Abstract
Tyrosine family recombinases (YRs) are widely utilized in genome engineering systems because they can easily direct DNA rearrangement. Cre recombinases, one of the most commonly used types of YRs, catalyze site-specific recombination between two loxP sites without the need for high-energy cofactors, other accessory proteins or a specific DNA target sequence between the loxP sites. Previous structural, analytical ultracentrifuge and electrophoretic analyses have provided details of the reaction kinetics and mechanisms of Cre recombinase activity; whether there are reaction intermediates or side pathways involved has been left unaddressed. Using tethered particle motion (TPM), the Cre-mediated site-specific recombination process has been delineated, from beginning to end, at the single-molecule level, including the formation of abortive complexes and wayward complexes blocking inactive nucleoprotein complexes from entering the recombination process. Reversibility in the strand-cleavage/-ligation process and the formation of a thermally stable Holliday junction intermediate were observed within the Cre-mediated site-specific recombination process. Rate constants for each elementary step, which explain the overall reaction outcomes under various conditions, were determined. Taking the findings of this study together, they demonstrate the potential of single-molecule methodology as an alternative approach for exploring reaction mechanisms in detail.
INTRODUCTION
Site-specific recombinases are categorized into the tyrosine family (YR) and the serine family (SR). These site-specific recombinases can integrate viral genomes into and excise them from host chromosomes. Moreover, they play a role in resolving multimeric chromosomes to ensure faithful partitioning, to maintain the correct 2µ plasmid copy number, and to turn a specific gene on or off (1–3). These recombinases possess the capability to catalyze the exchange of DNA strands, but their protein sequences do not share significant similarity (4). Based on sequence alignment, only five amino acid residues are conserved in the catalytic domains of YRs (5). It has been suggested that the similar catalytic properties of YRs are a result of their high degree of structural conservation, a similar distribution of α-helices and β-sheets and the conservation of residues within the catalytic domain (6). Unlike other YR family recombinases that require accessory proteins and their cognate binding sites, Cre-loxP and Flp-FRT have become useful engineering tools because of their simplicity (4). Both Cre-loxP and Flp-FRT have been widely used to generate conditional knockouts to study gene function in vivo (7,8).
YRs have been extensively studied using a variety of experimental approaches. For example, the recombination process mediated by λ-Int att and Cre-loxP has been studied using electron microscopy (9,10), electrophoretic mobility shift assay (EMSA) (11–13), atomic force microscopy (14,15), analytical ultracentrifuge (AUC) (12), structural analysis (13,16,17), surface plasmon resonance (SPR) (18) and single-molecule detection (19,20) analyses. Although Cre-loxP is the simplest system within the YR family that can catalyze excisions, inversions and translocations, the reaction pathway involved is complicated. It has been proposed that the recombination process consists of four main stages. These stages are the following: first, two recombinase monomers sequentially bind to half of an inverted repeat sequence (21–23); second, two of these nucleoprotein complexes (presynaptic complexes) form one synaptic complex via a Cre–Cre interaction (13,24–26); third, strand cleavage and relegation are executed twice, which is accompanied by a conformational change in the recombinase molecules (4); and, finally, the nucleoprotein complex decomposes into separate recombinases and recombinant products (27,28). Although the kinetics of each step have been extensively studied using EMSA (29–31), AUC (12) and SPR (18), whether there are other side pathways and intermediates remains unknown.
Recent single-molecule experiments performed by Mumm et al. (19) using tethered particle motion (TPM) have offered new insights into λ Int-mediated site-specific recombination events from start to finish. These experiments have shown that the formation of stable λ Int–DNA complexes requires both integrase and the accessory proteins integration host factor and excisionase. It was also observed that there are side pathways and different nucleoprotein complexes present that are able to prevent inactive synaptic complexes from entering the catalytic recombination process. TPM has been demonstrated to be a powerful method that can discriminate DNA length topological changes with a resolution of a few base pairs (32,33). In this study, TPM was adapted to monitor the interaction between an individual DNA molecule containing two repeated loxP sites and Cre recombinase from beginning to end. This detailed kinetic analysis explains the mechanistic events involved and their causes under different reaction conditions.
MATERIALS AND METHODS
Proteins and DNA substrates
The phage P1 site-specific recombinase Cre was purchased from New England Biolabs (NEB) and used without further purification. Two Cre mutants, Y324F and K201A, gifts from Professor Van Duyne, were expressed and purified as previously described (34). Parallel and inverse loxP site plasmids were constructed with pFOS1 (NEB) using sequential restriction enzyme digestion (EcoRI and XhoI, NEB) according to the NEB instruction manual. Then, 1267 bp DNA sequences containing either parallel loxP sites or inverse loxP sites were prepared via PCR using the appropriate plasmid construct as a template and digoxigenin and biotin-labeled primers. Control DNA molecules with a size of 1267 bp without loxP sites were prepared via PCR using pBR322 (NEB) as the template. DNA sequences 1267 or 580 bp in length containing single loxP sites were obtained via PCR using pFOS1 as the template. The primer sequences used in these assays are listed in Table 1.
Table 1.
Primer sequences used to obtain the experimental DNA sequences
| DNA substrate | Template | Primer sequence |
|---|---|---|
| Parallel LoxP sites: 1267 dsDNA | pFOS1(parallel) | 5′-DigN-GACGTGGTTTGATGGCCTC |
| 5′-Bio-CAGGACTGTCATTTGAGGG | ||
| Inverse LoxP sites: 1267 dsDNA | pFOS1(inverse) | 5′-DigN-GACGTGGTTTGATGGCCTC |
| 5′-Bio-CAGGACTGTCATTTGAGGG | ||
| Single LoxP site: 1267 ds DNA | pFOS1 | 5′-DigN-GACGTGGTTTGATGGCCTC |
| 5′-Bio-AATAAGATCACTACCGGG | ||
| 1267 dsDNA | pBR322 | 5′-DigN-CGTCACCCTGGATGCTGTAG |
| 5′-Bio-CGATGGATATGTTCTGC | ||
| 580 dsDNA | pFOS1(parallel) | 5′-DigN-TGGTGAAAACGGGGGCG |
| 5′-Bio-AGGACTGTCATTTGAGGG |
Single-molecule TPM measurements and data analysis
Streptavidin-coated polystyrene beads and the reaction chamber for these experiments were prepared as previously described (32). The Brownian motion (BM) of the tethered beads was observed using an inverted optical microscope (IX-71, Olympus, 100× objective with NA = 1.40) with a differential interference contrast (DIC) imaging system at 22°C. To enhance the contrast, a mercury lamp was used. Experimental images were acquired with an analog camera (Newvicon-70, frame interval: 33 ms) using custom software written in Labview. The centroid positions of chosen tethered beads were determined via a 2D Gaussian distribution. The subsequent 40 frames were used to calculate the x–y distribution scattering by means of the standard deviation (SD) to represent the BM amplitude. The 40-frame averaging window resulted in a time resolution of 1.33 s under our experimental setup. The data analysis followed previously described procedures (32). The relationship between the BM amplitude and the tethered DNA length is linear within ∼1.5 kbp using a 40-frame averaging window, as shown in Supplementary Figure S1. The resolution in the length determination for a 1267 bp DNA sequence with a 40-frame averaging window is ∼150 bp at a 95% confidence level (CL). A BM time trace was smoothed with a five-point adjacent averaging algorithm. Only molecules with a BM amplitude between 70 and 91 nm with a 40-frame averaging window prior to Cre addition were chosen as bare 1267 bp DNA molecules and were then used for further analyses. Molecules that stuck to the glass surface (with a BM of < 26 nm) for more than 5 data points or that exhibited distorted movement (the ratio of the BM for the x coordinate to the BM for the y coordinate is outside the range of 0.8–1.2) at any time were excluded from the analyses. The dwell time distribution histogram is fitted with either a single exponential or a bi-exponential decay algorithm with the following formulas:
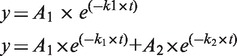 |
A1, A2, k1 and k2 are the fitting parameters determined by Origin 8.0.
DNA molecules containing both parallel and inverse loxP sites were labeled with digoxigenin and biotin at their 5′-ends. The DNA molecules were immobilized on an anti-digoxigenin-coated coverslip; then, the other end of each DNA molecule was tethered to streptavidin-coated 200 nm polystyrene beads. Cre-mediated site-specific recombination was initiated by adding the complete reaction solution containing 50 nM Cre prepared in reaction buffer (50 mM Tris–HCl, pH = 7.5, 33 mM NaCl, 10 mM MgCl2, 5 mM dithiothreitol and 1 mg/ml BSA). The recombination reaction was quenched with a 0.05% sodium dodecyl sulfate (SDS) solution after 30 min of reaction time. To verify the divalent ion, pH and salt concentration dependence of the recombination reaction, it was performed under different reaction conditions (Ca2+ replacement, divalent ion deprivation, high salt and high pH). All experiments were performed at room temperature (22°C).
RESULTS
The Cre-mediated site-specific recombination targeting parallel loxP sites
It has been shown that the Cre recombinase specifically binds to the loxP site and that formation of the synaptic complex includes multiple initiation steps (13,24–26). Once bound to the loxP sites, the Cre recombinase molecules bend the DNA strand sharply. Additional protein–protein interactions between the Cre recombinase molecules then stabilize the synaptic complex. Because the binding event between Cre recombinase and the loxP sites, which is followed by the inversion/excision processes, leads to apparent DNA length changes, a single-molecule TPM experiment was developed in this study to monitor Cre-mediated site-specific recombination in detail. Previous studies have demonstrated that TPM is a powerful tool that can be used to visualize DNA length and topological changes (32,33). Both continuous DNA length changes and conversions between different topological states can be monitored using the nearly force-free TPM method (35–39). In the present study, the BM amplitude of the 1267 bp DNA molecules without loxP sites (81.6 ± 8.6 nm, n = 53, as shown in Figure 1A) was measured initially, which served as a control. Only a small proportion of molecules, 9.4%, exhibited a decrease in BM amplitude after a 1-h incubation with 200 nM Cre recombinase, indicating a weak, non-specific interaction between Cre and the DNA (81.2 ± 9.6 nm and 62.9 ± 11.5 nm, n = 53), which is shown in Figure 1B as well as the BM time course (Figure 1C). Next, a similar experiment was performed on the 1267 bp DNA molecules containing a single loxP site (80.5 ± 12.0 nm, n = 85, as shown in Figure 1D). Only 15.3% of the DNA molecules showed a decrease in BM amplitude in response to Cre recombinase (80.5 ± 12.5 nm and 58.3 ± 9.4 nm, n = 85, Figure 1E). The 1267 bp DNA molecules with a single loxP site were analyzed under the same scenario; the BM time course seldom exhibited any change during the 30 min of reaction time (Figure 1F). It is expected that the interaction between Cre recombinase and DNA should lead to a decrease in DNA length because it has been reported that the overall bending of the DNA strand in the Cre-loxP synaptic complex occurs at an angle of ∼100° between the DNA arms (13). However, the BM histogram and BM time trace showed that the single interaction of Cre-loxP is too small to be discriminated by TPM because the resolution in the length determination for 1267 bp DNA molecules with a 40-frame averaging window is ∼150 bp in the range of the 95% CL (Supplementary Figure S1).
Figure 1.
Tethered particle motion experiment investigating the specific interaction between Cre recombinase and loxP sites. (A) Control experiment: the histogram of the BM amplitude was obtained using 1267 bp DNA molecules without loxP sites. The average BM value peaks at 81.6 ± 8.6 nm (n = 53). (B) A histogram of the BM amplitude obtained from 1267 bp DNA molecules without a loxP site after a 1-h incubation with 200 nM Cre recombinase. The histogram was fitted to a two-Gaussian distribution that represented two conditions, in which the centers of the peaks occur at BM values of 81.2 ± 9.6 nm and 62.9 ± 11.5 nm for naked DNA and Cre-bound DNA, respectively (n = 53). (C) The change in the BM amplitude of a single 1267 bp DNA molecule without a loxP site in response to the addition of Cre recombinase. The dashed line indicates the addition of Cre recombinase. (D) Control experiment: the histogram of the BM amplitude was obtained using 1267 bp DNA molecules containing one loxP site. The average BM value peaks at 80.5 ± 12.0 nm (n = 85). (E) A histogram of the BM amplitude obtained from 1267 bp DNA molecules containing one loxP site after 1-h incubation with 200 nM Cre recombinase. The histogram was fitted to a two-Gaussian distribution that represented two conditions, in which the centers of the peaks occur at BM values of 80.5 ± 12.5 and 58.3 ± 9.4 nm for naked DNA and Cre-bound DNA, respectively (n = 85). (F) The change in the BM amplitude of a single 1267 bp DNA molecule containing one loxP site in response to the addition of Cre recombinase. The dashed line indicates the addition of Cre recombinase. The cartoons shown below illustrate the experimental procedure. The white ball represents the streptavidin-labeled 200 nm polystyrene bead, the white spots represent the Cre recombinase molecules, the black curve represents the DNA molecules, the dashed arrowed line represents the Brownian motion amplitude, and the white line represents the loxP sequence. The error is the 95% CL.
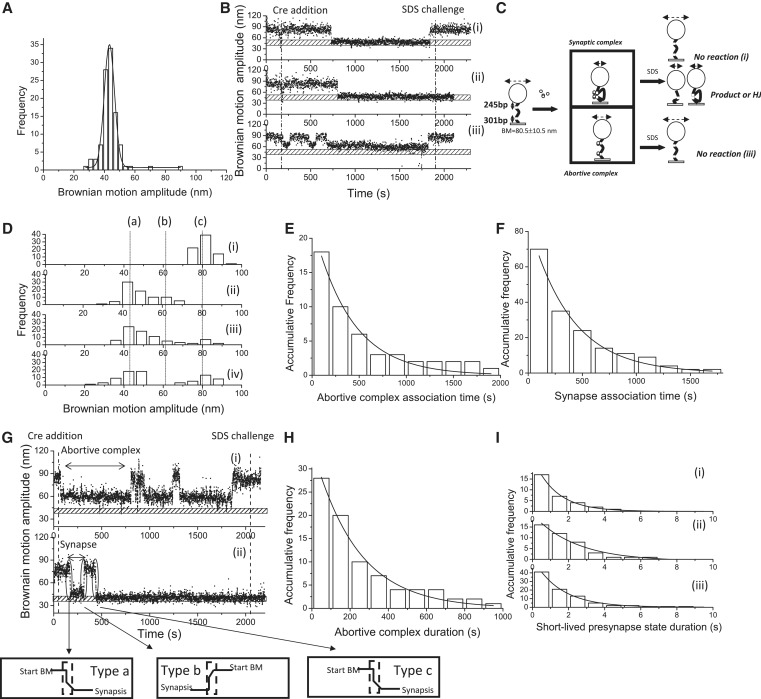
Next, the 1267 bp DNA molecules containing two parallel loxP sites separated by 653 bp were investigated to explore excision recombination. Previous studies have suggested that two Cre recombinases bind to one of the loxP sites, which consists of two recombinase-binding elements (RBEs), and that this process occurs with a high level of cooperation to form the presynaptic complex (13,22). One such Cre-loxP presynaptic complex then interacts with another Cre-loxP presynaptic complex to form a recombinant synaptic complex (13,26). The BM value of the excision product obtained using the 580 bp DNA molecule was determined (43.3 ± 6.8 nm, n = 103, Figure 2A). Examples of single-molecule time traces are shown in Figure 2B(i)–(iii). To confirm whether recombination proceeded or not, the DNA molecules were challenged with 0.05% SDS to disrupt non-covalent Cre–loxP interactions. The change in the BM amplitude of the DNA molecules containing parallel loxP sites can be categorized as either a synapse state or an abortive complex state which is deficient at the Cre–Cre interaction interface in response to the Cre recombinase. After the addition of SDS, there are two possible outcomes for molecules that have entered the synapse state. These outcomes are, first, a return to the original DNA substrate for the molecules that failed to proceed with the catalytic recombination process, as depicted in Figure 2C(i), and second, remaining at a short DNA length status, which corresponds to either the excision recombinant product or a stable Holliday junction intermediate, as depicted in Figure 2C(ii). In contrast, those molecules trapped at abortive complexes eventually returned to the original DNA substrate before the SDS challenge, as depicted in Figure 2C(iii). Among the 143 molecules analyzed with a starting BM amplitude between 70 and 91 nm based on a 40-frame averaging window, only 76 molecules showed a significant change in the BM time trace in response to the addition of Cre recombinase. Among these 76 molecules, 15 molecules showed a decrease in the BM amplitude to the average value of 59.3 ± 6.8 nm within the 95% CL and failed to synapse within the duration of the experiment [Figure 2B(iii) and 2D(ii)]. This magnitude of decrease in the BM amplitude is different from that observed for the excision recombinant product obtained using 580 bp DNA molecules. This scenario indicates the formation of abortive complexes in which two loxP sites are occupied, but with a defect at the Cre–Cre interaction interface. The remainder of the molecules showed a decrease in the BM amplitude to an average value of 43.8 ± 8.7 nm within the 95% CL, similar to that of the expected excision recombinant product, a 580 bp DNA molecule. This observation indicates the formation of synaptic complexes or the excision recombinant product [Figure 2D(ii)]. After 30 min of reaction time, the BM of the DNA molecules that failed to proceed in the catalytic recombination process returned to the starting BM value after the SDS challenge [Figure 2D(iv)]. However, the BM value of some DNA molecules (71.6%) remained low after the SDS challenge, indicating the formation of either excised products or a stable Holliday junction intermediate [Figure 2D(iv)]. The dwell times between the addition of Cre and the shortening of the DNA molecules containing parallel loxP sites were classified into two groups: one for molecules that will be trapped in an abortive complex and the other for molecules that will form the synaptic complex quickly with a short-lived presynaptic complex. Then, the dwell time histograms were fitted to a single exponential model (Figure 2E and 2F) with association rate constants of (6.8 ± 0.4) × 104 M−1 s−1 and (6.1 ± 0.6) × 104 M−1 s−1 for the formation of an abortive complex and the synapse state, respectively. A total of 15 of 76 DNA molecules containing parallel loxP sites were trapped in the abortive complex [Figure 2G(i)]. The distribution of the durations of abortive complexes was fitted to a single exponential model, and an abortive complex dissociation rate constant of (5.2 ± 1.0) × 10−3 s−1 was obtained (Figure 2H). For those molecules exhibiting BM transitions from the original substrate to the synapse state [Figure 2G(ii)], leading to unsuccessful recombination, the duration of the short-lived presynapse state is classified as type a, representing the formation of unstable synaptic complexes. For molecules exhibiting BM transitions from the synapse state to the original substrate, the duration of the short-lived presynapse state is classified as type b, representing the dissociation of short-lived presynaptic complexes. For molecules presenting decreased BM, even after an SDS challenge, indicating the formation of stable synaptic complexes, the duration of the short-lived presynapse state is classified as type c, which represents the formation of stable synaptic complexes. As a result, all the durations of the short-lived presynapse states were categorized and used to construct separate histograms with a bin size of 1 s [Figure 2I(i–iii)]. The histograms were fitted to a single exponential model with rate constants of (7.7 ± 0.1) × 10−1 s−1, (6.6 ± 0.4) × 10−1 s−1 and (4.2 ± 0.7) × 10−1 s−1, representing the formation of unstable synaptic complexes, the decomposition of unstable presynaptic complexes and the formation of stable synaptic complexes of DNA molecules that successfully synapse during the experimental period, respectively.
Figure 2.
Complex formation and recombination events of 1267 bp DNA molecules containing parallel loxP sites. (A) A histogram of the BM amplitude obtained from the 580 bp DNA molecules representing the recombinant products. The average BM value peaks at 43.3 ± 6.8 nm (n = 103). (B) (i)–(iii) The change in the BM amplitude of the 1267 bp DNA molecules containing parallel loxP sites in response to the addition of Cre recombinase. (i) An example of a molecule that synapsed but failed to continue through the recombination process. (ii) An example of a molecule that synapsed and continued through the strand-cleavage step to the strand-migration step and then to the strand-ligation steps. (iii) An example of a molecule that failed to synapse within the duration of the observations. The dashed lines indicate the addition of Cre recombinase and 0.05% SDS after 30 min of incubation time. The bar with the punctuate pattern labels the average BM value of the expected recombinant excision product. (C). Reaction scheme of the Cre-mediated site-specific recombination process involving 1267 bp DNA molecules containing parallel loxP sites. The initial substrate is the 1267 bp DNA molecule containing parallel loxP sites and has an average BM amplitude of 80.5 ± 10.5 nm. Two complexes are formed after interaction with Cre recombinase. The first is the synaptic complex that corresponds to the formation of a Cre tetramer with an average BM value of 43.8 ± 8.7 nm. The other complex is the abortive complex that failed to synapse within the duration of the observations, corresponding to the association of Cre dimers with loxP sites with an average BM value of 59.3 ± 6.8 nm. The reaction was stopped when 30 μl of 0.05% SDS was added after 30 min of incubation time. Three different outcomes were observed: (i) a molecule that synapsed but failed to complete the recombination process; (ii) a molecule that synapsed and formed either the recombinant product or a stable Holliday junction intermediate; and (iii) a molecule that failed to synapse within the duration of the observations. (D) (i) The distribution of the BM amplitude before the addition of Cre recombinase, which shows an average value of 80.5 ± 10.5 nm, is indicated with the letter (c). (ii) The distribution of the BM amplitude in response to the addition of Cre recombinase [59.3 ± 6.8 nm and 43.8 ± 8.7 nm for the abortive complex state and the synapse state, marked with (b) and (a), respectively]. (iii) The distribution of the BM amplitude after 30 min of incubation time following the addition of Cre recombinase. (iv) The distribution of the BM amplitude in response to the SDS challenge after 30 min of incubation time (n = 76). The distribution of the dwell times between recombinase addition and the change in the BM amplitude (E) to a value representing the abortive complex state and the association rate constant of (6.8 ± 0.4) × 104 M−1 s−1 (R2 = 0.96, n = 16) were obtained. (F) To a value representing the synapse state, and an association rate constant of (6.1 ± 0.6) × 104 M−1 s−1 (R2 = 0.99 n = 70) was obtained. The data were fitted to a single exponential decay algorithm (Origin 8.0). The N mentioned above is the number of molecules that were observed. The error is within the 95% CL. (G) BM time-trace of a 1267 bp DNA molecule containing parallel loxP sites in response to the addition of Cre recombinase. The dashed lines indicate the addition of Cre recombinase and the addition of 0.05% SDS after 30 min of incubation time. The bar with the punctuate pattern indicates the average BM value of the expected excision recombinant product. (i) The BM time trace of a molecule that has successfully formed a synaptic complex following a short-lived presynapse state. (ii) The BM time trace of a molecule that failed to synapse and was trapped in the abortive complex state. For those molecules undergoing BM transitions from the original substrate to the synapse state, leading to an unsuccessful recombination, the short-lived presynapse state duration is classified as type a. For molecules that experience BM transitions from the synapse state to the original substrate, the short-lived presynapse state duration is classified as type b. For those molecules showing decreased BM, even after the SDS challenge, indicating the formation of a stable synapse state, the short-lived presynapse state duration is classified as type c. (H) The dwell times in the abortive complexes formed with the DNA molecules containing parallel loxP sites that failed to synapse during observation were pooled and then fitted to a single exponential decay [R2 = 0.98, τ1 = (5.2 ± 1.0) × 10−3 s−1, n = 26]. (I) (i)–(iii) The dwell times in the short-lived presynapse state were pooled separately to build dwell-time histograms fitted to a single exponential decay algorithm with values of (7.7 ± 0.1) × 10−1 s−1 (n = 17, R2 = 0.99), (6.6 ± 0.4) × 10−1 s−1 (n = 16, R2 = 0.97) and (4.2 ± 0.7) × 10−1 s−1 (n = 40, R2 = 0.99), representing the association rate constant of unstable synaptic complexes, the dissociation rate constant of presynaptic complexes and the association rate constant of correct synaptic complexes, respectively. The N mentioned above is the number of events observed. The error is within a 95% CL.
The Cre-mediated site-specific recombination targeting inverse loxP sites
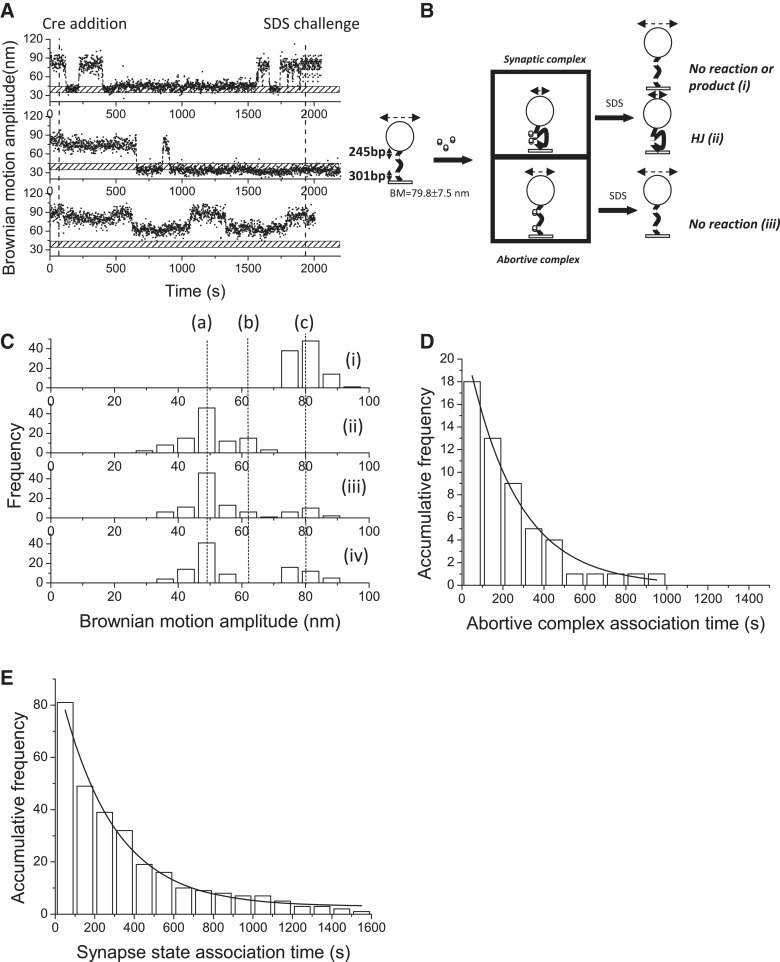
To distinguish whether Holliday junction intermediates or excision products are formed within the 30-min incubation time, the same experiments were performed on 1267 bp DNA molecules containing inverse loxP sites separated by 653 bp. The recombination process in the DNA molecules containing inverse loxP sites results in an inverted 653 bp DNA segment between the two sites and an unchanged DNA length for the recombinant product. Examples of single-molecule time traces are shown in Figure 3A(i)–(iii). For molecules that have proceeded to the synapse state, there are two possible outcomes following an SDS challenge: first, a return to the original DNA substrate or the formation of an inverse recombinant product [Figure 3B(i)], and second, remaining as a shortened DNA product, representing the stable Holliday junction intermediate [Figure 3B(ii)]. Molecules that are trapped at the abortive complex will return to the original DNA substrate prior to the SDS challenge. Of the 301 analyzed molecules with a starting BM value of 79.8 ± 7.5 nm, 101 molecules exhibited a change in their BM amplitude in response to the addition of Cre recombinase. Of the 101 analyzed molecules, 18 showed a decrease in BM amplitude to an average value of 64.3 ± 3.6 nm within the 95% CL and failed to synapse within the experimental duration [Figure 3A(iii) and 3C(ii)], indicating the formation of abortive complexes. When the remaining analyzed molecules were examined, the BM amplitude was found to decrease rapidly to an average value of 48.6 ± 10.2 nm within the 95% CL, indicating the formation of synaptic complexes [Figure 3C(ii)]. The BM amplitude of the synapse state for the DNA molecules containing inverse loxP sites is different from that of DNA molecules containing parallel loxP sites. The discrepancy in the BM amplitude between the synapse states occurs because the two loxP sites, when in the correct synapse state, are expected to be in an anti-parallel orientation. This constraint results in extra topological bending of DNA molecules containing inverse loxP sites (13,16,40). After 30 min of incubation with Cre recombinase, the percentage of DNA molecules in the abortive complex decreased because they were returning to the original DNA substrate state [Figure 3C(iii)]. Immediately upon the SDS challenge at 30 min reaction time, the BM amplitude of most of the DNA molecules (90%) remained at a low value of 48.6 ± 10.2 nm, indicating the formation of stable Holliday junction intermediates. Similar association constants of (8.1 ± 0.5) × 104 M−1 s−1 and (6.4 ± 0.3) × 104 M−1 s−1 for the abortive complexes and synaptic complexes, respectively, were calculated by fitting the dwell histograms to a single exponential model (Figure 3D and E).
Figure 3.
Complex formation and recombination events of 1267 bp DNA molecules containing inverse loxP sites. (A) (i)–(iii) The change in the BM amplitude of the 1267 bp DNA molecules containing inverse loxP sites in response to the addition of Cre recombinase. (i) An example of a molecule that synapsed and then returned to the original substrate, corresponding to either no reaction or completion of recombination. (ii) An example of a molecule that synapsed and was then trapped in a stable Holliday junction intermediate. (iii) An example of a molecule that failed to synapse within the duration of the observations. The dashed lines indicate the addition of Cre recombinase and 0.05% SDS after 30 min of incubation time. The bar with the punctuate pattern indicates the average BM value of the expected excision recombinant product. (B) Reaction scheme of the Cre-mediated site-specific recombination process involving 1267 bp DNA molecules containing inverse loxP sites. The starting substrates are 1267 bp DNA molecules containing inverse loxP sites and exhibit an average BM amplitude of 79.8 ± 7.5 nm. Two complexes are formed after interaction with Cre recombinase. The first complex is the synaptic complex, which corresponds to the formation of a Cre tetramer with an average BM value of 48.6 ± 10.2 nm. The other complex is the abortive complex that failed to synapse within the duration of the observations and corresponds to the association of Cre dimers with loxP sites with an average BM value of 64.3 ± 3.6 nm. The reaction was stopped when 30 μl of 0.05% SDS was added after 30 min of incubation time. Three different outcomes were observed: (i) a molecule that synapsed but either failed to complete recombination or formed the recombinant product after completion of the recombination process; (ii) a molecule that synapsed but was trapped in a stable Holliday junction intermediate; and (iii) a molecule that failed to synapse within the duration of the observations. (C) (i) The distribution of the BM amplitude before the addition of Cre recombinase showed an average value of 79.8 ± 7.5 nm, which is indicated with (c). (ii) The distribution of the BM amplitude in response to the addition of Cre recombinase (64.3 ± 3.6, 48.6 ± 10.2 nm for the abortive complex state and the synapse state, marked with (b) and (a), respectively). (iii) The distribution of the BM amplitude after 30 min of incubation time following the addition of Cre recombinase. (iv) The distribution of the BM amplitude in response to the SDS challenge after 30 min of incubation time (n = 101). The distribution of the dwell times between recombinase addition and the change in the BM amplitude (D) to a value representing the abortive complex state and an association rate constant of (8.1 ± 0.5) × 104 M−1 s−1 (R2 = 0.98, n = 18) were obtained. (E) To a value that represents the synapse state, and an association rate constant of (6.4 ± 0.3) × 104 M−1 s−1 (R2 = 0.98, n = 81) was obtained. The data were fitted to a single exponential decay algorithm (Origin 8.0). The N mentioned above is the number of molecules that were observed. The error is within the 95% CL.
Kinetic analysis and the detailed reaction mechanism of Cre-mediated site-specific recombination
For the 1267 bp DNA molecules containing parallel loxP sites, 25 of 76 molecules showed fluctuating BM transitions between the synapse state and the original substrate within the observation period (Figure 4A). Because excised DNA molecules are the expected recombinant products of DNA molecules containing parallel loxP sites, the synapse state duration is correlated with the stability of unstable synaptic complexes, i.e. wayward complexes. Therefore, the dwell time distribution can be interpreted in terms of single exponential decay. Similar fluctuation phenomena were observed for DNA molecules containing inverse loxP sites; the synapse state duration reflects both the stability of the wayward complex and the catalytic reactivity of recombination. Thus, in these circumstances, the synapse state duration distribution can be interpreted in terms of a bi-exponential model. Surprisingly, the duration histograms for both the parallel and inverse loxP site-containing DNA molecules could be fitted to a bi-exponential model, and similar rate constants were obtained (Figure 4B and Supplementary Figure S2 fitted via least square and maximum likelihood estimation, respectively). For the Cre-parallel loxP sites DNA system, the bi-exponential decay behavior can only be observed when the recombination process, i.e. strand cleavage/ligation, is reversible. In our experiments, the Holliday junction intermediate was very stable and served as a reaction trap, and the reaction could go forward, to form excision recombinant products, or go backward to the original substrates with an equal probability. The fast reaction process, with rate constants of (2.7 ± 0.5) × 10−2 s−1 and (2.7 ± 0.6) × 10−2 s−1 for DNA molecules containing parallel and inverse loxP sites, respectively, represents the dissociation rate constant of the wayward complex. The slow reaction process, with rate constants (2.0 ± 0.1) × 10−3 s−1 and (2.2 ± 0.2) × 10−3 s−1 for the DNA molecules containing parallel and inverse loxP sites, respectively, represents the catalytic reactivity of recombination.
Figure 4.
Kinetic analysis of the dwell time in the synapse state. (A) The BM time-trace of a 1267 bp DNA molecule containing parallel loxP sites in response to the addition of Cre recombinase. The dashed lines indicate the addition of Cre recombinase and 0.05% SDS after 30 min of incubation time. The bar with the punctuate pattern indicates the average BM value of the expected excision recombinant product. (B) (i) The distribution of the dwell time in the synapse state, indicated by the double-arrowed line in the above time trace, for DNA molecules containing parallel loxP sites was pooled to construct a histogram, and the rate constants were obtained by fitting to either a single exponential decay algorithm [R2 = 0.91, k1 = (4.4 ± 0.5) × 10−3 s−1, n = 118] or a bi-exponential decay algorithm [R2 = 1.00, k1 = (2.7 ± 0.5) × 10−2 s−1, A1 = 0.80; k2 = (2.0 ± 0.1) × 10−3 s−1, A2 = 0.20, n = 118]. Solid and dashed lines are used to represent single and bi-exponential fitting curves, respectively. (ii) The distribution of the dwell time in the synapse state, indicated by the double-arrowed line in the above time trace, for DNA molecules containing inverse loxP sites was pooled to construct a histogram, and the rate constants were obtained by fitting to either a single exponential decay algorithm [R2 = 0.91, k1 = (6.5 ± 0.9) × 10−3 s−1, n = 178] or a bi-exponential decay algorithm [R2 = 1.00, k1 = (2.7 ± 0.6) × 10−2 s−1, A1 = 0.85; k2 = (2.2 ± 0.2) × 10−3 s−1, A2 = 0.15, n = 178]. Solid and dashed lines are used to represent single and bi-exponential fitting curves, respectively. The N mentioned above is the number of events observed. The error is within the 95% CL.
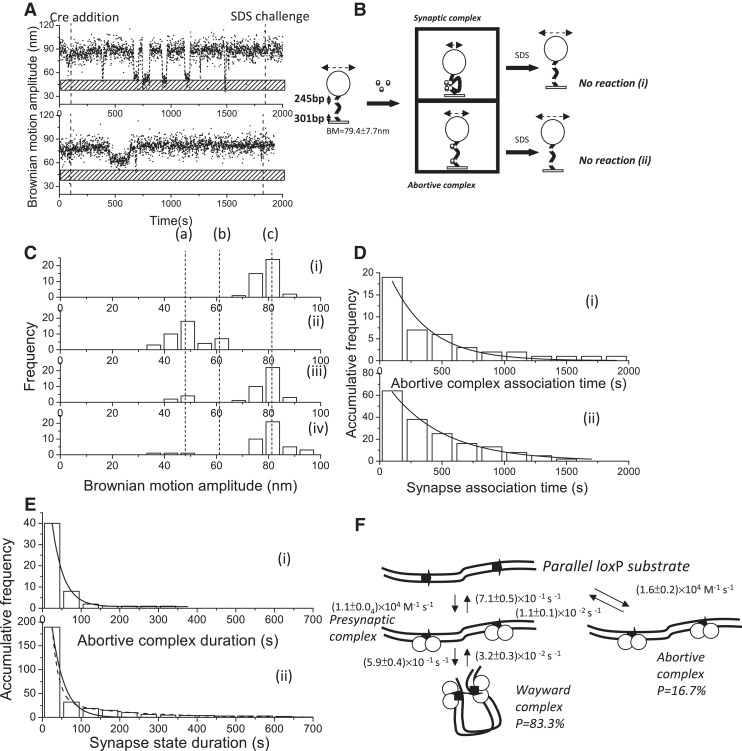
To verify whether the synapse state duration for the DNA molecules containing two loxP sites reflects both the decomposition of the wayward complex and the completion of recombination and whether cleavage is required for the formation of stable synaptic complexes, a Cre mutant (Y324F) was used to perform the same experiments and analyses. As expected, Tyr 324 is necessary for recombination because it serves as a nucleophile that attacks the phosphodiester bond within the active site of the synaptic complexes. Based on a previous study, it was noted that none of the 19 replacement residues showed any detectable recombination activity (41). Moreover, it has been shown that subsequent DNA cleavage by Int, a membrane of the tyrosine site-specific recombinases, will stabilize the synaptic complex; otherwise, the synaptic complex will easily decompose (19). Typical BM time traces are shown in Figure 5A(i–ii). The anticipated reaction scheme is depicted in Figure 5B, where the change in BM amplitude in response to CreY324F can be classified into either a synaptic complex or an abortive complex. As expected, the deficiency in strand cleavage will lead to no recombinant product being formed, as verified by the SDS challenge [Figure 5B(i)], and those molecules that formed abortive complexes will eventually return to the original DNA substrate [Figure 5B(ii)]. Among the 85 molecules with a starting BM amplitude between 70 and 91 nm, 42 molecules exhibited a significant change in the BM amplitude in response to the addition of CreY324F recombinase. A total of 7 of 42 molecules exhibited a decrease in the BM amplitude to an average value of 62.0 ± 4.2 nm and failed to synapse within the experimental period [95% CL, Figure 5C(ii)], indicating the formation of abortive complexes. For the remaining molecules, the BM amplitude decreased to an average value of 47.5 ± 9.4 nm, and a short-lived presynapse state was observed [95% CL, Figure 5C(ii)], indicating the formation of synaptic complexes. After 30 min of reaction time, most of the molecules returned to the original DNA substrates [Figure 5C(iii)]. After the SDS challenge, all the molecules were expected to return to the original DNA substrates because CreY324F lacks the ability to carry out strand cleavage. The fact that there were two molecules trapped at the lower BM value could be an experimental artifact or indicate purification contamination or sticking problems. With exception of the lower percentage of experimental artifacts, the TPM results shown here are consistent with those obtained by Gibb et al. (41). Smaller association rate constants of (1.6 ± 0.2) × 104 M−1 s−1 and (1.1 ± 0.0) × 104 M−1 s−1 for the formation of abortive complexes and synaptic complexes, respectively, were obtained by fitting the duration histograms to a single exponential model [Figure 5D(i–ii)]. An abortive complex dissociation rate constant of (1.1 ± 0.1) × 10−2 s−1 was obtained by fitting the abortive complex duration distribution to a single exponential model [Figure 5E(i)]. When the remaining molecules that formed a synaptic complex rapidly, with a short-lived presynapse state, were analyzed further, all of them (excluding experimental artifacts) exhibited fluctuating transitions between the synapse state and the original DNA substrate [Figure 5B(ii)]. The synapse duration histogram was fitted to a single exponential model with a rate constant of (3.2 ± 0.3) × 10−2 s−1, which represented the dissociation of wayward complexes. The kinetic reaction scheme of CreY324F-mediated recombination for DNA molecules containing parallel loxP sites is shown in Figure 5F. It can be inferred that CreY324F is deficient in the formation of a stable synaptic complex or lacks cleavage ability; therefore, the catalysis recombination pathway is blocked in the CreY324F-loxP system.
Figure 5.
Complex formation and recombination events of 1267 bp DNA molecules containing parallel loxP sites mediated by cleavage-deficient CreY324F. (A) (i)–(ii) A change in the BM amplitude of 1267 bp DNA molecules containing parallel loxP sites in response to the addition of CreY324F. (i) An example of a molecule that synapsed but failed to continue to the recombination process. (ii) An example of a molecule that failed to synapse within the duration of the observations. (B) The reaction scheme of the Cre-mediated site-specific recombination process in a 1267 bp DNA molecule containing parallel loxP sites. The initial substrate is a 1267 bp DNA molecule containing parallel loxP sites with an average BM value of 79.4 ± 7.7 nm. Two complexes are formed after the addition of Cre recombinase. The first complex is the synaptic complex, corresponding to the formation of a Cre tetramer with an average BM value of 47.5 ± 9.4 nm. The other complex is the abortive complex that failed to synapse within the duration of observations, corresponding to the association of Cre dimers with loxP sites, with an average BM value of 62.0 ± 4.2 nm. When 30 μl of 0.05% SDS was added after 30 min of incubation time, the reaction was stopped. Two different outcomes were observed: (i) a molecule that synapsed but lacked the strand-cleavage capability to complete recombination and returned to the original DNA substrate; and (ii) a molecule that failed to synapse within the duration of observations and eventually returned to the original DNA substrate. (C) (i) The distribution of the BM amplitude before the addition of Cre recombinase, with an average value of 79.4 ± 7.7 nm, which is indicated with (c). (ii) The distribution of the BM amplitude in response to the addition of Cre recombinase (62.0 ± 4.2 and 47.5 ± 9.4 nm for the abortive complex state and the synapse state, marked with (b) and (a), respectively). (iii) The distribution of the BM amplitude after 30 min of incubation time. (iv) The distribution of the BM amplitude in response to the SDS challenge after 30 min of incubation time (n = 42). The distribution of the dwell times between recombinase addition and the change in BM amplitude (D) to a value representing the abortive complex state and an association rate constant of (1.6 ± 0.2) × 104 M−1 s−1 (R2 = 0.96, n = 18) were obtained. (F) To a value that represents the synapse state, and an association rate constant of (1.1 ± 0.0) × 104 M−1 s−1 (R2 = 0.99, n = 64) was obtained. The data were fitted to a single exponential decay algorithm (Origin 8.0). The N mentioned above is the number of molecules that were observed. (E) (i) The dwell times in the abortive complex state for DNA molecules containing parallel loxP sites that failed to synapse during the observations were pooled and then fitted to a single exponential decay algorithm [R2 = 0.98, τ1 = (1.1 ± 0.1) × 10−2 s−1, n = 40]. (ii) The distribution of the dwell times in the synapse state was fitted to either a single exponential decay algorithm [R2 = 0.98, k1 = (3.2 ± 0.3) × 10−2 s−1, n = 189] or a bi-exponential decay algorithm [R2 = 1.00, k1 = (6.3 ± 0.5) × 10−2 s−1, A1 = 0.95; k2 = (5.7 ± 0.5) × 10−3 s−1, A2 = 0.05, n = 189]. The solid and dashed lines represent single and bi-exponential fitting curves, respectively. The N mentioned above is the number of events that were observed. The error is within the 95% CL. (F) The mechanism through which bacteriophage P1 CreY324F mediates site-specific recombination. The arrows on the DNA molecule indicate the parallel loxP sites. The empty circles represent Cre recombinases.
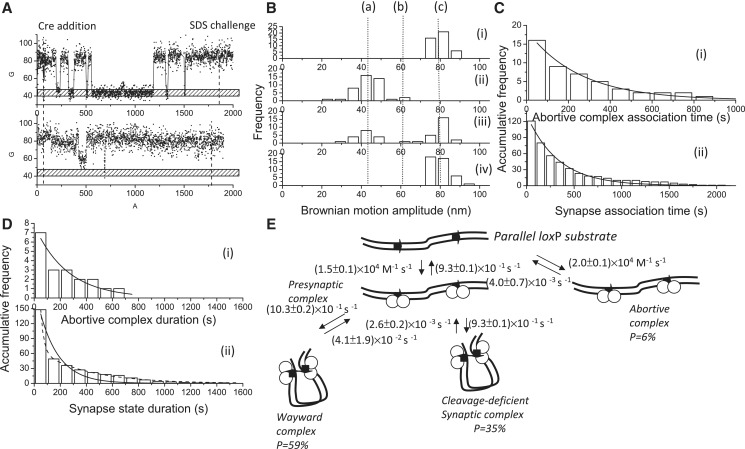
It has been known that the cleavage-deficient mutant CreK201A, from which the required general acid catalyst that activates O5' has been removed, possesses a similar synapsis ability as wild-type Cre, with Kd = 8.9 ± 0.6 nM (12). Therefore, CreK201A was used to perform the same experiments and analyses in order to verify whether cleavage is required for the formation of the stable synaptic complex in the Cre-loxP system. The BM time traces are shown in Figure 6A, and there are two typical reaction behaviors, similar to those of CreY324F. One reaction behavior is that a DNA molecule formed a synaptic complex, but there was no formation of a recombinant product verified by the SDS challenge, and the other is that a DNA molecule failed to synapse during the duration of the observations. Analysis of 59 molecules with a BM amplitude between 70 and 91 nm showed that 43 molecules exhibited a significant change in BM in response to the addition of CreK201A recombinase. The BM histograms are shown in Figure 6B, which are similar to those observed for CreY324F. Association rate constants of (2.0 ± 0.1) × 104 M−1 s−1 and (1.5 ± 0.1) × 104 M−1 s−1 for the formation of abortive complexes and synaptic complexes, respectively, were obtained by fitting the duration histograms to a single exponential model [Figure 6C(i–ii)]. The stability of the abortive complex was obtained by fitting the abortive complex duration histogram to a single exponential model with a dissociation rate constant of (4.0 ± 0.7) × 10−3 s−1 [Figure 6D(i)]. When the duration of the synaptic complex was analyzed further, it was found that the duration distribution was similar to that observed in the wild-type Cre-loxP system, and a bi-exponential decay model was used. The fast process, with a rate constant of (4.1 ± 1.9) × 10−2 s−1, represents the decomposition of the wayward complex. The same rate constants were obtained for both wild-type Cre and CreY324F, which reinforced the existence of a wayward complex, in the Cre-loxP system. On the other hand, it has been shown that the catalysis recombination process, i.e. strand cleavage/ligation, is reversible. Limited by the resolution of the TPM, the change in BM amplitude can only signal the decomposition of the synaptic complex, and the BM amplitudes observed for synaptic complexes, cleavage synaptic complexes and Holliday junction intermediates showed insignificant differences. Moreover, it has been reported that the rate-limiting step in the Cre-mediated recombination process is the dissociation of the recombined synapse (31). Therefore, the slow process observed in wild-type Cre and CreK201A, with a rate constant of (2.6 ± 0.2) × 10−3 s−1, represents the decomposition of the stable synaptic complex as well as the decomposition of the recombined synaptic complex.
Figure 6.
Complex formation and recombination events for 1267 bp DNA molecules containing parallel loxP sites mediated by cleavage-deficient CreK201A. (A) (i)–(ii) A change in the BM amplitude of 1267 bp DNA molecules containing parallel loxP sites in response to the addition of CreK201A. (i) An example of a molecule that synapsed but failed to continue to the recombination process. (ii) An example of a molecule that failed to synapse within the duration of the observations. (B) (i) The distribution of the BM amplitude before the addition of Cre recombinase, with an average value of 78.8 ± 10.2 nm, which is indicated with (c). (ii) The distribution of the BM amplitude in response to the addition of Cre recombinase (61.0 ± 1.3 nm and 44.0 ± 12.0 nm for the abortive complex state and the synapse state, marked with (b) and (a), respectively). (iii) The distribution of the BM amplitude at 30 min of incubation time. (iv) The distribution of the BM amplitude in response to the SDS challenge at 30 min of incubation time (n = 43). (C) The distribution of the dwell times between recombinase addition and the change in the BM amplitude to (i) a value that represented the abortive complex state and an association rate constant of (2.0 ± 0.1) × 104 M−1 s−1 (R2 = 0.98, n = 16) and to (ii) a value that represented the synapse state and an association rate constant of (1.5 ± 0.1) × 104 M−1 s−1 (R2 = 0.98, n = 121) were obtained. The data were fitted to a single exponential decay algorithm (Origin 8.0). The N mentioned above is the number of molecules observed. (D) (i) The dwell times in the abortive complex state for the DNA molecules containing parallel loxP sites that failed to synapse during the observations were pooled and then fitted to a single exponential decay algorithm [R2 = 0.98, τ1 = (4.0 ± 0.7) × 10−3 s−1, n = 7]. (ii) The distribution of the dwell times in the synapse state was fitted to either a single exponential decay algorithm [R2 = 0.98, k1 = (6.8 ± 0.8) × 10−3 s−1, n = 148] or a bi-exponential decay algorithm [R2 = 1.00, k1 = (4.1 ± 1.9) × 10−2 s−1, A1 = 0.90; k2 = (2.6 ± 0.2) × 10−3 s−1, A2 = 0.10, n = 148]. Solid and dashed lines represent single and bi-exponential fitting curves, respectively. The N mentioned above is the number of events observed. The error is within the 95% CL. (E) The mechanism through which bacteriophage P1 CreK201A mediates site-specific recombination. The arrows on the DNA molecule indicate the parallel loxP sites. The empty circles represent Cre recombinases. The arrows on the DNA molecule indicate the parallel loxP sites.
Effects of pH, divalent ions and monovalent ions on the Cre-mediated site-specific recombination process
Previous studies have shown that the Cre-loxP synapsis is pH dependent and that the presence of divalent ions or polyamines will increase the recombination efficiency by facilitating cooperative Cre-loxP binding (16,18,25). However, the causes of the occurrence of unstable synaptic complex breakdown at a high pH and how divalent ions facilitate the recombination process remain unclear (12). In this study, the potential of TPM to delineate the detailed kinetic steps involved in this process has been demonstrated, which allows a more quantitative understanding of the phenomena involved. To expand our knowledge regarding the effects of pH and ions, Cre-mediated site-specific recombination was studied using TPM under five different reaction conditions: in the presence of Mg2+, in the presence of Ca2+, in the absence of divalent ions, in the presence of a high concentration of monovalent ions and at a high pH. The Mg2+ condition was defined as the normal condition and has been explored above. The same experiments and analyses were then performed under the various conditions described above, and the resulting kinetic parameters are listed in Table 2. The recombination efficiency in these circumstances is defined as the ratio of the number of molecules forming either recombinant products or Holliday junction intermediates (with a BM amplitude of 43.8 ± 8.7 nm after the SDS challenge at 30 min of incubation time) to the number of molecules responding to Cre recombinase. The detailed kinetic analyses are presented in Supplementary Figures S3–S6.
Table 2.
Kinetic and thermodynamic information related to Cre-mediated site-specific recombination under different conditions
| Conditions | [NaCl] mM | [M2+] mM | pH | PAbortive (%) | PWayward (%) | kAssociation (M−1 s−1)a | kDissociation (s−1) | kshort-lived presynapse (s−1)b | ksynapse decomposition (s−1) | kRecombined synapse decomposition (s−1) | Efficiency (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal (n = 76) | 33 | 2 | 7.5 | 19.7 | 33.7 | (6.8 ± 0.4) × 104 | (5.2 ± 1.0) × 10−3 |
|
(2.7 ± 0.5) × 10−2 | (2.0 ± 0.1) × 10−3 | 71.6 |
| (6.1 ± 0.6) × 104 | |||||||||||
| Ca2+ (n = 37) | 33 | 2 | 7.5 | 18.2 | 31.8 | (5.9 ± 0.7) × 104 | (5.4 ± 0.7) × 10−3 |
|
(2.6 ± 0.4) × 10−2 | (2.0 ± 0.2) × 10−3 | 65.7 |
| (5.0 ± 0.3) × 104 | |||||||||||
| No divalent cations (n = 61) | 33 | 0 | 7.5 | 17.3 | 53.3 | (6.4 ± 0.5) × 104 | (8.0 ± 0.8) × 10−3 |
|
(2.2 ± 0.2) × 10−2 | (1.7 ± 0.1) × 10−3 | 37.1 |
| (4.4 ± 0.3) × 104 | |||||||||||
| High salt (n = 47) | 150 | 2 | 7.5 | 13.5 | 54.1 | (5.9 ± 0.7) × 104 | (5.4 ± 0.7) × 10−3 |
|
(1.8 ± 0.2) × 10−2 | (2.2 ± 0.2) × 10−3 | 44.8 |
| (5.0 ± 0.3) × 104 | |||||||||||
| pH = 8.5 (n = 82) | 33 | 2 | 8.5 | 17.2 | 44.8 | (5.7 ± 0.1) × 104 | (5.4 ± 0.7) × 10−3 |
|
(3.4 ± 0.9) × 10−2 | (2.5 ± 0.1) × 10−3 | 54.6 |
| (4.0 ± 0.2) × 104 |
akAssociation: the two listed constants represent the association rate constants of the presynaptic complex and the synaptic complex.
bkshort-lived presynapse: the three listed constants represent the rate constants for the formation of unstable synaptic complexes, the decomposition of unstable presynaptic complexes and the formation of stable synaptic complexes of DNA molecules that successfully synapse during the experimental period.
It is known that the catalytic activity, but not the DNA binding affinity of some endonucleases is affected when Mg2+ ions are replaced by Ca2+ ions (42,43). According to previous studies and gel electrophoresis results, it has been shown that there is no significant difference in the Cre-mediated recombination process under divalent ion replacement conditions (12). Therefore, TPM experiments and kinetic analyses were performed using different reaction buffer conditions to explore the effect of two divalent ion species, Mg2+ and Ca2+, on the Cre-mediated recombination process; in addition, the effect of the absence of divalent ions was also explored. When the TPM results were examined in depth, the affinity of Cre-loxP and the stability of the synaptic complex were found to be similar in the presence of either Mg2+ or Ca2+ (Table 2, Supplementary Figure S3). These results reinforced the hypothesis that Mg2+ and Ca2+ have the same positive effect on the Cre-mediated recombination process (25). When divalent ions were removed from the reaction buffer, it was found that the recombination efficiency was lower than under normal conditions. A detailed kinetic analysis showed that the lower recombination efficiency could be attributed to a lower probability (29.4%) of forming the correct synaptic complex leading into a successful catalytic recombination process. In addition, based on kinetic analysis, it was found that not only was the Cre–loxP interaction weaker, but the stability of the synaptic complex was also reduced in the absence of divalent ions (Table 2, Supplementary Figure S4). Structural data indicated that the enhancement of the stability of the synaptic complex was due to the formation of divalent-cationic species stabilized by close contacts (∼4 Å) between the phosphate backbones within the Cre-loxP synaptic complex (12,16). Based on the experimental results obtained under divalent ion replacement and divalent ion deprivation, the requirement for divalent ion species during the Cre-mediated recombination process has been confirmed.
To explore the effect of high concentrations of monovalent ions, the same experiments and kinetic analyses were performed at a concentration of 150 mM Na+. The electrostatic interaction between the recombinases and the loxP site appears to be reduced at a high ionic strength; hence, the association rate constants have been found to be smaller (44,45). However, the kinetic analysis showed that the high concentration of monovalent ions appeared to only affect the formation and stability of abortive complexes and not the formation and stability of synaptic complexes (Table 2, Supplementary Figure S5). These results are consistent with previous studies in which it was found that divalent cationic species alone are sufficient to stabilize the close contacts between the phosphate backbones within the synaptic complexes (12). Based on the TPM kinetic analysis, it can be concluded that the lower recombination efficiency observed at high monovalent ionic strengths is due to a lower probability (32.4%) of forming the correct synaptic complex, rather than a reduction of the catalytic ability (Table 2).
Previous studies have indicated that the Cre-mediated site-specific recombination reaction is robust over a pH range of 5.5–8.0 but is inhibited once the pH is raised to 8.5. A possible explanation for the pH dependence of synapsis and recombination is that the cysteine residues located within the helical core binding domain of Cre recombinase tend to ionize when the pH is greater than 8.33 and that these ionized residues affect the stability of synaptic complexes (12). The TPM results obtained at a high pH showed that a lower association rate occurred, which appears to confirm that increased electrostatic repulsion is the main factor that weakens the Cre-loxP interaction. The kinetic analysis showed that the increased electrostatic repulsion at a high pH weakens the Cre–Cre interaction [kAssociation = (4.0 ± 0.2) × 104 M−1s−1], reducing the probability of forming the correct synaptic complex (38%, compared to 46.6% under normal conditions) (Table 2, Supplementary Figure S5). The results regarding the pH dependence of synapsis obtained using TPM together with detailed kinetic analysis reinforced the hypothesis that increased electrostatic repulsion is the main factor that affects synaptic complex stability at a high pH.
DISCUSSION
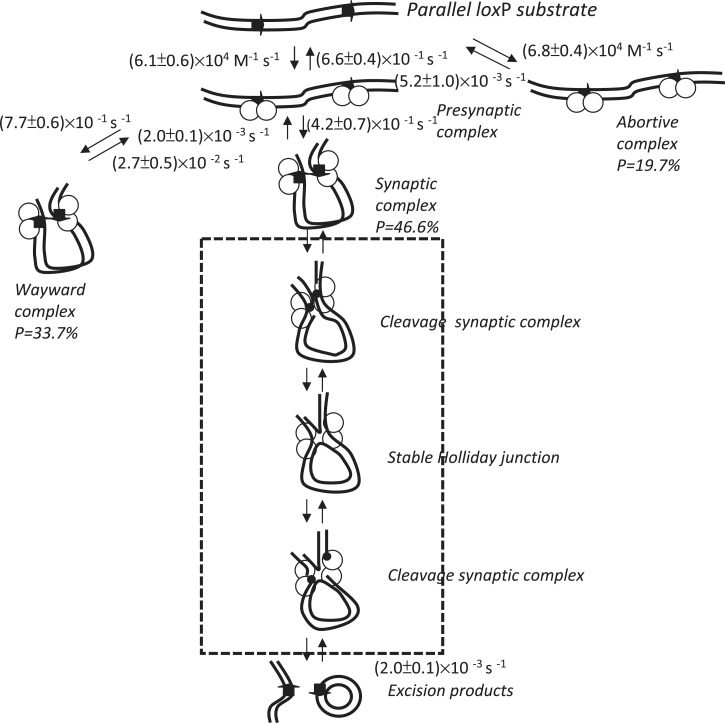
Using the above kinetic results, the kinetic details of the reaction pathway from beginning to end can be delineated (Figure 7). In a previous single-molecule study on the tyrosine family recombinase λ Int, it was found that presynaptic complexes formed at a rate constant >0.05 s−1, and no detectable dissociation process can be observed within 60 min of reaction time (19). Moreover, it has been found that the interaction between Cre and loxP occurs very rapidly, with a rate constant of ∼108 M−1 s−1 (31). However, the association rate constant of (6.8 ± 0.4) × 104 M−1 s−1 obtained via TPM is slower compared to previous results. It is believed that Cre recombinase is monomeric in solution and binds to the loxP site one by one to form a dimer (21–23). However, only the presynapse state in which two loxP sites are occupied by Cre recombinases can be differentiated because of the limited resolution of TPM. According to our data, the kAssociation value observed is the rate constant of abortive complex formation, rather than that of DNA binding. Therefore, it cannot be ruled out that the measured association rate constant is underestimated. For these abortive complexes, the dissociation rate constant of Cre recombinase from the loxP site is (5.2 ± 1.0) × 10−3 s−1, which is consistent with previous results (31). The reason for the formation of abortive complexes could be either that there are too few Cre recombinases at the loxP site or that there are one or more inactive Cre recombinases occupying the loxP site. However, an alternative interpretation that cannot be ruled out is that this 19.7% of the population of abortive complexes, results from inactive Cre molecules formed during preparation of the Cre enzyme. In spite of the observed formation of abortive complexes, 61 of 76 molecules formed synaptic complexes rapidly from short-lived presynaptic complexes, and rate constants of (7.7 ± 0.1) × 10−1 s−1 and (4.2 ± 0.7) × 10−1 s−1 were obtained for the formation of wayward complexes and stable synaptic complexes, respectively. It has been reported that this process, which requires a collision between DNA partners, takes place relatively slowly (31,46). However, rapid synaptic behavior has been observed in both the λ Int and Cre recombinase systems (12,19,31). The similarity between these two tyrosine family recombinases reinforces the hypothesis that there are no intrinsic mechanistic properties that cause synapsis to be a step that occurs slowly in vitro. However, the discrepancy between the TPM results and in vivo observations could be explained by the in vivo synapsis process being either intermolecular or involving a much larger separation between the recombining sites, making synapsis a slower process in vivo.
Figure 7.
Mechanism by which bacteriophage P1 Cre recombinase mediates site-specific recombination. The rate constant for each step was obtained via either single exponential decay fitting or bi-exponential decay fitting to the duration distribution histograms. The arrows on the DNA molecule indicate the parallel loxP sites. The empty circles represent the Cre recombinases. The small black dots represent the covalent phosphotyrosine linkage that is created after DNA cleavage. All the processes within the frame—from strand cleavage to strand migration to strand ligation—were assigned as catalytic recombination processes.
Among the DNA molecules that formed synaptic complexes rapidly via a short-lived presynapse state, 25 of 61 DNA molecules were subjected to a pathway ending in a wayward complex. These wayward complexes, in which the Cre–Cre interactions are relatively unstable, readily decompose at a rate constant of (2.7 ± 0.5) × 10−2 s−1. It has been shown that CreY324F is deficient in synapsis, showing a lower synapsis Kd > 200 nM, and lacks in the strand cleavage (12). That only the wayward complex, decomposing at a rate constant of (3.2 ± 0.3) × 10−2 s−1, was observed in CreY324F-loxP system reinforces the hypothesis that there are side pathways and futile intermediates in Cre-loxP recombination process. Because of the limited resolution of TPM, it is not possible to differentiate whether the observed decomposition of these inactive synaptic complexes is actually due to the dissociation of Cre recombinase from one RBE of a single loxP site or is simply the decomposition of the wayward complex as a whole.
Reversibility in strand cleavage/ligation is an expected property of the Cre-mediated site-specific recombination process because this reaction is isoenergetic, and no high-energy cofactors are required (4,25). If the reaction were irreversible, the synapse state duration analysis of DNA molecules containing parallel loxP sites would only reflect the decomposition of wayward complexes; in contrast, the synapse state duration analysis for DNA molecules containing inverse loxP sites will represent two reaction pathways, i.e. the decomposition of the wayward complex and the decomposition of the recombined synaptic complex. Based on kinetic analysis of the synapse state duration, similar rate constants of (2.0 ± 0.1) × 10−3 s−1 and (2.2 ± 0.2) × 10−3 s−1 were obtained for the rate-limiting step in the catalytic recombination process, i.e. the decomposition of the recombined synaptic complex, for DNA molecules containing parallel loxP sites and inverse loxP sites, respectively. All the experimental results support the hypothesis that strand cleavage/ligation is reversible in the Cre-mediated site-specific recombination process.
This reaction pathway, strand cleavage/ligation, is abolished once the Tyr324 has been replaced with Phenylalanine, blocking the opportunity to entering the catalytic recombination process. An alternative explanation for the single reaction pathway observed in the decomposition of synaptic complexes is that CreY324F is simply defective in synapsis, and cleavage is not required for the formation of a stable synaptic complex. In order to verify the requirement of strand cleavage in the formation of stable synaptic complexes, same experiments were performed in CreK201A-loxP system. Based on kinetic analysis of the synapse state duration for the synapsis-capable CreK201A, even though it lacks in strand-cleavage ability, similar rate constant of (2.6 ± 0.2) × 10−3 s−1, representing the decomposition of the synaptic complex, was obtained. It supported the statement that strand cleavage is not required for the formation of the stable synaptic complex in the Cre-loxP system.
CONCLUSION
Using a novel single-molecule method, TPM, the detailed reaction mechanisms and kinetics involved in the Cre-mediated site-specific recombination process have been revealed. The proposed reversible reaction mechanism and the presence of a thermally stable Holliday junction intermediate during the Cre-mediated site-specific recombination process are consistent with previous experimental results. Similar to the λ Int system, wayward pathways and various reaction intermediates have been observed in the Cre-loxP system too (19). Moreover, the detailed kinetic analysis performed in this study reinforces the importance of divalent cations and electrostatic interactions for stabilizing the close contact that maintains the Cre–Cre interface in the synapse state. These effects under different conditions were explored in detail using a single-molecule tool approach, which now offers a novel means of exploring reaction mechanisms other than conventional tools.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6, Supplementary Methods and Supplementary Reference [47].
FUNDING
Funding for open access charge: National Science Council of Taiwan and Yang Ming University (to H.F.F.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
The author is grateful for the generosity of Van Duyne labs in providing the Cre mutant constructs. The author thanks Hung-Wen Li for the helpful discussion about experimental data. The author thanks Po-Yu Tsai for helpful discussion about MLE data analysis. The author appreciates the reviewers’ helpful comments.
REFERENCES
- 1.Futcher AB. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J. Theor. Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 2.Sherratt DJ, Arciszewska LK, Blakely G, Colloms S, Grant K, Leslie N, McCulloch R. Site-specific recombination and circular chromosome segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995;347:37–42. doi: 10.1098/rstb.1995.0006. [DOI] [PubMed] [Google Scholar]
- 3.Segall AM, Nash HA. Architectural flexibility in lambda site-specific recombination: three alternate conformations channel the attL site into three distinct pathways. Genes Cells. 1996;1:453–463. doi: 10.1046/j.1365-2443.1996.d01-254.x. [DOI] [PubMed] [Google Scholar]
- 4.Stark WM, Boocock MR, Sherratt DJ. Catalysis by site-specific recombinases. Trends Genet. 1992;8:432–439. [PubMed] [Google Scholar]
- 5.Esposito D, Scocca JJ. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 1997;25:3605–3614. doi: 10.1093/nar/25.18.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolb AF. Genome engineering using site-specific recombinases. Cloning Stem Cells. 2002;4:65–80. doi: 10.1089/153623002753632066. [DOI] [PubMed] [Google Scholar]
- 7.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 8.Le Y, Sauer B. Conditional gene knockout using cre recombinase. Methods Mol. Biol. 2000;136:477–485. doi: 10.1385/1-59259-065-9:477. [DOI] [PubMed] [Google Scholar]
- 9.Better M, Lu C, Williams RC, Echols H. Site-specific DNA condensation and pairing mediated by the int protein of bacteriophage lambda. Proc. Natl Acad. Sci. USA. 1982;79:5837–5841. doi: 10.1073/pnas.79.19.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton DL, Abremski K. Site-specific recombination by the bacteriophage P1 lox-Cre system. Cre-mediated synapsis of two lox sites. J. Mol. Biol. 1984;178:481–486. doi: 10.1016/0022-2836(84)90154-2. [DOI] [PubMed] [Google Scholar]
- 11.Segall AM, Nash HA. Synaptic intermediates in bacteriophage lambda site-specific recombination: integrase can align pairs of attachment sites. EMBO J. 1993;12:4567–4576. doi: 10.1002/j.1460-2075.1993.tb06145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh K, Guo F, Van Duyne GD. Synapsis of loxP sites by Cre recombinase. J. Biol. Chem. 2007;282:24004–24016. doi: 10.1074/jbc.M703283200. [DOI] [PubMed] [Google Scholar]
- 13.Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 14.Vetcher AA, Lushnikov AY, Navarra-Madsen J, Scharein RG, Lyubchenko YL, Darcy IK, Levene SD. DNA topology and geometry in Flp and Cre recombination. J. Mol. Biol. 2006;357:1089–1104. doi: 10.1016/j.jmb.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 15.Cassell G, Moision R, Rabani E, Segall A. The geometry of a synaptic intermediate in a pathway of bacteriophage lambda site-specific recombination. Nucleic Acids Res. 1999;27:1145–1151. doi: 10.1093/nar/27.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo F, Gopaul DN, Van Duyne GD. Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proc. Natl Acad. Sci. USA. 1999;96:7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ennifar E, Meyer JE, Buchholz F, Stewart AF, Suck D. Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation. Nucleic Acids Res. 2003;31:5449–5460. doi: 10.1093/nar/gkg732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rüfer A, Neuenschwander PF, Sauer B. Analysis of Cre-loxP interaction by surface plasmon resonance: influence of spermidine on cooperativity. Anal. Biochem. 2002;308:90–99. doi: 10.1016/s0003-2697(02)00247-6. [DOI] [PubMed] [Google Scholar]
- 19.Mumm JP, Landy A, Gelles J. Viewing single lambda site-specific recombination events from start to finish. EMBO J. 2006;25:4586–4595. doi: 10.1038/sj.emboj.7601325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai H, Sun M, Ghosh P, Hatfull GF, Grindley ND, Marko JF. Single-molecule analysis reveals the molecular bearing mechanism of DNA strand exchange by a serine recombinase. Proc. Natl Acad. Sci. USA. 2011;108:7419–7424. doi: 10.1073/pnas.1018436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews BJ, Beatty LG, Sadowski PD. Isolation of intermediates in the binding of the FLP recombinase of the yeast plasmid 2-micron circle to its target sequence. J. Mol. Biol. 1987;193:345–358. doi: 10.1016/0022-2836(87)90223-3. [DOI] [PubMed] [Google Scholar]
- 22.Hoess RH, Abremski K. Interaction of the bacteriophage P1 recombinase Cre with the recombining site loxP. Proc. Natl Acad. Sci. USA. 1984;81:1026–1029. doi: 10.1073/pnas.81.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack A, Sauer B, Abremski K, Hoess R. Stoichiometry of the Cre recombinase bound to the lox recombining site. Nucleic Acids Res. 1992;20:4451–4455. doi: 10.1093/nar/20.17.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beatty LG, Babineau-Clary D, Hogrefe C, Sadowski PD. FLP site-specific recombinase of yeast 2-micron plasmid. Topological features of the reaction. J. Mol. Biol. 1986;188:529–544. doi: 10.1016/s0022-2836(86)80003-1. [DOI] [PubMed] [Google Scholar]
- 25.Abremski K, Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J. Biol. Chem. 1984;259:1509–1514. [PubMed] [Google Scholar]
- 26.Adams DE, Bliska JB, Cozzarelli NR. Cre-lox recombination in Escherichia coli cells. J. Mol. Biol. 1992;226:661–673. doi: 10.1016/0022-2836(92)90623-r. [DOI] [PubMed] [Google Scholar]
- 27.Gates CA, Cox MM. FLP recombinase is an enzyme. Proc. Natl Acad. Sci. USA. 1988;85:4628–4632. doi: 10.1073/pnas.85.13.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waite LL, Cox MM. A protein dissociation step limits turnover in FLP recombinase-mediated site-specific recombination. J. Biol. Chem. 1995;270:23409–23414. doi: 10.1074/jbc.270.40.23409. [DOI] [PubMed] [Google Scholar]
- 29.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single-stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. doi: 10.1038/nature08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrova V, Chitteni-Pattu S, Drees JC, Inman RB, Cox MM. An SOS inhibitor that binds to free RecA protein: the PsiB protein. Mol. Cell. 2009;36:121–130. doi: 10.1016/j.molcel.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringrose L, Lounnas V, Ehrlich L, Buchholz F, Wade R, Stewart AF. Comparative kinetic analysis of FLP and cre recombinases: mathematical models for DNA binding and recombination. J. Mol. Biol. 1998;284:363–384. doi: 10.1006/jmbi.1998.2149. [DOI] [PubMed] [Google Scholar]
- 32.Fan HF, Li HW. Studying RecBCD helicase translocation along Chi-DNA using tethered particle motion with a stretching force. Biophys. J. 2009;96:1875–1883. doi: 10.1016/j.bpj.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan HF, Cox MM, Li HW. Developing single-molecule TPM experiments for direct observation of successful RecA-mediated strand exchange reaction. PLoS One. 2011;6:e21359. doi: 10.1371/journal.pone.0021359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh K, Van Duyne GD. Cre-loxP biochemistry. Methods. 2002;28:374–383. doi: 10.1016/s1046-2023(02)00244-x. [DOI] [PubMed] [Google Scholar]
- 35.Manghi M, Tardin C, Baglio J, Rousseau P, Salomé L, Destainville N. Probing DNA conformational changes with high temporal resolution by tethered particle motion. Phys. Biol. 2010;7:046003. doi: 10.1088/1478-3975/7/4/046003. [DOI] [PubMed] [Google Scholar]
- 36.Manzo C, Finzi L. Quantitative analysis of DNA-looping kinetics from tethered particle motion experiments. Methods Enzymol. 2010;475:199–220. doi: 10.1016/S0076-6879(10)75009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson PC, Zurla C, Brogioli D, Beausang JF, Finzi L, Dunlap D. Tethered particle motion as a diagnostic of DNA tether length. J. Phys. Chem. B. 2006;110:17260–17267. doi: 10.1021/jp0630673. [DOI] [PubMed] [Google Scholar]
- 38.Vanzi F, Broggio C, Sacconi L, Pavone FS. Lac repressor hinge flexibility and DNA looping: single molecule kinetics by tethered particle motion. Nucleic Acids Res. 2006;34:3409–3420. doi: 10.1093/nar/gkl393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin H, Landick R, Gelles J. Tethered particle motion method for studying transcript elongation by a single RNA polymerase molecule. Biophys. J. 1994;67:2468–2478. doi: 10.1016/S0006-3495(94)80735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitts PA, Nash HA. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987;329:346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- 41.Gibb B, Gupta K, Ghosh K, Sharp R, Chen J, Van Duyne GD. Requirements for catalysis in the Cre recombinase active site. Nucleic Acids Res. 2010;38:5817–5832. doi: 10.1093/nar/gkq384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vipond IB, Halford SE. Specific DNA recognition by EcoRV restriction endonuclease induced by calcium ions. Biochemistry. 1995;34:1113–1119. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- 43.Rouzina I, Bloomfield VA. DNA bending by small, mobile multivalent cations. Biophys. J. 1998;74:3152–3164. doi: 10.1016/S0006-3495(98)78021-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor–operator interaction: kinetic measurements and conclusions. Biochemistry. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 45.Lohman TM. Kinetics of protein-nucleic acid interactions: use of salt effects to probe mechanisms of interaction. CRC Crit. Rev. Biochem. 1986;19:191–245. doi: 10.3109/10409238609084656. [DOI] [PubMed] [Google Scholar]
- 46.Bankhead TM, Etzel BJ, Wolven F, Bordenave S, Boldt JL, Larsen TA, Segall AM. Mutations at residues 282, 286, and 293 of phage lambda integrase exert pathway-specific effects on synapsis and catalysis in recombination. J. Bacteriol. 2003;185:2653–2666. doi: 10.1128/JB.185.8.2653-2666.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandt S. Data Analysis: Statistical and Computational Methods for Scientists and Engineers. 3rd edn. Berlin: Springer; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.