Abstract
Mitochondrial ribosomes and translation factors co-purify with mitochondrial nucleoids of human cells, based on affinity protein purification of tagged mitochondrial DNA binding proteins. Among the most frequently identified proteins were ATAD3 and prohibitin, which have been identified previously as nucleoid components, using a variety of methods. Both proteins are demonstrated to be required for mitochondrial protein synthesis in human cultured cells, and the major binding partner of ATAD3 is the mitochondrial ribosome. Altered ATAD3 expression also perturbs mtDNA maintenance and replication. These findings suggest an intimate association between nucleoids and the machinery of protein synthesis in mitochondria. ATAD3 and prohibitin are tightly associated with the mitochondrial membranes and so we propose that they support nucleic acid complexes at the inner membrane of the mitochondrion.
INTRODUCTION
Proteins associate with mitochondrial DNA (mtDNA) to form nucleoprotein complexes, known as nucleoids (1). Mitochondrial nucleoid proteins perform a variety of functions, including organizing and protecting mtDNA. When mtDNA is copied or expressed, ancillary factors are recruited to the nucleoid; therefore, the mitochondrial nucleoid is dynamic, it is not a single discrete entity. Consequently, the complete characterization of the protein repertoire of mitochondrial nucleoids will probably require the application of several different approaches. Previously, we isolated nucleoids from lysates of rat liver mitochondria by affinity capture of a generic DNA binding protein, and identified five known or candidate nucleoid proteins (2). They were TFAM, the mitochondrial transcription factor A (3); ATAD3, a protein with displacement loop binding properties, which has also been implicated in processes in mitochondria other than DNA metabolism (4); hydroxyacyl dehydrogenase A; NIPSNAP1, a mitochondrial protein linked to amino acid metabolism, (5) and TUFM, the mitochondrial translation elongation factor (6). More recently, we identified proteins that are more tightly associated with mtDNA than TFAM, they included two cytoskeletal proteins, β-actin and non-muscle myosin IIA that contribute to mtDNA maintenance (7). Enriched mitochondrial nucleoprotein preparations have been isolated independently by immunocapture with antibodies to two known mtDNA binding proteins, SSBP1 or mitochondrial single-stranded DNA binding protein, and TFAM (8) and by chemically cross-linking proteins to human mtDNA (9). While these procedures identified many plausible and established DNA interacting proteins, they also contained probable contaminants, including 24 cytosolic ribosomal proteins in the latter case. In this article, TFAM and SSBP1 were tagged and expressed in human cells and candidate protein partners identified by mass spectrometry, after tandem affinity chromatography. Among the most frequently identified proteins associated with mtDNA were components of mitochondrial ribosomes and other proteins known to be involved in protein synthesis in mitochondria. Three other proteins identified in the mitochondrial nucleoprotein preparations, ATAD3, prohibitin and CRIF1 (GADD45GIP1), were investigated and each was demonstrated to contribute to mitochondrial protein synthesis in proliferating human cells.
MATERIALS AND METHODS
Cell culture
Human osteosarcoma (HOS 143B cells) and human embryonic kidney cells (HEK293T) were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) and 10% fetal bovine serum. In the case of transgenic HEK293T cells (control, TFAM, SSBP1, ATAD3A and ATAD3B), the serum was tetracycline-free and the medium included 15 µg/ml of blastcidin and 100 µg/ml of Zeocin or 100 µg/ml of hygromycin and 15 µg/ml of blastcidin for control and transgene expressing cells, respectively.
Affinity purification of nucleoids
Human cDNA, specifying TFAM, SSBP1, ATAD3B or ATAD3A, was introduced into HEK293T cells (Invitrogen) to establish inducible, transgenic cell lines. Each transgene carried a carboxy-terminal linker sequence (LEGTGGAG), followed by Strep II and FLAG tags. Transgene expression was induced with 2–20 ng/ml doxycycline for 24–48 h. Mitochondria were isolated by a method modified from 1 × 109 HEK293T cells (8). Briefly, cells were disrupted by homogenization in hypotonic buffer (20 mM HEPES pH 8, 5 mM KCl, 1.5 mM MgCl2 and 2 mM DTT), and mixed with a mannitol–sucrose buffer to final concentrations of 210 mM mannitol, 70 mM sucrose, 20 mM HEPES pH 8 and 2 mM EDTA (1 × MSB), prior to purification of mitochondria by differential centrifugation. 2 mg/mL mitochondria were treated with 100 µg/ml DNase I (Sigma) for 30 min at 4°C in 1 × MSB; digestion was terminated by the addition of 12 mM EDTA, and the mitochondria were washed three times with 1 × MSB. The mitochondria were further purified by differential and (1.0 M/1.5 M) sucrose step-gradient centrifugation. Mitochondria recovered from the interface of the sucrose gradient were washed three times with 1 × MSB. In some purifications, sucrose gradient-purified mitochondria were incubated with trypsin (50 µg/ml; BDH) at 30°C for 30 min and then washed four times. Digestion was terminated by resuspending the final pellet in 20 µg/ml bovine pancreatic trypsin inhibitor (Sigma).
Mitochondria lysed in 20 mM HEPES pH 7.6, 5 mM EDTA, 150 mM NaCl, 2 mM DTT, 0.2 mM PMSF, 1v/25v Roche protease inhibitor solution and 0.4% n-dodecyl-β-d-maltoside at a final concentration of 5 mg/ml were rotated for 20 min, centrifuged at 1000gmax for 10 min and the supernatant was loaded onto a 0.25 ml Strep II-tag gravity column (IBA), which was flushed with 5 column volumes (CV) of wash buffer (20 mM HEPES pH 7.6, 1 mM EDTA, 150 mM NaCl, 2 mM DTT, 0.2 mM PMSF, 1v/50v Roche protease inhibitor and 0.05% dodecylmaltoside). Bound protein was eluted with 6 × 0.5 CV elution buffer (wash buffer and 10 mM desthiobiotin). After addition of the third aliquot of elution buffer, the column was capped and incubated overnight at 4°C. For tandem affinity purifications (TAPs), DTT was omitted during the StrepII purification steps, and eluted fractions were pooled and loaded onto a 0.5 ml FLAG tag agarose gravity column (Sigma). The column was flushed with 20 CV of wash buffer, followed by 5 CV of elution buffer (wash buffer plus 100 µg/ml 3 × FLAG peptide, Sigma). Eluted fractions from TAP (Strep II and FLAG purifications) were pooled and layered onto 20% sucrose in 20 mM HEPES–KOH pH 7.6, 1 mM EDTA, 150 mM NaCl, 2 mM DTT, 0.2 mM PMSF, 0.05% dodecylmaltoside and 1v/50v Roche protease inhibitor tablet and centrifuged (100 000gmax, 2 h). The pellet of mitochondrial nucleoprotein was dissolved in 4% SDS, 20 mM Tris–HCl pH 7.4, 1 mM EDTA, 2 mM DTT and stored at −20°C.
Mass spectrometry
Protein samples were fractionated by SDS–PAGE, stained with Commassie blue dye, excised and subjected to in-gel tryptic digestion and then analysed in a MALDI–TOF–TOF mass spectrometer (model 4800; ABI-Sciex), using matrix α-cyano-4-hydroxycinnamic acid. Acquired peptide mass data were compared with sequence databases using MASCOT, and proteins were identified from a significant peptide mass fingerprint (P < 0.05) or from peptide fragmentation data (P < 0.05). Most proteins were identified by both criteria (see Supplementary Table S1).
For SILAC experiments, cells were grown in SILAC medium (Dundee Cell Products) containing 13C6-lysine and 13C6-arginine (K6R6) for six or more passages. Lysed mitochondria from 1 × 108 HEK293T cells that had been induced to express ATAD3B.FLAG.StrepII, with 5 ng/ml of doxycycline for 24 h, were mixed with an equal amount (w/w) of lysed mitochondria from cells grown in 12C-lysine and 12C-arginine. The mitochondrial 1000gmax supernatant was loaded onto a column of StrepII Sepharose (IBA GmBH), as already described. The material eluted with desthiobiotin was precipitated overnight with 20 × ethanol, resuspended in 20 µl of 4% SDS, 20 mM Tris–HCl pH 7.4, 2 mM EDTA containing 10 mM DTT and alkylated in the dark for 30 min with 30 mM iodoacetamide (Sigma). Excess reagent was quenched by the addition of DTT to 40 mM. Samples were fractionated by SDS–PAGE. Bands were excised, digested in-gel with trypsin and analysed by LC/MS/MS using an LTQ OrbiTrap XL mass spectrometer (Thermo), following chromatography on a nanoscale reverse-phase column (75 µm i.d. × 60 mm; Nanoseparations, Nieukoop, The Netherlands) with an acetonitrile gradient in 0.1% (v/v) formic acid, 250 nl/min. Proteins were identified by comparison of both peptide mass and fragmentation data with National Center for Biotechnology Information (NCBI) sequence database using the MASCOT algorithm (10). Relative quantification of peptides containing ‘heavy’ and ‘light’ isotopes was performed with the MAXQUANT suite of algorithms (11).
Cell transfection
For RNAi experiments, 143B osteosarcoma cells were transfected once or twice with lipofectamine 2000 (Invitrogen) and 5 or 10 nM double-stranded (ds) RNA, as previously described (2). After 48, 72 or 144 h, mitochondrial translation assays were performed (see below). Double-stranded RNAs were 5′-UCAAUGAGGAGAAUUUACGGAAGCA-3′ and 5′-UGCUUCCGUAAAUUCUC CUCAUUGA-3′ (ATAD3 dsRNA1) and 5′-UGGUGAGACUGCAUUUUGA CAACUG-3′ and 5′-CAGUUGUCAAAAUGCAGUCUCACCA-3′ (ATAD3. dsRNA2). PHB1 siRNA experiments employed 5′-CACAGCCUUCCUUCUGC UCUU-3′ and 5′-GAGCAGAAGGAAGGCUGUGUU-3′. CRIF1 siRNA target sequences were G1 5′-ACAGATGATTGTGAACTGGCA-3′ and G5 5′-AAGAACGCGAATGGTACCCGA-3′. Qiagen: allstar negative control siRNA was used as a control dsRNA. Plasmid DNA transfection was similar except that 1.5 µg plasmid DNA replaced dsRNA, and antibiotic selection was applied after 24 or 48 h.
Estimation of transcript and mtDNA copy number
Mitochondrial mRNA abundance was estimated relative to the mRNA for GAPDH. Total RNA was extracted from cells with TRIzol reagent (Invitrogen) and cDNA was generated with an Ominiscript reverse transcription kit with random hexamer primers (Qiagen) according to the manufacturer’s instructions. mtDNA copy number and the relative abundance of mRNA were estimated by quantitative PCR, as described before (12). Primers with the following sequences were employed: APP, forward 5′-TTTTTGTGTGCTCTCCCAGGTCT-3′, reverse 5′-TGGTCACTGGTTGGTTGGC-3′, APP probe 5′-CCCTGAACTGCAGATCACCAATGCGGTAG-3′; COXII, forward 5′-CGTCTGAACTATCCTGCCCG-3′, reverse 5′-TGGTAAGGGAGGG ATCGTTG-3′, probe 5′-CGCCCTCCCATCCCTACGCATC-3′; Cytb, forward 5′-GCCTGCCTGATCCTCCAAAT-3′, reverse 5′-AAGGTAGCGGATGATTCAGCC-3′, probe 5′-CACCAGACGCCTCAACCGCCTT-3′; ND1, forward 5′-CCCTAAAA CCCGCCACATCT-3′, reverse 5′-AGAGCGATGGTGAGAGCTAAGG-3′, probe 5′-ATCACCCTCTACATCACCGCCCCG-3′; 16S rRNA, forward 5′-TTTGCAAGG AGAGCCAAAGC-3′, reverse 5′-AGACGGGTGTGCTCTTTTAGC T-3′, probe 5′-AGACCCCCGAAACCAGACGAGCTACC-3′; 12S rRNA forward 5′-CCCCAGGTTGGTCAATTTC-3′, reverse 5′-CGGCTTCTATGACTTGGGTTAA-3′, probe 5′-TGCAGCCACCGCGGTCA-3′; 18S, forward 5′-TCGAGGCCCTGT AATTGGAA-3′, reverse 5′-CCCTCCAATGGATCCTCGTT-3′, probe 5′-AGTC CACTTTAAATCCTT-3′; GFP, forward 5′-GGAGAGGGTGAAGGTGATGC-3′, reverse 5′-ATCCGGATAACGGGAAAAGC-3′, probe 5′-CCTGTTCCATGGCC AACACTTG-3′. Probes contained a 5′-FAM fluorophore and a 3′-TAMRA quencher (Sigma Genosys). Cycle conditions were the default setting on the ABI sequence detection system 7700.
Iodixanol gradient analysis of mitochondrial lysates
Iodixanol gradient fractionation was as previously described (7). Briefly, sucrose gradient-purified HEK293T cell mitochondria were treated with trypsin, as described above, lysed with 0.4% n-dodecyl-β-d-maltoside and a 1000gmax supernatant loaded on a 20–42.5% iodixanol gradient and centrifuged at 100 000gmax for 14 h. Fractions of 0.5 ml were collected from the base of the tube, and the associated nucleic acid and proteins were resolved by 1% agarose and 4–12% polyacrylamide gel electrophoresis, respectively.
DNA analysis
DNA extraction and Southern blotting were performed as described previously (2). Mitochondrial DNA copy number was estimated by comparing the signal from a radiolabelled probe with major non-coding region, nucleotides 16343-151 of human mtDNA with that of a nuclear 18S rRNA probe.
Mitochondrial translation
The translation of proteins encoded in mtDNA was performed as described before (13). Briefly, cells were incubated with emetine for 10 min to inhibit cytosolic protein synthesis, before adding 35S-methionine for 30 min, after which cells were lysed and fractionated by SDS–PAGE (4–20% Tris–glycine PAG or as indicated in the figure legends). Gels were dried and exposed to phosphor plates, and the signals quantified with a Typhoon™ phosphorimager (GE Healthcare).
Immunoblotting
Immunodetection utilized antibodies to mouse anti-TUFM (1:1000, Abnova), mouse monoclonal anti-Flag M2 (1:1000, Sigma), rabbit anti-POLMRT (1:1000, Abcam), rabbit anti-TOM20 (1:20 000, Santa Cruz), rabbit anti-MRPL11 (1:1000, cell signaling), rabbit anti-prohibitin (1:500 BioLegend), mouse anti-MRPS2 (1:500 Abcam), rabbit anti-PTCD3 1:500 (Abcam), rabbit anti-HSPD1 1:5000 (Abcam), rabbit anti-UQCRC1 (complex III): 1:3000 (Sigma), mouse anti-SDH (SDHB, complex II), 1:500 (Mitoscience), rabbit anti-cytochrome oxidase II 1:200 (Abcam), rabbit anti-ERAL1 (1:2000) (ProteinTech), mouse anti-GAPDH (1:20 000) (Abcam), rabbit anti-H2A and anti-H2B (1:2000) (Abcam) and anti-PAPM (1:1000) (Abcam). Rabbit anti-SSBP1 (1:500) and rabbit anti-TFAM (1:20 000) were kind gifts of Drs M. Zeviani and R. Wiesner, respectively. Rabbit anti-ATAD3, 1:50 000; anti-CRIF1, 1:2000, anti-ATPase g subunit, 1:5000; anti-NDUFS1, 1:4000 and chicken anti-FA11 (1:10 000) were raised against recombinant proteins produced in-house. Secondary antibodies were anti-rabbit or anti-mouse HRP 1:1000 (Promega).
Confocal microscopy
Mitochondrial DNA was visualized using mouse anti-DNA (1:200) (Novus Biologicals) and goat anti-mouse-Alex-Fluro®488 (1:1000) (Invitrogen); ATAD3.HA was detected using rat anti-HA (1:1000) (Roche) and goat anti-rat-Alex-Fluro®647; Crif1-Flag was detected using mouse anti-Flag (1:1000, Sigma) and goat anti-mouse Alex-Fluro®488; endogenous ATAD3 was detected using rabbit anti-ATAD3 (1:2000, made in-house). Slides were mounted using ProLong_Gold antifade. Cells were imaged with an LSM 510 Meta confocal microscope (Zeiss) used in conjunction with the LSM 510 software. Images were acquired with a Zeiss 63× /1.40 oil immersion objective, set at zoom 2.4. MitoTracker orange signal was pseudo-coloured red to improve contrast. Images were edited in Photoshop Elements (Adobe).
ATAD3 Immunoprecipitation
Two hundred microlitre protein A sepharose (GE Healthcare) pre-incubated with rabbit anti-ATAD3 serum (or an equal volume of rabbit preimmune serum) was cross-linked by BS3 (bis-sulphosuccinimidyl suberate, Thermo Scientific), according to the manufacturer’s protocol. The beads were incubated with 1 ml of the mitochondrial 1000gmax supernatant prepared as described above for overnight at 4°C. Beads were washed five times with 200 µl mitochondria lysis buffer before removal of bound proteins by boiling in SDS sample buffer. Samples were resolved by SDS–PAGE, transferred to nitrocellulose and probed with various antibodies.
Deposition of data
Mass spectrometric data associated with SILAC-labelled affinity purification of ATAD3 have been deposited at the TRANCHE repository (https://proteomecommons.org) and may be downloaded using the following hash: DqM6cArdfZugsbmp7VqMhCN10zEgPrrrDjppdB + yV66a1QDIUS9sbDP1N + e VOZlhPHzG1DTeV + j7i1oGchGl6YfKot4AAAAAAAAVPA==
RESULTS AND DISCUSSION
Isolation and characterization of human mitochondrial nucleoids
In five independent TAP experiments, nucleoids were isolated from the mitochondria of human embryonic kidney (HEK293T) cells expressing StrepII and FLAG-tagged forms of TFAM or SSBP1 and identified by mass spectrometry (see ‘Materials and Methods’ section). The procedure enriched the bait protein greatly and simplified the protein profile considerably (Supplementary Figure S1). The identified proteins are listed in Table 1 with details in Supplementary Table S1. Several of the candidate nucleoid proteins, ATAD3, prohibitin, TUFM, β-actin and TFAM, were also present in enriched mitochondrial nucleoprotein preparations from rat liver isolated by other methods (2,7), whereas none of the proteins that co-purified with tagged TFAM or SSBP1 were identified by mass spectrometry when mitochondrial lysates of control HEK293T cells were subjected to the first affinity purification step (14). ATAD3 has been implicated in mtDNA metabolism previously, as it binds preferentially to DNA structures with a displacement loop in vitro, and its abundance in cells affects mtDNA topology, the dimensions of the mitochondrial nucleoid and the abundance of 7S DNA, which forms displacement loops (2,15,16). Its presence in all TAP preparations of nucleoids (Table 1) supports the view that ATAD3 is associated with many, if not all, mitochondrial nucleoids, and, as shown below, contributes to protein synthesis in the mitochondrion. The identification of β-actin in three of the five TAPs (Table 1) supports the earlier assignment of β-actin as a protein that interacts with the mitochondrial nucleoid (7). CKAP4, cytoskeletal-associated protein 4, which has hitherto been linked to the endoplasmic reticulum (17), was identified in two TAPs, and so is a candidate for linking mitochondrial nucleoids to the cytoskeleton. Histones appeared in all the preparations; however, their abundance was reduced considerably by treatment of isolated mitochondria with trypsin, prior to lysis and TAP (Supplementary Figure S2). It is not known if the residual histones that survive trypsin treatment are genuine mitochondrial proteins.
Table 1.
Proteins identified by MALDI–TOF–TOF mass spectrometry from five StrepII and FLAG tag tandem affinity purifications of TFAM or SSBP1, from DNA-enriched mitochondrial supernatants of HEK293T cells (see ‘Materials and Methods’ section)
| A: TFAM-Flag-StrepII | B: SSBP1-Flag-StrepII | C: TFAM-Flag-StrepII | D: SSBP1-Flag-StrepII | E: TFAM-Flag-StrepII |
|---|---|---|---|---|
| ATAD3A | ATAD3A | ATAD3A | ATAD3A | ATAD3A |
| SSBP1 | TFAM | SSBP1 | TFAM | SSBP1 |
| TUFM | TUFM | TUFM | DHX30 | TUFM |
| DHX30 | DHX30 | DHX30 | PTCD3 | PTCD3 |
| PTCD3 | β-actin | PTCD3 | PHB2 | β-actin |
| PHB1 | PHB2 | LRP59 | ||
| CKAP4 | β-actin | CRIF1 | ||
| LRP59 | CKAP4 | |||
| CRIF1 | LRP59 | |||
| MRPS5, MRPS7, MRPS9, MRPS10, MRPS11, MRPS14, MRPS16, MRPS17, MRPS22, MRPS23, MRPS24, MRPS27, MRPS29, MRPS31, MRPS35 | MRPS5, MRPS7, MRPS9, MRPS10, MRPS14, MRPS23, MRPS26, MRPS27, MRPS28, MRPS29, MRPS31, MRPS34 | MRPS5, MRPS9, MRPS10, MRPS14, MRPS22, MRPS26, MRPS27, MRPS28, MRPS29, MRPS31 | MRPS9, MRPS18B, MRPS26, MRPS30 | MRPS5, MRPS9, MRPS10, MRPS22, MRPS23, MRPS26, MRPS27, MRPS28, MRPS29, MRPS31, MRPS34 |
| MRPL4a, MRPL22, MRPL23, MRPL35, MRPL38, MRPL39, MRPL41, MRPL43, MRPL49, ICT1 | MRPL4a, MRPL22, MRPL23, MRPL35, MRPL38, MRPL39, MRPL41, MRPL43, MRPL49, ICT1 | MRPL11, MRPL23, MRPL43 | MRPL4, MRPL11, MRPL15, MRPL19, MRPL28, MRPL37, MRPL38, MRPL44 | MRPL3, MRPL4, MRPL4a, MRPL15, MRPL22 |
| Histones: | Histone: | Histones: | Histones: | Histones: |
| H2B, H4F | H2A2 | H2A2, H2B.1, H3, H4 | H2A, H2B, H4 | H2A, H2B, H3.3, H4 |
The list comprises proteins identified in two or more mitochondrial nucleoid preparations. Proteins identified once only, which do not appear in the table were: leucine rich PPR-motif containing protein (LRP130, or LRPPRC), trifunctional enzyme subunit alpha (HADHA), RNA methyltransferase-like 1, Splicing factor-2 associated protein p32 (SF2P32), dihydrolipoamide branch chain transacylase (DBT), serine hydroxymethyl transferase (SHMT), the mitochondrial phosphate carrier (SLC25A3), C4orf14 (NOA1)—see accompanying article, and protein RPL18a of the 60S cytosolic ribosome. The bait protein was identified in all TAPs (Supplementary Table S1). Abbreviations are as follows: ATAD3, ATPase family AAA domain-containing protein 3, DHX30 (putative ATP-dependent RNA helicase), TUFM (mitochondrial translation elongation factor), PHB 1 or 2 (prohibitin), PTCD3 (pentatricopeptide repeat-containing protein 3), LRP59 (leucine-rich protein 59), CRIF1 or Gadd45GIP1 (growth arrest and DNA damage-inducible proteins-interacting protein 1), the peptidyl tRNA hydrolase, ICT1, CKAP4 (cytoskeletal associated protein 4). Further details of the identification of proteins appear in Supplementary Table S1, and the mitochondrial ribosomal proteins (MRPs) identified in the TAPs are collated in Supplementary Table S2, and shown in parallel with those MRPs identified in subsequent analyses. Histones were for the most part degraded when mitochondria were incubated with trypsin prior to lysis (Supplementary Figure S2). HSPA9 was identified in four TAPs but is not listed in the table, as it was also associated with affinity-purified mitochondrially targeted GFP (Supplementary Figure S3-A).
Many components of mitochondrial ribosomes were identified in the TFAM and SSBP1 affinity-purified preparations (Table 1). However, no mitochondrial ribosome component, or candidate nucleoid protein, was identified when the FLAG and StrepII tags were linked to a mitochondrially targeted form of GFP (mtGFP) and affinity purified from HEK293T cells (Supplementary Figure S3-A). However, HSPA9 (HSP75) was present as before, suggesting that it is induced in response to high-level expression of many transgene products. Nor were there any mitochondrial nucleoid proteins or translation factors, with the possible exception of SF2P32 (18), when tagged NIPSNAP2 was affinity purified from mitochondria of HEK293T cells (Supplementary Figure S3-B). Thus, the expression and purification system used does not systematically isolate mitochondrial ribosomes and translation factors, and so the co-purification of such factors with TFAM and SSBP1 implies bona fide interactions.
Nucleoid components involved in protein translation in mitochondria
The presence in TAP nucleoprotein complexes of 12–25 protein components of mitochondrial ribosomes and of pentatricopeptide protein 3 (19,20) and TUFM (6) (Table 1; Supplementary Tables S1 and S2), suggested a link between mitochondrial nucleoids and the mitochondrial translation apparatus. This interpretation was supported by immunoblot analysis of the StrepII affinity purified material, which contained a substantial amount of a small ribosome assembly protein, ERAL1 (21,22) and two mitochondrial ribosomal proteins, MRPS2 and MRPL11, yet very little respiratory chain protein (Supplementary Figure S4). Moreover, analysis of all the proteins in the gel, with an OrbiTrap mass spectrometer, after additional TAPs of TFAM, identified 31 and 56 mitochondrial ribosomal subunits in two experiments (Supplementary Table S2).
Mitochondrial mRNAs are associated with high-density nucleoprotein complexes
The co-purification of mitochondrial ribosomes and ancillary factors with mitochondrial nucleoids (Table 1), suggested an intimate association between the two types of nucleic acid-containing complex, and a previous study identified a number of candidate nucleoid proteins among those proteins immunoprecipitated with mtRRF, a mitochondrial ribosome recycling factor (23). Mitochondrial RNA and DNA also co-fractionate on 20–42.5% iodixanol gradients (Figure 1a), although this could reflect identical buoyant densities, rather than a physical interaction. In any case, both complexes depend on protein for their position on the gradients, as mitochondrial DNA and RNA stripped of protein barely entered 20–42.5% iodixanol gradients, and so they were found near the top of the density gradient (Figure 1b). The vast majority of messenger RNAs were contained in the ribonucleoprotein complexes that coincided with mitochondrial ribosomes and nucleoids (Figure 1a, c and d), suggesting that mitochondrial mRNAs are not free in the matrix; an arrangement that may be designed to avoid degradation by RNases and damage by free radicals generated in the mitochondrion.
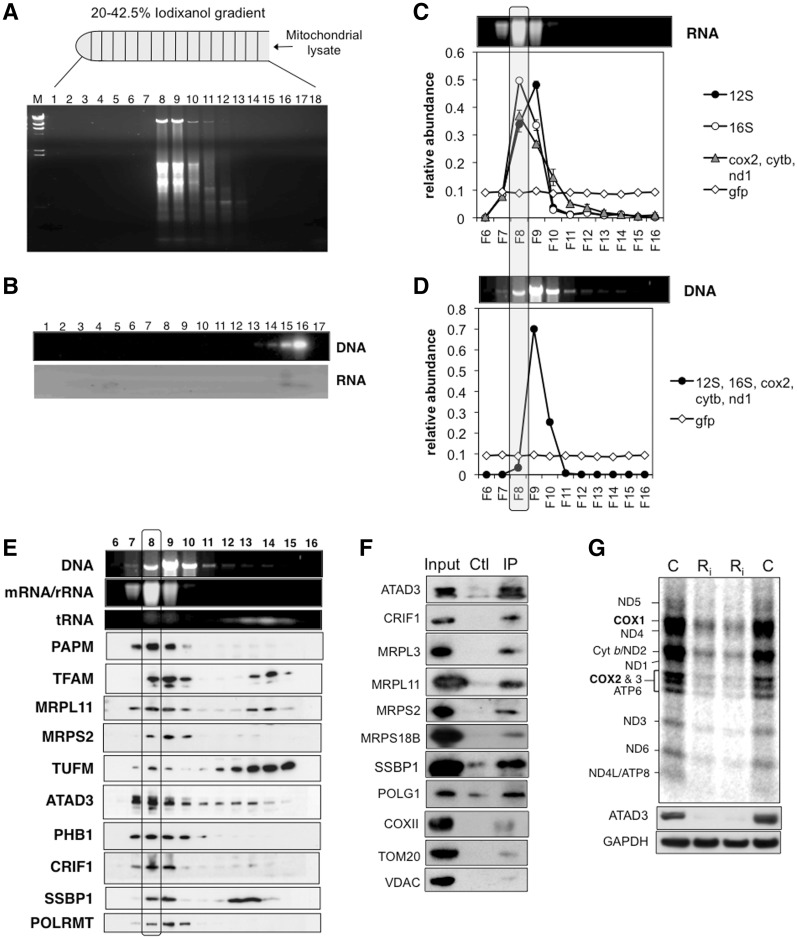
Figure 1.
Fractionation of mitochondrial nucleoprotein complexes on iodixanol gradients. (A) Mitochondrial 1000gmax supernatants from HEK293T cells were fractionated on 20–42.5% iodixanol gradients, and extracted nucleic acids from each fraction were separated by agarose gel electrophoresis and stained with ethidium bromide. (B) Protease-treated mitochondrial nucleic acids fractionated on a 20–42.5% iodixanol gradient. (C) The chart shows the relative abundance of five mitochondrial RNAs (12S rRNA, 16S rRNA, cox2, cytb and nd1) based on quantitative RT-PCR (see ‘Materials and Methods’ section); above the chart is a transilluminator image of the ethidium bromide-stained RNA from each fraction. The qPCR results for the three mRNAs (cox2, cytb and nd1) were similar and so they were grouped together (blue line). Each fraction was ‘spiked’ with an equal amount of synthetic GFP transcript (green line), as a control. Quantitative RT-PCR of 18S rRNA was performed in parallel, but none was detected, indicating that any contamination with RNA of cytosolic ribosomes was below the limits of detection. (D) Ethidium bromide staining and qPCR detection of mtDNA based on the amplification of five mitochondrial genes (see ‘Materials and Methods’ section). (E) The DNA and RNA detected by transillumination from panels (C) and (D) are displayed above a series of immunoblots for a range of mitochondrial proteins (see text for details). The boxed area includes all the results for fraction 8, in which mRNA peaked. (F) Immunoblots of mitochondrial 1000gmax supernatants of HEK cells before (input) and after IP with anti-ATAD3 antibody. The control was instead incubated with preimmune serum. (G) The products of mitochondrial protein synthesis were labelled with 35S-methionine in 143B cells subjected to one or two rounds of transfection with dsRNA1 targeting ATAD3 (Ri) or mock transfected (C) cells. Large gel image: mitochondrial translation products at the base of the panel are immunoblots of the same cell lysates probed with antibodies to ATAD3 or GAPDH.
For unknown technical reasons possibly related to trace RNase contamination, mtDNA and mtRNA were partly resolved in some experiments (Figure 1c and d). Messenger RNAs and 16S rRNA peaked in one fraction (No. 8, in the example shown), whereas 12S rRNA was most abundant in an adjacent fraction (No. 9) (Figure 1c). The partial resolution of DNA and RNA protein complexes enabled specific proteins to be assigned to particular complexes. Messenger RNAs displayed a similar distribution to the 39S subunit (16S rRNA and MRPL11) on the iodixanol gradients (Figure 1e), suggesting that mRNAs are more tightly bound to the large, as opposed to the small, subunit of the mitochondrial ribosome, or that they associate with an unknown complex of similar buoyant density to the 39S subunit. As expected, TFAM, SSBP1 and the mitochondrial RNA polymerase (POLRMT) co-fractionated with mtDNA, whereas a known RNA transacting protein, mitochondrial polyA polymerase, PAPM showed a similar distribution to mRNA and the 39S subunit (Figure 1c–e). The majority of mitochondrial proteins were located in fractions near the top of the gradient, including proteins of the oxidative phosphorylation system (Supplementary Figure S5).
ATAD3 co-fractionates with mitochondrial RNA, co-purifies with mitochondrial ribosomes and contributes to mitochondrial protein synthesis
In those iodixanol gradients where mitochondrial RNA and DNA were not coincident, ATAD3 protein peaked with the mRNAs, the 39S subunit and the mitochondrial polyA polymerase (Figure 1c and e), suggesting that it might play a role in mitochondrial translation, or RNA metabolism. The co-purification of only a small fraction of ATAD3 with tagged TFAM and SSBP1 (Supplementary Figure S4) also implied that the protein might have partners other than the mitochondrial nucleoid. In an effort to identify new partners of ATAD3, a FLAG.StrepII tagged version of ATAD3B (ATAD3B.FS) was expressed in HEK293T cells. Although mitochondrial ribosomal proteins MRPS18B and MRPL11 co-purified with ATAD3B.FS based on immunoblotting (Supplementary Figure S6), the enrichment of ATAD3B.FS was not as great as that of other tagged proteins (Supplementary Figure S4). Therefore, as a more sensitive screen for protein partners of ATAD3, stable isotope labelling of proteins in cell culture (SILAC) coupled to Orbitrap MS analysis was performed (24). Heavy (or light) labelled mitochondria, isolated from HEK293T cells expressing ATAD3B.FS, were mixed with an equal amount of light (or heavy) labelled mitochondria from uninduced HEK293T cells. In the interpretation of these experiments, affinity-purified proteins from the mixed mitochondrial lysates were considered to be associated with the bait protein when they were present in a 2-fold, or greater, excess over proteins in control mitochondria. Forty-six proteins fulfilled these criteria (Table 2 and Supplementary Table S3); the list was dominated by components of the mitochondrial ribosome and other proteins involved in mitochondrial RNA metabolism. Thirty-five of the proteins were MRPs, and five others (PTCD3, DHX30, LRP130, OXA1 and FASTKD2) have been implicated in mitochondrial translation or RNA metabolism (25–28). Little is known about human GTP-binding protein 10 (GTPBP10 or OBGH2), but its bacterial homologue obg 2, has been linked to ribosome maturation in prokaryotes (29). GTPBP5 (OBGH1) has been reported to localize to mitochondria, whereas GTPBP10 (OBGH2) was found exclusively in the nuclei of HeLa cells in a previous study (30). The other candidate partners of ATAD3B: HADHA, LRP130, DHX30, ATAD3A and TiD1 (Table 2), are all potential nucleoid-interacting proteins based on earlier reports (2,8,31).
Table 2.
ATAD3B partner proteins based on SILAC analysis
| Mitochondrial translation associated proteins | |
| MRPs | 35 mitochondrial ribosomal proteins (20 MRPL, 15 MRPS) |
| PTCD3 | Pentatricopeptide repeat-containing protein 3 |
| FASTKD2 | FAST kinase domain-containing protein 2 |
| CRIF1 | Growth arrest and DNA damage-inducible proteins-interacting protein 1 |
| GTPBP10 | GTP binding protein 10; Protein obg homolog 2 |
| ATAD3A | ATPase family AAA domain-containing protein 3A |
| OXA1 | Mitochondrial inner membrane protein OXA1L |
| Nucleoid-interacting proteins | |
| DHX30 | Putative ATP-dependent RNA helicase DHX30 |
| LRP130 | Leucine-rich PPR motif-containing protein; LRPPRC |
| DNAJA3 (TID1) | DnaJ homolog subfamily A member 3, mitochondrial |
| HADHA | Trifunctional enzyme subunit alpha |
The proteins listed were identified in SILAC experiments as the most likely protein partners of ATAD3B, based on mass spectrometry of affinity captured ATAD3B.Flag.StrepII (see main text and Supplementary Table S3). Two chaperones that were also found associated with mtGFP (Supplementary Figure S3-A) were disregarded.
A SILAC experiment using ATAD3A as the bait protein also yielded many components of the 55S ribosome (Table 3 and Supplementary Table S4). Forty-five of the top 54 proteins were 55S ribosome components (including ICT1), and 65 MRPs achieved a better than 2:1 ratio (Supplementary Table S4), accounting for ∼80% of the mitochondrial ribosome. Although both 28S and 39S subunits were well represented, there was a bias towards the large subunit: the first 31 proteins on the list included 23 components of the 39S subunit, whereas the highest scoring protein of the 28S subunit was 32nd on the list (Supplementary Table S4). This bias suggests that ATAD3A interacts (directly or indirectly) with the 39S subunit, as opposed to the 28S subunit. This detail aside, the mitochondrial ribosome is the prime candidate partner of both ATAD3A and ATAD3B (Table 4). Of the other proteins co-purifying with recombinant ATAD3A (Table 3 and Supplementary Table S4), GTPBP5 and GTPBP10 rise to seventh and ninth on the list of candidate protein partners of ATAD3A, respectively, if the mitochondrial ribosomal proteins are conflated as the 55S ribosome, rather than being scored individually. GTPBP10 was also a candidate partner of ATAD3B (Table 4), and GTPBP5 is another homologue of the Era/Obg family (29), as is the proposed 28S assembly factor, ERAL1 (21), which was also among the potential partners of ATAD3A (Table 3 and Supplementary Table S4). A fourth member of the family, C4orf14 associates specifically with the 28S subunit (see accompanying article). Thus, it appears that at least four members of the Era/Obg family (GTPBP5/OBGH1, GTPBP10/OBGH2, ERAL1 and C4orf14) have been retained in eukaryotes to enable translation of proteins in mitochondria.
Table 3.
ATAD3A partner proteins based on SILAC analysis
| Proteins with links to mitochondrial translation or RNA metabolism | |
| MRPs | 69 mitochondrial ribosomal proteins (41 MRPL, 28 MRPS) |
| MRP63 | mitochondrial ribosomal protein 63 |
| ICT1 | Peptidyl-tRNA hydrolase ICT1, mitochondrial |
| ARL2 | ADP-ribosylation factor-like protein 2 |
| GTPBP5 | GTP-binding protein 5;Protein obg homolog 1 |
| GTPBP10 | GTP binding protein 10; Protein obg homolog 2 |
| IARS2 | Isoleucine–tRNA ligase |
| PTCD3 | Pentatricopeptide repeat-containing protein 3 |
| RPUSD4 | RNA pseudouridylate synthase domain-containing protein 4 |
| NUBPL | Nucleotide-binding protein-like |
| ERAL1 | Conserved ERA-like GTPase |
| TUFM | Mitochondrial elongation factor Tu |
| CRIF1 | Growth arrest and DNA damage-inducible proteins-interacting protein 1 |
| FASTKD5 | FAST kinase domain-containing protein 5 |
| DDX28 | Mitochondrial DEAD box protein 28;probable ATP-dependent RNA helicase |
| TRUB2B | Probable tRNA pseudouridine synthase 2 |
| LRP130 | Leucine-rich PPR motif-containing protein; LRPPRC |
| MTERFD1 | mTERF domain-containing protein 1 |
| GUF1 | GTP-binding protein GUF1 homolog |
| POLRMT | Mitochondrial DNA-directed RNA polymerase |
| MRPP3 | Mitochondrial ribonuclease P protein 3 |
| METT11D1 | Methyltransferase 11 domain-containing protein 1 |
| PHB2 | Prohibitin 2;BAP37 |
| Proteins with links to lipid metabolism | |
| STARD9 | StAR-related lipid transfer protein 9 |
| HADHA | Trifunctional enzyme subunit alpha |
| HADHB | Trifunctional enzyme subunit beta |
| SLC25A1 | Tricarboxylate transport protein |
| SC2 | trans-2,3-enoyl-CoA reductase |
| SPTLC1 | Serine palmitoyltransferase 1 |
The proteins listed were identified in one SILAC experiment as the most likely protein partners of ATAD3A, based on mass spectrometry of affinity captured ATAD3A.Flag.StrepII (see main text and Supplementary Table S4). The proteins are grouped by known or inferred function. The complete list of proteins appears in Supplementary Table S4.
Table 4.
Shared candidate partner proteins of ATAD3A and ATAD3B
| MRPs | Mitochondrial ribosomal proteins (69 for ATAD3A, 35 for ATAD3B) |
| PTCD3 | Pentatricopeptide repeat-containing protein 3 |
| GTPBP10 | GTP binding protein 10; Protein obg homolog 2 |
| CRIF1 | Growth arrest and DNA-damage-inducible proteins-interacting protein 1 |
| LRP130 | Leucine-rich PPR motif-containing protein; LRPPRC |
| DNAJA3 (TID1) | DnaJ homolog subfamily A member 3, mitochondrial |
| HADHA | Trifunctional enzyme subunit α |
The list is conflated from Supplementary Tables S3 and S4.
STARD9 (START domain-containing protein 9) was high on the list of putative binding partners of ATAD3A. This interaction is plausible, since STAR-domain containing proteins are involved in cholesterol and lipid channelling, and ATAD3A has been implicated in cholesterol uptake by mitochondria (4). Also, the storage of intestinal fat is reduced in nematodes when ATAD3 is scarce (32). ATAD3A studies may thus offer insight into lipid metabolism and the, as yet poorly understood, role of mitochondrial cholesterol in non-steroidogenic cells. C18orf19 was the single strongest candidate partner protein of ATAD3A (Supplementary Table S4); however, its function is not known.
Immunoprecipitation (IP) of endogenous ATAD3 from HEK293T cells, using an anti-ATAD3 antibody corroborated the SILAC-affinity purification analyses, inasmuch as four subunits of the 55S ribosome were captured by the antibody (Figure 1f). CRIF1 and the mitochondrial DNA polymerase, POLG1, produced the strongest IP to input signals. However, POLG1 displayed the smallest difference between ATAD3 IP and mock precipitation. Hence, CRIF1 was the single most likely partner of ATAD3, of the proteins tested, based on IP (Figure 1f).
Since the candidate protein partners of ATAD3A and ATAD3B were suggestive of interactions with the mitochondrial ribosome, we next assessed the contribution of endogenous ATAD3 to mitochondrial translation by silencing ATAD3A and ATAD3B in human osteosarcoma cells. Gene silencing of ATAD3 for 48 or 72 h severely impaired mitochondrial protein synthesis, compared to mock transfected cells, or cells transfected with a scrambled dsRNA, based on 35S-methionine labelling of mitochondrial translation products (Figure 1g and Supplementary Figure S7).
ATAD3 and mtDNA metabolism
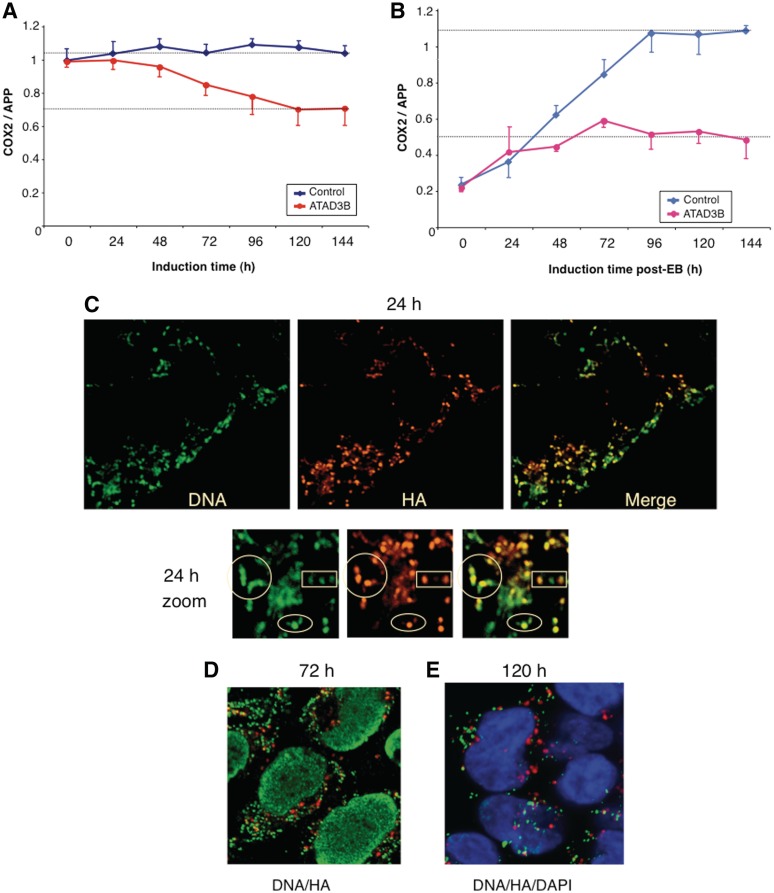
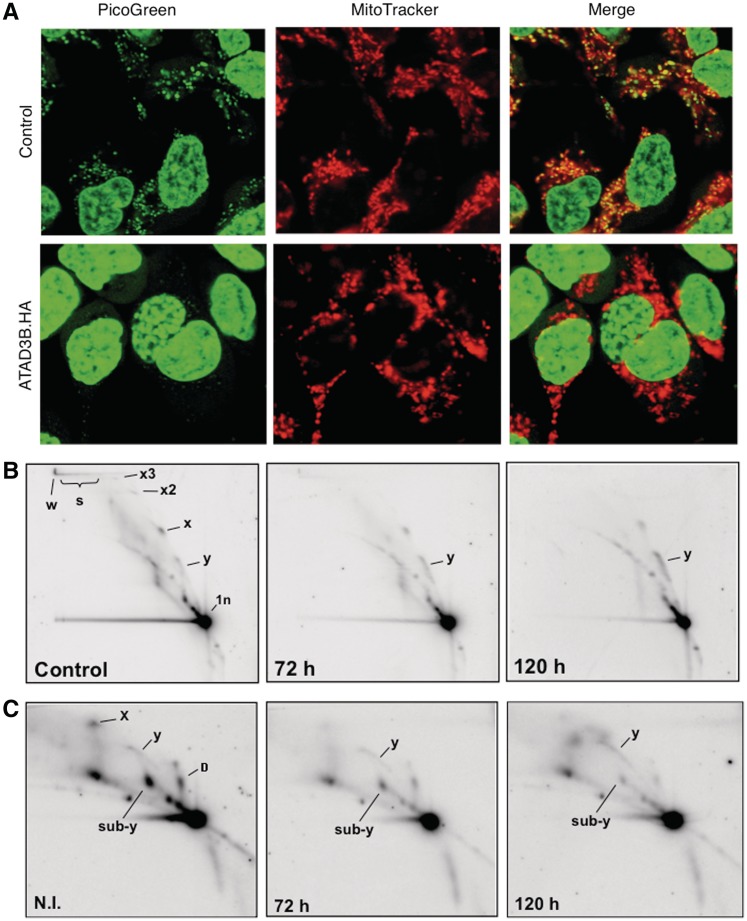
ATAD3 was found previously in highly enriched mitochondrial nucleoprotein preparations that were largely free of ribosomes (2,7), and in this study it was identified in all the TAPs of two mtDNA binding proteins (Table 1). However, SILAC analysis suggested the major partner of recombinant ATAD3 to be the mitochondrial ribosome (Table 4). The co-fractionation of endogenous ATAD3 primarily with mitochondrial RNA (Figure 1e), and the pronounced impairment of mitochondrial protein synthesis in response to ATAD3 gene-silencing (Figure 1g) also argued in favour of a role for the protein in mitochondrial translation. To investigate ATAD3’s role in mtDNA metabolism further, a haemagglutinin (HA)-tagged form of ATAD3B was established in HEK293T cells (Supplementary Figure S8). Expression of the transgene led to a 30% reduction in mtDNA copy number, over the course of 6 days (Figure 2a). This was a marginally stronger phenotype than that observed for ATAD3 gene-silencing, which caused a 20% decrease in mtDNA copy number (2). In contrast, elevated expression of HA-tagged POLG2 in HEK293T cells had no effect on mtDNA copy number (data not shown). The impact of ATAD3B.HA on mtDNA was more pronounced when mtDNA copy number was first reduced to 20% of normal with ethidium bromide. In control cells, mtDNA copy number returned to normal within 4 days of removal of the drug, whereas there was a marked impairment of mtDNA replication in cells expressing ATAD3B.HA (Figure 2b), or ATAD3B.FS (Supplementary Figure S9-A). Analysing the distribution of ATAD3.HA and mtDNA in HEK293T cells suggested the underlying problem for mtDNA might be a shortage of interacting ATAD3. There was good co-localization 24 h after the induction between mtDNA and ATAD3B.HA (Figure 2c), and restoration of mtDNA copy number was similar to controls at this time point (Figure 2b). However, at later time points, when copy number was lagging appreciably behind that of controls (Figure 2b), ATAD3B.HA formed foci that did not co-localize with mtDNA (Figure 2d and e), and these foci appeared to include the endogenous protein, as immunocytochemistry with an antibody to ATAD3 produced similar results (Supplementary Figure S9-B). ATAD3 gene-silencing diminished picogreen staining of mtDNA and depleted cells of detergent-resistant mtDNA multimers held together by protein (2), and elevated ATAD3B expression produced the same two phenotypes (Figure 3a and b). Thus, three analyses of mtDNA, copy number, picogreen staining and protein-dependent multimers, suggest that over-expression of ATAD3B.HA results in loss of function of endogenous ATAD3, in relation to mtDNA transactions, which might be explained by the aggregation of the protein, separate from mtDNA (Figure 2d and e). Expression of ATAD3B.HA also altered the features of mitochondrial replication intermediates (Figure 3c). Specifically, there was an increase in duplex DNA intermediates (resolving on the replication fork, or y, arc), and a decrease in the sub-y arc of RNA-containing intermediates (Figure 3c), which are indicative of a change in the mode or mechanism of mtDNA replication [for details, see Ref. (33)]. Therefore, native ATAD3 may well participate in mtDNA replication.
Figure 2.
Elevated expression of HA tagged ATAD3B impedes mtDNA replication by concentrating the protein in foci separate from mitochondrial nucleoids. (A) HEK293T cells carrying an ATAD3B transgene with an HA tag were induced to express the recombinant protein with 50 ng/ml doxycyline for 6 days. DNA was harvested daily and the mtDNA copy number assayed by qPCR. Controls were HEK293T cells transformed with an empty vector and maintained in the same dose of drug. (B) qPCR estimation of mtDNA copy number after harvesting DNA from cells expressing ATAD3B.HA protein, or control cells, at daily intervals, having first reduced mtDNA copy number to 20% of normal by exposure to 50 ng/ml ethidium bromide for 72 h. The cells were maintained in 50 ng/ml doxycycline throughout the experiment. For charts (A) and (B), n = 3 experiments, error bars are 1 SD from the mean. The distribution of mitochondrial nucleoids (anti-DNA antibody) and ATAD3B.HA (anti-HA antibody) was determined by confocal microscopy 24, 72 and 120 h after induction of the transgene with 50 ng/ml doxycycline [panels (C–E), respectively].
Figure 3.
Elevated expression of ATAD3B.HA perturbs mtDNA topology, protein-dependent mtDNA multimers and mitochondrial replication intermediates. (A) HEK293T cells expressing ATAD3B.HA for 5 days were DNA stained with picogreen and the mitochondrial network was labelled with mitotracker orange (pseudo-coloured red for contrast). (B) Mitochondrial nucleoprotein was isolated from HEK293T cells expressing ATAD3B.HA for 3 or 5 days (50 ng/ml doxycycline), and from (control) cells cultured without the drug. The DNA was digested with AccI, separated by 2D-AGE and hybridized to a probe detecting the major non-coding region of human mtDNA [see Ref. (41) for a detailed exposition of the protein-dependent multimers of mtDNA labelled x2, x3, d and w]. (C) Mitochondrial replication intermediates of cells induced to express ATAD3B.HA and controls. Samples and processing were identical to panel B except that the DNA was treated additionally with proteinase K during the isolation procedure. Note the enhanced standard replication fork (y) arc after 120 h expression of ATAD3B.HA in panels (B) and (C).
If some native ATAD3 interacts directly with mtDNA, as implied by the results already described (Figures 2 and 3), why was TFAM not among the candidate partner proteins identified in the SILAC experiments (Tables 2 and 3)? Comparison of the results for the affinity capture of mtDNA binding proteins (TFAM and SSBP1) and ATAD3 suggests that the isolation procedure identifies only the most abundant partners of the bait protein. In the case of nucleoid capture, DNA polymerase γ was enriched based on immunoblotting (Supplementary Figure S4), yet not sufficiently to be visible by Coomassie staining and so it was not identified by mass spectrometry (Table 1), presumably because only a subset of molecules is undergoing replication at any one time. In the same vein, ribosomes are numerically superior to nucleoids and so more ATAD3 will be associated with 55S ribosomes than with mtDNA, assuming it binds to both. Furthermore, in the SILAC experiments, heavy and light forms of the various proteins were present on the same column, and so there may have been some transfer of free TFAM, POLG1 and SSBP1 (Figure 1e) from the control mitochondrial lysates to the recombinant ATAD3 bound nucleoids, which would have the effect of reducing the bait-associated protein to control ratio.
Prohibitin and CRIF1 contribute to mitochondrial protein synthesis
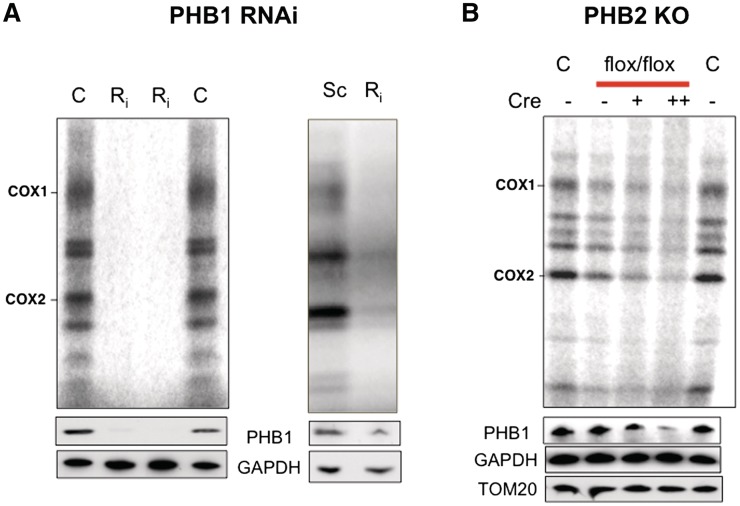
In addition to being identified in multiple TAPs of mtDNA binding proteins (Table 1), prohibitin co-fractionated with mtDNA and mitochondrial ribosomes (Figure 1e). These results support the earlier assignment of prohibitin as a mitochondrial nucleoid interacting factor (7,8,34), and suggest that prohibitin is associated with ribonucleoprotein complexes, as well as mtDNA. Given these similarities, prohibitin might, like ATAD3, contribute to mitochondrial translation. Prohibitin forms oligomeric rings composed of two proteins, prohibitin 1 and 2 in equimolar amounts; neither protein can form a complex alone, and each is rapidly turned over in the absence of the other (35). Therefore, targeting either gene should be equally effective. Prohibitin 1 was targeted with dsRNA in human cultured cells and found to severely impair mitochondrial protein synthesis (Figure 4a), and excision of the PHB2 gene in mouse embryonic fibroblasts (MEFS) also impaired mitochondrial protein synthesis (Figure 4b), albeit to a lesser extent. The apparently milder impairment of protein synthesis after (PHB2) gene excision, as compared to (PHB1) siRNA, can be explained by the former treatment causing more cell death (based on visual inspection) and the retention of the gene by some of the MEFs (Supplementary Figure S10-A). Moreover, the loss of prohibitin is accompanied by a marked inhibition of cell growth in MEFs (35), which may attenuate the mitochondrial translation phenotype.
Figure 4.
Decreased expression of prohibitin impairs mitochondrial protein synthesis. (A) Mitochondrial translation was assayed in mock transfected 143B cells (C) or cells transfected with a scrambled dsRNA (Sc), or with dsRNA targeting PHB1 (Ri) and separated by SDS–PAGE (4–12% Nu-PAG, Invitrogen). Decreased expression of the target protein was confirmed by immunoblotting of PHB1 with GAPDH as the reference protein. (B) The PHB2 gene was ablated in 90% of cells by incubating MEFs, whose sole copy of PHB2 is flanked by flox sites, with cre-recombinase [Supplementary Figure S10-A and Ref. (42)]. C, control MEFs, PHBflx/flx MEFs with flox sites flanking the prohibitin 2 (PHB2) gene. Cre, cre-recombinase that excises the PHB2 gene; −, no cre added; +, one dose of cre; ++, two doses of cre (see Supplementary Figure S10-A).
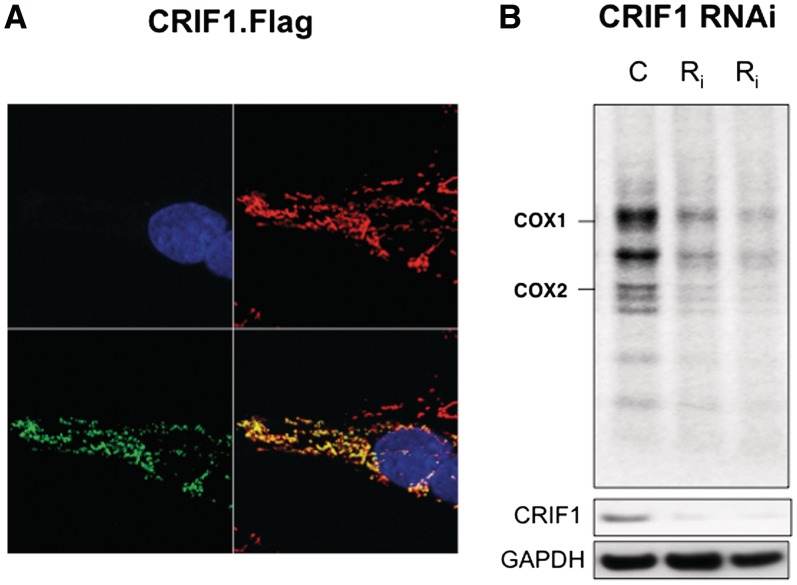
CRIF1 appeared to be an unlikely nucleoid interacting protein (Table 1) or partner of ATAD3 (Table 4), because it was reported to be a nuclear protein (36). However, the recombinant CRIF1 studied previously carried a tag at its amino terminus, and tags so positioned are well known to mask mitochondrial targeting signals (MTS). In contrast, CRIF1 tagged at the carboxy-terminus was targeted exclusively to mitochondria (Figure 5a), suggesting that the protein has an archetypal amino terminal MTS, as predicted by several algorithms (PSORTII, Target P, Mitoprot and Predator), and a carboxy-terminal nuclear localization signal. A CRIF1-GFP fusion protein was also targeted to mitochondria (37). Mitochondrial targeting of CRIF1 (Figure 5a) explains the identification of the protein in two of five TAP experiments (Table 1). Moreover, the protein co-fractionated with mtRNA on iodixanol gradients (Figure 1e) and gene silencing of CRIF1 with either of two siRNAs resulted in markedly decreased mitochondrial protein synthesis (Figure 5b and Supplementary Figure S10-B). Therefore, we conclude that CRIF1 is a mitochondrial protein with a role in protein synthesis in the organelle, which furthermore might act as the link between ATAD3 and the mitochondrial ribosome (Figure 1f).
Figure 5.
CRIF1, with C-terminal FLAG-StrepII tags, is targeted exclusively to mitochondria and gene-silencing of CRIF1 impairs mitochondrial protein synthesis. (A) After transient transfection of HOS cells with a CRIF1.FLAG.StepII cDNA cloned in pcDNA5 (Invitrogen), the cells were labelled with DAPI, mitotracker orange (pseudo-coloured red for contrast) and anti-CRIF1 (green). (B) Mitochondrial translation products in mock transfected HOS cells (C) or cells transfected with 10 nM dsRNA, G1 or G5 targeting CRIF1 (Ri), as per ATAD3 (see Figure 1g and ‘Materials and Methods’ section).
ATAD3 and prohibitin are linked to nucleoids and the mitochondrial translation machinery
Based on this report, ATAD3 has roles in DNA metabolism and protein synthesis in mitochondria, and it appears to be physically and functionally allied to prohibitin. ATAD3 and prohibitin co-purify with TFAM or SSBP1 affinity-captured nucleoids (Table 1); they co-fractionate with mitochondrial nucleic acids on iodixanol gradients (Figure 1e) and gene silencing of PHB or ATAD3 has the same effect on mtDNA (2,38). Both ATAD3 and the putative scaffold rings of prohibitin are required for normal mitochondrial morphology (4,35) and mitochondrial translation (Figures 1g and 4). Collectively, these results suggest that the two proteins have multiple, overlapping roles in mitochondrial biogenesis and mitochondrial nucleic acid metabolism.
GTPases are used extensively to drive ribosome biogenesis in bacteria (39), and the ATPase of ATAD3 might be employed to similar effect, although this would imply the protein has fundamentally different roles in translation and mtDNA organization. Alternatively, the protein might have a structural role. The first report of ATAD3 found that only a fraction of the protein co-localizes with mtDNA. It also forms ‘threads’ throughout much of the mitochondrial network and both ATAD3 and mtDNA are tightly associated with mitochondrial membranes (2). Therefore, ATAD3 might form a filament to which mitochondrial nucleoids and mitochondrial ribosomes attach, perhaps aided by prohibitin. This hypothesis ‘while tentative’ is compatible with the distribution of ATAD3 in cells and its tight association with mitochondrial membranes and mtDNA (2,7); the co-purification of ATAD3, prohibitin and components of the 55S ribosomes with mtDNA binding proteins (Table 1), and the mtDNA related phenotypes associated with ATAD3 gene-silencing and over-expression (Figures 2 and 3) (2); while also accommodating the findings that a majority of endogenous ATAD3 co-fractionates with mtRNA (Figure 1e), the major binding partner of recombinant ATAD3 is the mitochondrial ribosome (Table 4), and RNA interference of ATAD3 markedly impairs mitochondrial translation (Figure 1g). The more marked effect of ATAD3 siRNA on mitochondrial translation (Figure 1g) compared to mtDNA copy number (2) implies that when ATAD3 is scarce, nucleoids take precedence; and because mitochondrial translation is particularly important during periods of maximal growth (40), ATAD3 (or prohibitin) deficiency is expected to be felt most keenly in proliferating cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–10 and Supplementary References [43,44].
FUNDING
Medical Research Council (MRC) and the Biotechnology and Biological Sciences Research Council (BBSRC); Intramural program of the National Institutes of Health, National Heart, Lung and Blood Institute (to K.N.); Academy of Finland (to H.M.C.). Funding for open access charge: MRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for the help and encouragement of numerous members of the Mitochondrial Biology Unit, especially Drs Shujing Ding, Joe Carroll and Michal Minczuk. PHB2 knockout MEFs were kindly provided by Prof. T. Langer. We are also indebted to Dr Hans Spelbrink for many useful discussions and the provision of TFAM and SSBP1 cDNAs. T.L. is a NIH-CamGrad Scholar.
REFERENCES
- 1.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 2.He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, et al. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J. Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 4.Gilquin B, Taillebourg E, Cherradi N, Hubstenberger A, Gay O, Merle N, Assard N, Fauvarque MO, Tomohiro S, Kuge O, et al. The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol. Cell Biol. 2010;30:1984–1996. doi: 10.1128/MCB.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islam MM, Nautiyal M, Wynn RM, Mobley JA, Chuang DT, Hutson SM. Branched-chain amino acid metabolon: interaction of glutamate dehydrogenase with the mitochondrial branched-chain aminotransferase (BCATm) J. Biol. Chem. 2010;285:265–276. doi: 10.1074/jbc.M109.048777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai YC, Bullard JM, Thompson NL, Spremulli LL. Interaction of mitochondrial elongation factor Tu with aminoacyl-tRNA and elongation factor Ts. J. Biol. Chem. 2000;275:20308–20314. doi: 10.1074/jbc.M001899200. [DOI] [PubMed] [Google Scholar]
- 7.Reyes A, He J, Mao CC, Bailey LJ, Di Re M, Sembongi H, Kazak L, Dzionek K, Holmes JB, Cluett TJ, et al. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011;39:5098–5108. doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 9.Bogenhagen DF, Rousseau D, Burke S. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 2008;283:3665–3675. doi: 10.1074/jbc.M708444200. [DOI] [PubMed] [Google Scholar]
- 10.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 12.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 13.Dunbar DR, Moonie PA, Zeviani M, Holt IJ. Complex I deficiency is associated with 3243G:C mitochondrial DNA in osteosarcoma cell cybrids. Hum. Mol. Genet. 1996;5:123–129. doi: 10.1093/hmg/5.1.123. [DOI] [PubMed] [Google Scholar]
- 14.Minczuk M, He J, Duch AM, Ettema TJ, Chlebowski A, Dzionek K, Nijtmans LG, Huynen MA, Holt IJ. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebelo AP, Williams SL, Moraes CT. In vivo methylation of mtDNA reveals the dynamics of protein–mtDNA interactions. Nucleic Acids Res. 2009;37:6701–6715. doi: 10.1093/nar/gkp727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN. Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion. 2007;7:311–321. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Vedrenne C, Hauri HP. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic. 2006;7:639–646. doi: 10.1111/j.1600-0854.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- 18.Fogal V, Richardson AD, Karmali PP, Scheffler IE, Smith JW, Ruoslahti E. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol. Cell Biol. 2010;30:1303–1318. doi: 10.1128/MCB.01101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies SM, Rackham O, Shearwood AM, Hamilton KL, Narsai R, Whelan J, Filipovska A. Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 2009;583:1853–1858. doi: 10.1016/j.febslet.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 20.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 21.Dennerlein S, Rozanska A, Wydro M, Chrzanowska-Lightowlers ZM, Lightowlers RN. Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit. Biochem. J. 2010;430:551–558. doi: 10.1042/BJ20100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchiumi T, Ohgaki K, Yagi M, Aoki Y, Sakai A, Matsumoto S, Kang D. ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res. 2010;38:5554–5568. doi: 10.1093/nar/gkq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, Kühl I, Gaisne M, Bonnefoy N, Smeitink JA, et al. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res. 2008;36:5787–5799. doi: 10.1093/nar/gkn576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gohil VM, Nilsson R, Belcher-Timme CA, Luo B, Root DE, Mootha VK. Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J. Biol. Chem. 2010;285:13742–13747. doi: 10.1074/jbc.M109.098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simarro M, Gimenez-Cassina A, Kedersha N, Lazaro JB, Adelmant GO, Marto JA, Rhee K, Tisdale S, Danial N, Benarafa C, et al. Fast kinase domain-containing protein 3 is a mitochondrial protein essential for cellular respiration. Biochem. Biophys. Res. Commun. 2010;401:440–446. doi: 10.1016/j.bbrc.2010.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haque ME, Elmore KB, Tripathy A, Koc H, Koc EC, Spremulli LL. Properties of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L and its interactions with mammalian mitochondrial ribosomes. J. Biol. Chem. 2010;285:28353–28362. doi: 10.1074/jbc.M110.148262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato A, Kobayashi G, Hayashi H, Yoshida H, Wada A, Maeda M, Hiraga S, Takeyasu K, Wada C. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- 30.Hirano Y, Ohniwa RL, Wada C, Yoshimura SH, Takeyasu K. Human small G proteins, ObgH1, and ObgH2, participate in the maintenance of mitochondria and nucleolar architectures. Genes Cells. 2006;11:1295–1304. doi: 10.1111/j.1365-2443.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- 31.Lu B, Garrido N, Spelbrink JN, Suzuki CK. Tid1 isoforms are mitochondrial DnaJ-like chaperones with unique carboxyl termini that determine cytosolic fate. J. Biol. Chem. 2006;281:13150–13158. doi: 10.1074/jbc.M509179200. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann M, Bellance N, Rossignol R, Koopman WJ, Willems PH, Mayatepek E, Bossinger O, Distelmaier F. C. elegans ATAD-3 is essential for mitochondrial activity and development. PLoS One. 2009;4:e7644. doi: 10.1371/journal.pone.0007644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogenhagen DF, Wang Y, Shen EL, Kobayashi R. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol. Cell. Proteomics. 2003;2:1205–1216. doi: 10.1074/mcp.M300035-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Merkwirth C, Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta. 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 36.Chung HK, Yi YW, Jung NC, Kim D, Suh JM, Kim H, Park KC, Song JH, Kim DW, Hwang ES, et al. CR6-interacting factor 1 interacts with Gadd45 family proteins and modulates the cell cycle. J. Biol. Chem. 2003;278:28079–28088. doi: 10.1074/jbc.M212835200. [DOI] [PubMed] [Google Scholar]
- 37.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasashima K, Sumitani M, Satoh M, Endo H. Human prohibitin 1 maintains the organization and stability of the mitochondrial nucleoids. Exp. Cell Res. 2008;314:988–996. doi: 10.1016/j.yexcr.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 39.Strunk BS, Loucks CR, Su M, Vashisth H, Cheng S, Schilling J, Brooks CL, 3rd, Karbstein K, Skiniotis G. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs HT, Fernández-Ayala DJ, Manjiry S, Kemppainen E, Toivonen JM, O'Dell KM. Mitochondrial disease in flies. Biochim. Biophys. Acta. 2004;1659:190–196. doi: 10.1016/j.bbabio.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Di Re M, Sembongi H, He J, Reyes A, Yasukawa T, Martinsson P, Bailey LJ, Goffart S, Boyd-Kirkup JD, Wong TS, et al. The accessory subunit of mitochondrial DNA polymerase gamma determines the DNA content of mitochondrial nucleoids in human cultured cells. Nucleic Acids Res. 2009;37:5701–5713. doi: 10.1093/nar/gkp614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT, von Kleist-Retzow JC, Waisman A, Westermann B, Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22:476–488. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sondheimer N, Fang JK, Polyak E, Falk MJ, Avadhani NG. Leucine-rich pentatricopeptide-repeat containing protein regulates mitochondrial transcription. Biochemistry. 2010;49:7467–7473. doi: 10.1021/bi1008479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirum-Connolly K, Mason TL. Functional requirement of a site-specific ribose methylation in ribosomal RNA. Science. 1993;262:1886–1889. doi: 10.1126/science.8266080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.