradient purified mitochondria from HEK293T cells were lysed and fractionated on sucrose gradients. Antibodies to MRPS2 and MRPS18, and MRPL3 and MRPL11 were used as markers of the 28S and 39S subunits, respectively. (C) 143B cells were transfected with dsRNA (c3 or c6) targeting C4orf14 mRNA and the effects on mitochondrial protein synthesis (i); selected proteins in mitochondria (ii) and mitochondrial ribosomal RNAs (panel D and Supplementary Figure S2) were examined 72 h later. GAPDH: glyceraldehyde-3-phosphate dehydrogenase, the outer mitochondrial membrane protein TOM20, a putative mitochondrial RNA helicase DHX30 and components of the 55S ribosome (MRPS2, MRPS29, MRPL3 and MRPL11).
As a more sensitive and less error prone test of C4orf14’s major protein partners, SILAC coupled to Orbitrap liquid chromatography (LC)/MS/MS analysis (31) was performed. Heavy labelled proteins purified on a streptactin column from mitochondria of HEK293T cells expressing C4orf14.FLAG.StrepII tagged protein, were mixed with an equal amount of material isolated by the same procedure from control HEK293T cells grown in medium without heavy isotopes. Affinity purified proteins were considered to be associated with the bait protein when they were present in a 2-fold, or greater, excess over corresponding proteins in control mitochondria, in both of the experiments performed. Forty-five proteins fulfilled these criteria, and the list of candidate partners of C4orf14 was dominated by proteins of the mitochondrial small ribosomal subunit (Table 1). Several of the other proteins have also been linked to mitochondrial protein synthesis, or mtRNA metabolism (see ‘Discussion’ section).
Table 1.
Protein interactions with C4orf14 identified by Orbitrap LC/MS/MS analysis and quantified by SILAC labelling
| Protein names | Gene name(s) | Isotope ratios |
|
|---|---|---|---|
| Experiment 1 | Experiment 2 | ||
| Uncharacterized protein C4orf14 | C4orf14 | 33.3 | 30.1 |
| 28S ribosomal protein S22, mitochondrial | MRPS22 | 30.8 | 30.0 |
| 28S ribosomal protein S27, mitochondrial | MRPS27 | 28.3 | 27.0 |
| 28S ribosomal protein S28, mitochondrial | MRPS28; MRPS35 | 27.5 | 26.4 |
| 28S ribosomal protein S17, mitochondrial | MRPS17 | 27.1 | 30.1 |
| 28S ribosomal protein S23, mitochondrial | MRPS23 | 26.0 | 25.8 |
| 28S ribosomal protein S15, mitochondrial | MRPS15 | 24.4 | 15.7 |
| 28S ribosomal protein S2, mitochondrial | MRPS2 | 24.4 | 36.3 |
| 28S ribosomal protein S29, mitochondrial | MRPS29 | 23.9 | 20.9 |
| 28S ribosomal protein S31, mitochondrial | MRPS31 | 23.5 | 23.36 |
| 28S ribosomal protein S7, mitochondrial | MRPS7 | 23.1 | 13.5 |
| 28S ribosomal protein S10, mitochondrial | MRPS10 | 23.0 | 24.9 |
| Pentatricopeptide repeat-containing protein 3, mitochondrial | PTCD3 | 22.2 | 24.0 |
| 28S ribosomal protein S5, mitochondrial | MRPS5 | 21.4 | 24.4 |
| 28S ribosomal protein S6, mitochondrial | MRPS6 | 21.0 | 17.1 |
| 28S ribosomal protein S34, mitochondrial | MRPS34 | 20.9 | 14.9 |
| 28S ribosomal protein S11, mitochondrial | MRPS11 | 19.8 | 32.4 |
| 28S ribosomal protein S9, mitochondrial | MRPS9 | 19.7 | 21.8 |
| 28S ribosomal protein S13, mitochondrial | MRPS26 | 19.5 | 9.23 |
| 28S ribosomal protein S25, mitochondrial | MRPS25 | 19.3 | 20.5 |
| 28S ribosomal protein S14, mitochondrial | MRPS14 | 17.1 | 21.9 |
| 28S ribosomal protein S21, mitochondrial | MRPS21 | 17.0 | 22.8 |
| Malonyl-CoA-acyl carrier protein transacylase, mitochondrial | MCAT | 16.7 | 9.17 |
| 28S ribosomal protein S18-2, mitochondrial | MRPS18B | 16.2 | 9.63 |
| Conserved ERA-like GTPase | ERAL1 | 14.9 | 3.94 |
| 130 kDa leucine-rich protein;GP130; leucine-rich PPR motif-containing protein, mitochondrial | LRP130;LRPPRC | 13.8 | 5.18 |
| 28S ribosomal protein S24, mitochondrial | MRPS24 | 12.6 | 17.2 |
| Methyltransferase 11 domain-containing protein 1; protein RSM22 homolog, mitochondrial | METT11D1 | 11.4 | 7.38 |
| Coiled–coil-helix-coiled–coil-helix domain-containing protein 1 | CHCHD1 | 11.2 | 6.85 |
| Complement component 1 Q subcomponent-binding protein, mitochondrial | C1QBP | 10.7 | 5.54 |
| Pentatricopeptide repeat-containing protein 1 | PTCD1 | 10.7 | 2.86 |
| Probable serine carboxypeptidase CPVL | CPVL;PSEC0124 | 10.3 | 9.08 |
| RNA methyltransferase-like protein 1 | RNMTL1 | 10.1 | 2.12 |
| 28S ribosomal protein S33, mitochondrial | MRPS33 | 9.34 | 19.4 |
| Mitochondrial ATP-dependent protease Lon | LONP1 | 9.30 | 6.38 |
| 39S ribosomal protein L12, mitochondrial | MRPL12 | 9.29 | 2.65 |
| Mitochondrial 12S rRNA dimethylase 1 | TFB1M | 9.22 | 5.44 |
| DNA-directed RNA polymerase, mitochondrial | POLRMT | 6.73 | 2.66 |
| Mitochondrial transcription factor 1 | TFAM | 4.12 | 3.72 |
| 3-hydroxy-2-methylbutyryl-CoA dehydrogenase | MRPP2;HADH2 | 2.76 | 2.79 |
| Leucine-rich repeat-containing protein 59 | LRRC59 | 2.70 | 5.18 |
| Pyrroline-5-carboxylate reductase 2 | PYCR2 | 2.65 | 2.74 |
| Aldehyde dehydrogenase family 18 member A1 | ALDH18A1 | 2.14 | 3.15 |
| Caseinolytic peptidase B protein homologue | CLPB | 2.09 | 2.60 |
| Dihydrolipoamide succinyltransferase component of 2-oxoglutarate dehydrogenase complex | DLST | 2.02 | 4.82 |
C4orf14.FLAG.StrepII protein was induced with 2 ng/ml doxycyline for 24 h, and the protein affinity purified from isolated mitochondria, prior to MS analysis. The bait protein (C4orf14) and putative partners returned a ratio of ≥2 : 1 over control proteins. The data are derived from the MaxQuant outputs from duplicate affinity purifications performed with SILAC labels and features proteins with test (T) to reference (R) ratios of >2.0, where the test was mitochondrial lysates of HEK cells expressing tagged C4orf14 and the reference was mitochondrial lysates of control HEK cells without the transgene (see ‘Materials and methods’ section and main text for details). The proteins identified are listed in decreasing ratios over proteins in control samples in the first experiment. The ratios have been corrected to three significant figures and the names (gene and protein) for each entry have been simplified, many synonyms were excluded for clarity.
Centrifugation on sucrose gradients is widely used to fractionate mitochondrial and other ribosomal subunits. Analysis of the distribution of C4orf14 on such gradients indicated that the endogenous protein co-fractionates with the components of the small mitochondrial ribosomal subunit (Figure 2B); thereby corroborating the affinity capture data for recombinant C4orf14 (Table 1 and Supplementary Table S1 and Figure 2A). The recombinant protein also resolved on sucrose gradients in the same region as the 28S subunit (Supplementary Figure S1). Next, gene silencing with either one of two dsRNAs (c3 and c6) targeting C4orf14 was performed on human osteosarcoma cells, to determine the effects on mitochondrial translation and mitochondrial ribosomes. Both dsRNAs greatly reduced the expression of C4orf14 (Supplementary Figure S2A), and this was associated with a marked decrease in mitochondrial protein synthesis [Figure 2C (i)], confirming an earlier report that demonstrated decreased mitochondrial translation in a C4orf14 (NOA1) knockout mouse (24). In addition, we found that RNAi of C4orf14 decreased the steady-state levels of two components of the 28S subunit (MRPS2 and MRPS29), but not the 39S subunit components MRPL3 and MRPL11 [Figure 2C (ii)], and one of the two dsRNAs (c3) specifically decreased the expression of 12S rRNA (Figure 2D and Supplementary Figure S2B). The other dsRNA, c6, induced a decrease in the steady-state level of all five mitochondrial transcripts tested (Figure 2D and Supplementary Figure S2B), which may be well due to the off-target effects of the dsRNA. Whatever the cause of the broader effects of dsRNA c6 compared to c3, they did not exacerbate the defect in protein synthesis or the decrease in the levels of 28S subunit components that were equally marked for the two dsRNAs (Figure 2C). Thus, several lines of evidence suggest that C4orf14 physically and functionally interacts with the mitochondrial small ribosomal subunit (Table 1 and Supplementary Table S1, Figure 2A–D), rather than the 39S subunit as was previously suggested (24). Moreover, our reassignment of C4orf14 as a 28S subunit interacting protein is consistent with earlier studies that linked C4orf14’s prokaryote homologue (YqeH) to biogenesis of, the small ribosomal subunit (22).
C4orf14 interacts with the small mitochondrial ribosomal subunit via a GTP-binding mechanism
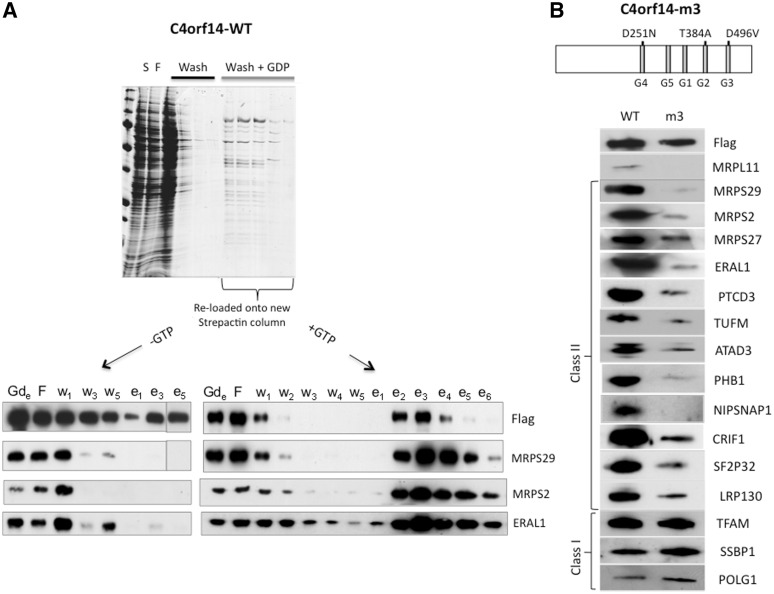
Because C4orf14 has a predicted GTPase domain and GTPases play multiple roles in ribosomal biogenesis in other systems (1), GTP was deemed likely to play a critical role in the interaction of C4orf14 with components of the small mitochondrial ribosomal subunit. Therefore, the purification procedure for tagged C4orf14 was modified by adding GDP to the wash buffer, as a competitor of GTP. This resulted in the release of a considerable amount of the mitochondrial small ribosomal subunit from the streptactin column (Figure 3A). It is inferred that an excess of GDP leads to many copies of C4orf14 binding to the nucleotide diphosphate in place of GTP, inducing a conformational change, which results in the release of the 28S subunit. The fact that GDP also released some tagged C4orf14 suggests that multiple copies of the protein form a complex with the 28S subunit. The tagged protein together with components of the 28S subunit and ERAL1 (a 28S subunit assembly factor) (17,32) were re-captured on a fresh column when the eluted material was first dialysed and then supplemented with GTP, whereas in the absence of GTP tagged-C4orf14 was retained without 28S subunit components, or ERAL1 (Figure 3A). These results indicate that C4orf14’s interaction with the 28S subunit, and ancillary factors, is reversible and dependent on GTP.
Figure 3.
Dependence of the interaction of C4orf14 with the 28S ribosomal subunit on its GTPase being functional and GTP. (A) A strepactin column with bound recombinant C4orf14 was washed first with 150 mM NaCl (black bar), and then with buffer containing 20 mM GDP pH 7.6 (grey bar) (see ‘Materials and methods’ section). Proteins from the different stages of the purification procedure were stained with Coomassie blue after fractionation by SDS–PAGE (upper gel image). After dialysis, the ‘GDP-eluted’ material (Gde) was incubated with (+GTP) or without (−GTP) and then re-loaded on new strepactin columns; thereafter the standard washes and desthiobiotin elution procedure were applied, and proteins from various steps of the procedure were analysed by immunoblotting to detect recombinant C4orf14 (anti-FLAG antibody), the 28S subunit (MRPS2 and MRSP29) and the 28S subunit assembly factor ERAL1. (B) The upper part of the panel shows a schematic representation of C4orf14 indicating the positions of the three mutated residues in the m3 mutant version of the transgene. G1–G5 are sequence motifs that are characteristic of circularly permuted GTPases. The lower portion of the panel comprises a series of immunoblots of eluted fraction 3 (see Figure 4C), indicating the relative abundance of established and candidate nucleoid proteins isolated with the wild-type (WT) and m3 versions of C4orf14. Of the proteins tested, the m3 version of C4orf14 co-purified exclusively with established mtDNA-binding proteins (class I); many of the other (class II) nucleoid proteins are known to contribute to mitochondrial protein synthesis (see ‘Discussion’ section).
Three residues are predicted to be critical for C4orf14’s GTPase activity (25), and their mutation abolishes the GTPase activity of the plant homologue of C4orf14 (26). The introduction of the mutations D251N, T384A and D496V in the G4, G2 and G3 motifs, respectively (25), led to the mutant form of C4orf14, known as m3, which was no longer co-purified with the 28S subunit, or other predicted or known mitochondrial translation factors (Figure 3B and Supplementary Figure S3); nor did it co-fractionate with the 28S subunit on sucrose gradients, unlike the endogenous protein and recombinant GTPase active C4orf14 (Supplementary Figure S4). Collectively, these data suggest that C4orf14’s interaction with the small mitochondrial ribosomal subunit (and/or its associates) is dependent on GTP, and that GTP hydrolysis is the mechanism by which the 28S subunit is released by C4orf14, presumably in a process that leads to assembly of the monosome. Logically, GTP hydrolysis and 28S subunit release by C4orf14 must precede assembly of the 55S monosome, as no 39S subunit co-purified with recombinant C4orf14, whereas purification of ATAD3 via the same procedure yielded many components of both the 28S and the 39S subunits, without C4orf14 (19). Thus, there is nothing to suggest the purification procedure disrupts the association between small and large subunits of the mitochondrial ribosome. The absence of any hint of 39S subunit in our preparations of C4orf14 (MRPL12 aside, see ‘Discussion’ section) strongly suggests the protein does not participate in protein synthesis, but instead is involved in 28S subunit maturation. It might additionally serve to prevent premature translation initiation (1).
C4orf14 is associated with mtDNA and contributes to its maintenance
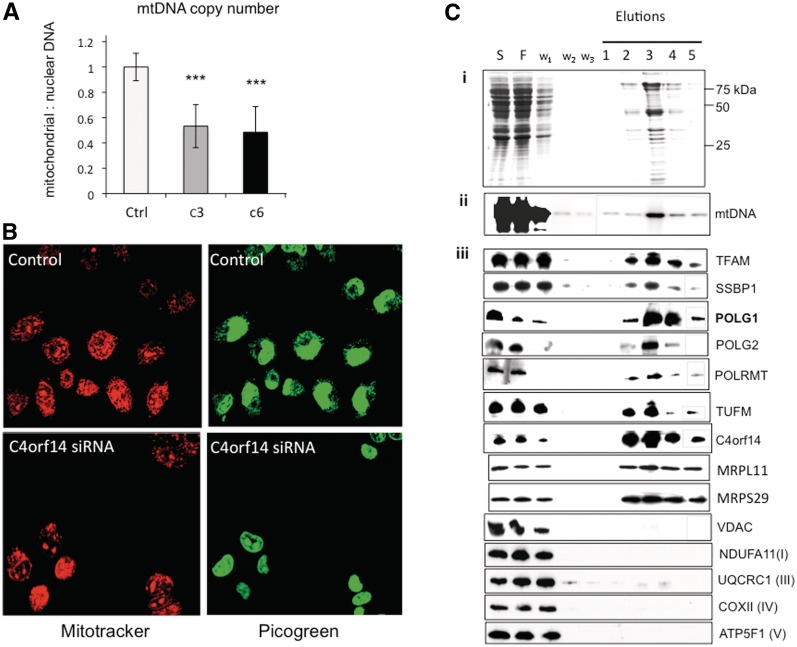
YqeH, the bacterial homologue of C4orf14, has been implicated in DNA replication (23), and C4orf14 co-purified with recombinant tagged TFAM (Figure 1A). Therefore, we analysed the effects of C4orf14 gene silencing on picogreen staining of mtDNA and copy number. Treating osteosarcoma cells for 72 h with either one of two siRNAs targeting C4orf14 (c3 or c6) halved mtDNA copy number, based on Southern hybridization and qPCR (Figure 4A). Both methods recorded a significant decrease in mtDNA copy number as a result of C4orf14 gene silencing (P < 0.05, P < 0.01, unpaired student’s t-test, respectively), with a combined P < 0.001 for both dsRNAs.
Figure 4.
C4orf14 gene silencing decreases mtDNA copy number and PicoGreen staining of mitochondrial nucleoids; and affinity purification of tagged C4orf14 co-purifies mtDNA-binding proteins. (A) Combined Southern hybridization and qPCR estimates of mtDNA copy number, from 143B cells exposed for 72 h to dsRNAs c3 or c6, compared to mock transfected cells (n = 6 independent experiments, three qPCR and three Southern hybridizations). (B) mitochondrial nucleoids (and the nucleus) were stained with picogreen, and the mitochondrial network was stained with MitoTracker orange (pseudo-coloured red for contrast) in mock-transfected 143B osteosarcoma cells, and cells transfected with dsRNAs targeting C4orf14. Cell images were captured 72 h after transfection with a Zeiss LSM510 confocal microscope. (C) mtDNA and mitochondrial nucleoid proteins co-purified with affinity captured C4orf14.FLAG.StrepII. (i) SDS–PAGE showing Coomassie stained proteins from various stages of the affinity purification process: S, 1000gmax supernatant after mitochondrial lysis; F, flow-through; w, wash. (ii) Southern blotting of PvuII digested DNA from various stages of the affinity purification process. (iii) Immunoblotting of specific mitochondrial proteins from various stages of the affinity purification process. Some samples (gray-lined) were fractionated on a separate gel, simultaneously and under identical conditions. NDUFA11, a subunit of respiratory complex I; UQCRC1, a subunit of respiratory complex III; COXII, a subunit of respiratory complex IV; ATP5F1, a subunit of respiratory complex V; VDAC, voltage-dependent anion channel of the outer mitochondrial membrane; other proteins are described in the text.
RNA interference of C4orf14 also inhibited picogreen staining of mtDNA (Figure 4B) that is attributable to perturbed mtDNA topology [Supplementary Figure S5 and (9)]. In addition to the 28S subunit, affinity purification of tagged C4orf14 yielded TFAM (Figure 2A and Table 1) and some mtDNA (Figure 4C). Although some well-established mtDNA-binding proteins were not identified by MS, they did co-purify with tagged C4orf14, based on immunoblotting (Figure 4C). In particular, mtDNA polymerase γ was considerably enriched in preparations of affinity purified C4orf14, whereas highly abundant proteins of the outer and inner mitochondrial membrane were markedly depleted during the affinity purification process (Figure 4C). And, as detailed above, endogenous C4orf14 was enriched when TFAM was the bait protein (Figure 1A), suggesting it is bound, directly or indirectly, to many mitochondrial nucleoids. C4orf14’s GTPase activity is not required for its interaction with mitochondrial nucleoids, as the m3 variant co-purified mitochondrial nucleoid proteins, POLG1, TFAM and SSBP1 (Figure 3B), as well as mtDNA itself (Supplementary Figure S6), nor is it linked to nucleoids via the translation apparatus (Figure 3B).
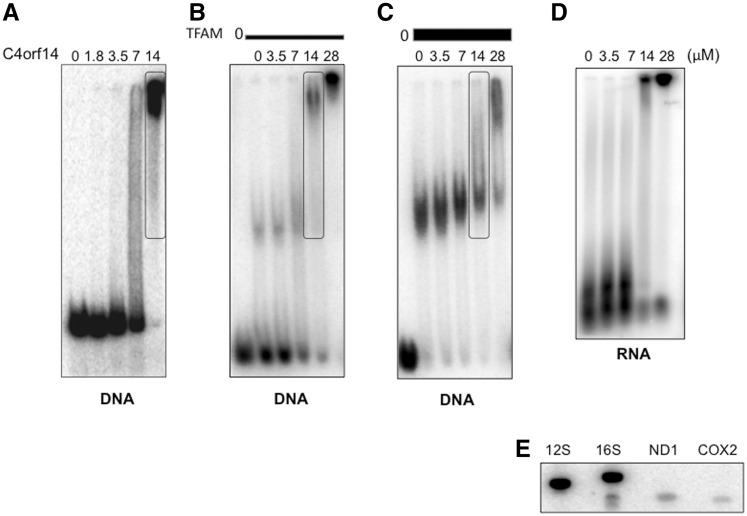
To determine if C4orf14 binds to nucleic acids, a recombinant form of the protein was purified from E. coli (see ‘Materials and methods’ section). Incubation with DNA produced a substantial electrophoretic mobility shift (Figure 5A), indicating that C4orf14 is capable of interacting directly with mtDNA in vitro. Nevertheless, protein–protein interactions could also contribute to C4orf14’s association with mitochondrial nucleoids. However, this does not appear to be the case for TFAM, as it inhibited, rather than enhanced, C4orf14’s interaction with DNA, in vitro (Figure 5A–C). Recombinant C4orf14 also bound to RNA, in vitro, based on electrophoretic mobility shift assays (Figure 5D). To determine if C4orf14 had any preference for 12S rRNA the protein was also incubated with purified mtRNA, and the captured RNA was used to probe fragments of mtDNA; it labelled fragments of mtDNA corresponding to the 12S and 16S genes to similar extents (Figure 5E), suggesting that it is a non-specific RNA-binding protein, and so its interaction with the 28S subunit most probably depends additionally on protein–protein interactions.
Figure 5.
Recombinant C4orf14 binds to DNA and to RNA non-specifically. (A–D) Gel-retardation assays using 53 nM radiolabelled 170-bp DNA incubated with different amounts of recombinant C4orf14 as indicated, without (A), or pre-mixed with 2.8 -µM TFAM (B) or 5.6 µM TFAM (C) for 20 min at 37°C. The boxed lanes each contained the same amount of C4orf14 (14 µM). (D) Radiolabelled mtRNA was incubated without protein or with 3.5, 7, 14 or 28 µM C4orf14-MBP for 20 min at 37°C, prior to separation by 1D-AGE. (E) C4orf14-MBP captured mtRNA (see ‘Materials and methods’ section) was end-labelled and incubated with a Southern blot of amplified fragments of mtDNA corresponding to 12S and 16S rRNAs, cytochrome c oxidase subunit II (COX2) and NADH dehydrogenase subunit 1 (ND1). Ribosomal RNAs are of much greater abundance than mRNAs, which accounts for the disparity in signal between the two types of gene.
DISCUSSION
Based on this study C4orf14 is involved in 28S subunit biogenesis and mtDNA maintenance. Recombinant and endogenous C4orf14 not only co-purified and co-fractionated with the 28S ribosomal subunit, mtDNA and mtDNA-binding proteins (Table 1 and Supplementary Table S1, Figures 2–4), its absence was associated with depletion of components of the 28S subunit [Figure 2C (ii)], presumably due to an inability to assemble the small ribosomal subunit, and with mtDNA depletion (Figure 4A). It is possible that the interactions of C4orf14 with the 28S subunit and mitochondrial nucleoids are independent of one another; however, assembly of the 28S subunit (and subsequently the 55S ribosome) at the mitochondrial nucleoid would offer a number of advantages: it would provide a means of co-regulating mtDNA copy number and mitochondrial translation (in order to satisfy respiratory demand), and would enable direct transfer of mitochondrial mRNAs from the nucleoid to mitochondrial ribosomes. Direct transfer is implied by the fact that all detectable mRNAs are to be found in complexes with ribosomes or mtDNA (19). Once mRNAs have transferred to the translation machinery it could disengage from the nucleoid, although the accompanying study of ATAD3 and allied proteins suggests ribosomes remain in close proximity to nucleoids after their assembly. A more detailed assessment of the candidate partner proteins of C4orf14 follows, which supports the view that the protein is involved in the assembly of the 28S subunit and cooperates with proteins at the interface of transcription and translation.
The co-affinity purification of recombinant C4orf14 with a set of proteins associated with one particular complex, the 28S subunit, and others allied to mitochondrial protein synthesis, suggests the interactions are highly specific and therefore likely to be legitimate. In the first SILAC experiment, proteins of the mitochondrial 28S subunit occupied 20 of the top 22 positions (Table 1), the other two were PTCD3, which has previously been linked to MRPS (29,30), and the bait protein. The second SILAC experiment was much the same (Table 1). The ability of the recombinant protein to re-capture the 28S subunit only if supplied with GTP, and the absence of appreciable amounts of 28S subunit co-purifying with the GTPase disabled variant of C4orf14 provided further strong evidence of a meaningful interaction between the proteins. Nevertheless it was important to establish links between the endogenous protein and the 28S subunit. Gene silencing of C4orf14 indicated that the 28S subunit is dependent on C4orf14, as it led to depletion of components of the 28S, but not the 39S, subunit, and the endogenous protein co-fractionated with the 28S subunit on sucrose gradients. Thus, there can be little doubt that C4orf14 interacts specifically with the 28S subunit, directly or indirectly. With that in mind the list of candidate partners of C4orf14 warrants close inspection. ERAL1 is involved in assembly of the small mitochondrial ribosomal subunit (17,32), and it is a candidate partner of C4orf14 based on the SILAC analysis (Table 1). Moreover, it was enriched in TFAM affinity capture experiments along with C4orf14 (Figure 1A), and it was captured by GTPase active, but not GTPase disabled, C4orf14 (Figure 3B). Finally, ERAL1’s interaction with C4orf14 is dependent on GTP and disturbed by GDP (Figure 3A). Therefore, the data are consistent with C4orf14 and ERAL1 acting in concert, as part of a complex, to engineer 28S subunit assembly. Although no protein assigned to the 39S subunit was identified by MALDI-TOF-TOF mass spectrometry (Figure 2A and Supplementary Table S1), the more sensitive Orbitrap analysis identified MRPL12 as a putative partner of C4orf14. It is noteworthy that no other MRPL was identified in either of the two long lists of 121 and 115 proteins (data not shown), and only traces of MRPL components could be detected by immunoblotting (Figure 4), and so MRPL12 is the only assigned component of the 39S subunit that potentially interacts with C4orf14. Independent of its place in the mitochondrial ribosome, MRPL12 interacts with POLRMT (it also co-purified with C4orf14, see Table 1), and so MRPL12 might couple transcription and translation (18). Specifically, MRPL12 may be involved in the handover of mRNA from the transcription machinery to the translation apparatus, which initially comprises the 28S subunit and other components of the translation apparatus, but not the 39S subunit. LRP130, another candidate partner of C4orf14 (Table 1), interacts with both mitochondrial RNA and DNA (33–35), and so it also might contribute to the transfer of RNA from the nucleoid to the translation apparatus. The function of TFB1M is to modify the ribosomal RNA of the 28S subunit (12S rRNA), to make it fit for assembly with its protein partners (36), which suggests C4orf14 is involved in the earliest stages of 28S subunit assembly. HADH2 or MRRP2 is a component of the mitochondrial RNase P complex, which has the ability to process the polycistronic mitochondrial transcripts (37), and so this suggests a system in which RNA is processed shortly after synthesis and transferred directly to the translation machinery.
The known functions of the other proteins on the list offer few obvious clues as to potentially meaningful interactions with C4orf14, although given the high purity of the preparations they should be considered as serious candidate partners. None more so than MCAT, the mitochondrial malonyl-CoA-acyl carrier protein transacylase, which was the highest scoring protein, aside from the 28S subunit (Table 1), and so it may well moonlight in mitochondrial translation.
C4orf14 recruits components of the mitochondrial protein synthesis apparatus, in addition to the 28S ribosome subunit
Several factors involved in mitochondrial translation (PTCD3, MRPS2, MRPS27, MRPS29, ERAL1 and TUFM) were found to co-purify with wild-type C4orf14, but not the GTPase disabled (m3) variant (Figure 3B). These data suggest that C4orf14 is required for the recruitment of a variety of components of the mitochondrial translation apparatus, and therefore any protein co-purifying with wild-type C4orf14, and not with GTPase disabled C4orf14, potentially has a role in mitochondrial translation. In our parallel study, we found ATAD3, prohibitin and CRIF1 in enriched mtDNA preparations and showed that they contribute to mitochondrial protein synthesis (19). Affinity capture of ATAD3, prohibitin and CRIF1 was adversely affected by disabling the GTPase of C4orf14, whereas mtDNA and some other nucleoid proteins were affinity captured with both C4orf14 m3 and wild-type proteins (Figure 3B and Supplementary Figure S3). Therefore, the different binding properties of GTPase active and disabled C4orf14 suggest a new classification scheme for mitochondrial nucleoid interacting proteins. Class I nucleoid proteins are those that associate with mtDNA independent of the apparatus of mitochondrial protein synthesis; they include TFAM, SSBP1 and POLG1. Class II nucleoid proteins contribute to mitochondrial protein synthesis and functional C4orf14 is proposed as the means by which they are recruited to mitochondrial nucleoids. Candidate class II mitochondrial nucleoid proteins with established links to mitochondrial protein synthesis are the 28S subunit (based on MRPS29, S27 and S2), ERAL1, PTCD3, TUFM and SF2P32 (Figure 3B). To these can be added, ATAD3, prohibitin and CRIF1, as they fully satisfy the definition of class II nucleoid proteins [Figure 3 and (19)]. Some ATAD3 might instead be recruited to mtDNA directly, possibly via the D-loop (9), in which case it would belong to both classes of nucleoid proteins. NIPSNAP1 is present in enriched preparations of mitochondrial nucleoproteins (9,10) and it behaves like a class II nucleoid protein (Figure 3B) and so it may well play a role in mitochondrial translation. Further more detailed comparisons of proteins associated with GTPase-active and GTPase-disabled C4orf14 can be expected to reveal additional factors in mitochondria that contribute to protein synthesis.
This study implicates C4orf14 in DNA metabolism and protein synthesis in mitochondria. The accompanying article suggests that the same is true for ATAD3, and possibly prohibitin too. Thus, the two studies point to a new category of proteins operating at the interface of DNA maintenance and expression. Such proteins might be pivotal to balancing mtDNA copy number and expression, as it is well known that mtDNA numbers are closely related to energy demand in cells and tissues. An analogous coordination of DNA replication and protein synthesis has been proposed for prokaryotes based on gene organization (38), and so the mitochondrial version may have been inherited from its prokaryote ancestor.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–6.
FUNDING
Medical Research Council (MRC); Biotechnology and Biological Sciences Research Council (BBSRC); European Union; Academy of Finland (to H.M.C.). Funding for open access charge: MRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for the help and encouragement of members of the Mitochondrial Biology Unit, especially Drs Shujing Ding, Juan Fan kindly provided MBP.
REFERENCES
- 1.Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, 3rd, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 3.McKee EE, Ferguson M, Bentley AT, Marks TA. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob. Agents Chemother. 2006;50:2042–2049. doi: 10.1128/AAC.01411-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smits P, Smeitink J, van den Heuvel L. Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J. Biomed. Biotechnol. 2010;2010:737385. doi: 10.1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai YC, Bullard JM, Thompson NL, Spremulli LL. Interaction of mitochondrial elongation factor Tu with aminoacyl-tRNA and elongation factor Ts. J. Biol. Chem. 2000;275:20308–20314. doi: 10.1074/jbc.M001899200. [DOI] [PubMed] [Google Scholar]
- 6.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 7.Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 8.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 9.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes A, He J, Mao CC, Bailey LJ, Di Re M, Sembongi H, Kazak L, Dzionek K, Holmes JB, Cluett TJ, et al. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011;39:5098–5108. doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 12.Bateman JM, Perlman PS, Butow RA. Mutational bisection of the mitochondrial DNA stability and amino acid biosynthetic functions of ilv5p of budding yeast. Genetics. 2002;161:1043–1052. doi: 10.1093/genetics/161.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman BA, Kolesar JE, Perlman PS, Butow RA. A function for the mitochondrial chaperonin Hsp60 in the structure and transmission of mitochondrial DNA nucleoids in Saccharomyces cerevisiae. J. Cell Biol. 2003;163:457–461. doi: 10.1083/jcb.200306132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufman BA, Newman SM, Hallberg RL, Slaughter CA, Perlman PS, Butow RA. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc. Natl Acad. Sci. USA. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, Kühl I, Gaisne M, Bonnefoy N, Smeitink JA, et al. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res. 2008;36:5787–5799. doi: 10.1093/nar/gkn576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchiumi T, Ohgaki K, Yagi M, Aoki Y, Sakai A, Matsumoto S, Kang D. ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res. 2010;38:5554–5568. doi: 10.1093/nar/gkq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Cotney J, Shadel GS. Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J. Biol. Chem. 2007;282:12610–12618. doi: 10.1074/jbc.M700461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, et al. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 2012;40:6109–6121. doi: 10.1093/nar/gks266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anand B, Surana P, Bhogaraju S, Pahari S, Prakash B. Circularly permuted GTPase YqeH binds 30S ribosomal subunit: Implications for its role in ribosome assembly. Biochem. Biophys. Res. Commun. 2009;386:602–606. doi: 10.1016/j.bbrc.2009.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loh PC, Morimoto T, Matsuo Y, Oshima T, Ogasawara N. The GTP-binding protein YqeH participates in biogenesis of the 30S ribosome subunit in Bacillus subtilis. Genes. Genet. Syst. 2007;82:281–289. doi: 10.1266/ggs.82.281. [DOI] [PubMed] [Google Scholar]
- 22.Uicker WC, Schaefer L, Koenigsknecht M, Britton RA. The essential GTPase YqeH is required for proper ribosome assembly in Bacillus subtilis. J. Bacteriol. 2007;189:2926–2929. doi: 10.1128/JB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto T, Loh PC, Hirai T, Asai K, Kobayashi K, Moriya S, Ogasawara N. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology. 2002;148:3539–3552. doi: 10.1099/00221287-148-11-3539. [DOI] [PubMed] [Google Scholar]
- 24.Kolanczyk M, Pech M, Zemojtel T, Yamamoto H, Mikula I, Calvaruso MA, van den Brand M, Richter R, Fischer B, Ritz A, et al. NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol. Biol. Cell. 2011;22:1–11. doi: 10.1091/mbc.E10-07-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudhamsu J, Lee GI, Klessig DF, Crane BR. The structure of YqeH. An AtNOS1/AtNOA1 ortholog that couples GTP hydrolysis to molecular recognition. J. Biol. Chem. 2008;283:32968–32976. doi: 10.1074/jbc.M804837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreau M, Lee GI, Wang Y, Crane BR, Klessig DF. AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 2008;283:32957–32967. doi: 10.1074/jbc.M804838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 28.Dunbar DR, Moonie PA, Zeviani M, Holt IJ. Complex I deficiency is associated with 3243G:C mitochondrial DNA in osteosarcoma cell cybrids. Hum. Mol. Genet. 1996;5:123–129. doi: 10.1093/hmg/5.1.123. [DOI] [PubMed] [Google Scholar]
- 29.Davies SM, Rackham O, Shearwood AM, Hamilton KL, Narsai R, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 2009;583:1853–1858. doi: 10.1016/j.febslet.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 30.Koc EC, Spremulli LL. RNA-binding proteins of mammalian mitochondria. Mitochondrion. 2003;2:277–291. doi: 10.1016/S1567-7249(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 31.Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dennerlein S, Rozanska A, Wydro M, Chrzanowska-Lightowlers ZM, Lightowlers RN. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem. J. 2010;430:551–558. doi: 10.1042/BJ20100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gohil VM, Nilsson R, Belcher-Timme CA, Luo B, Root DE, Mootha VK. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J. Biol. Chem. 2010;285:13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sondheimer N, Fang JK, Polyak E, Falk MJ, Avadhani NG. Leucine-rich pentatricopeptide-repeat containing protein regulates mitochondrial transcription. Biochemistry. 2010;49:7467–7473. doi: 10.1021/bi1008479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat. Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- 37.Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Berthon J, Fujikane R, Forterre P. When DNA replication and protein synthesis come together. Trends Biochem. Sci. 2009;34:429–434. doi: 10.1016/j.tibs.2009.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.