Abstract
Genetic factors underlie dental fluorosis (DF) susceptibility/resistance. The A/J (DF susceptible) and 129P3/J (DF resistant) strains have been previously used to detect quantitative trait loci (QTL) associated with DF on chromosomes (Chr) 2 and 11. In the present study increased marker density genotyping followed by interval mapping was performed to narrow the QTL intervals and improve the LOD scores. Narrower intervals on Chr 2 where LOD ≥ 6.0 (57–84 cM or ~51 Mb), LOD ≥ 7.0 (62–79 cM or ~32 Mb), and LOD ≥ 8.0 (65–74 cM or ~17 Mb); and on Chr 11 where LOD ≥ 6.0 the interval was 18–51 cM (~53 Mb), LOD ≥ 7.0 (28–48 cM or ~34 Mb), and LOD ≥ 8.0 (31–45 cM or~22 Mb) were obtained. Haplotype analysis between A/J and 129P3/J further reduced QTL intervals. Accn1 was selected as a candidate gene based upon its location near the peak LOD score on Chr 11 and distant homology with the C. elegans fluoride resistance gene flr1. DF severity between Accn1−/− and wildtype mice was not different. The loss of ACCN1 function does not modify DF severity in mice. Narrowing the DF QTL intervals will facilitate additional candidate gene selections and interrogation.
Keywords: dental fluorosis, genetics, QTL, mice, ACCN1, ASIC2
Fluoride (F) in various chemical forms and exposures has been repeatedly shown to have actions on the development and homeostasis of mineralized tissues (1, 2). Excessive systemic fluoride during critical periods of tooth enamel development (amelogenesis) can result in an undesirable developmental defect of tooth enamel, dental fluorosis (enamel fluorosis). Dental fluorosis (DF) is characterized by increased porosity (subsurface hypomineralization) with a loss of enamel translucency and increased opacity (3). It is generally accepted that increasing DF severity correlates with increasing F exposure. DF remains highly prevalent world-wide. As recent as 2005 23% of persons in the United States aged 6–39 years had very mild or greater enamel fluorosis (4). We have demonstrated that genetic background plays an integral role in responses of mineralized tissues to systemic fluoride (5–10). Genetically diverse inbred strains of mice when provided equivalent exposures to F in their environment show strain specific DF severities (5, 9). DF can be considered a complex condition influenced by environmental (F exposure) and host factors (genetics). The later is determined by multiple genes with additive effects (11). Quantitative trait locus (QTL) detection mapping has been extensively used in mapping loci associated with complex traits. Generally large panels of animals and large QTL effects improve the power to detect and map QTLs. Two traditional QTL mapping approaches that can yield large panels are F2 and backcross strategies. In mice F2 panels are created from a two generation cross between two parental inbred strains which have measurable differences for a particular trait. In the case of DF we used the 129P3/J (DF resistant strain) and the A/J (DF susceptible strain)(5, 11). Recently we identified QTLs on chromosomes (Chr) 2 and 11 in mice that are strongly associated with DF susceptibility (11). On Chr 2 a QTL designated as Dfs1 (MGI Accession ID: MGI: 4418088) was detected with an interval of approximately 77.62 Mb and flanked by markers rs13476589 and UT_2_156.443943. A second QTL on Chr 11 designated Dfs2 (MGI Accession ID: MGI: 4418089) was detected with an interval of approximately 74.29 Mb and flanked by markers rs3708339 and rs6161623. The QTLs Dfs1 and Dfs2 combined contain over 2600 genes. The present study had two goals. First, to refine the two DF QTL intervals through the use of increased marker densities on Chr 2 and Chr 11 and by two strain haplotype analysis between the parental A/J and 129P3/J strains. Second, to test a candidate gene Accn1 which encodes an amiloride-sensitive cation channel 1, neuronal (degenerin) and resides on Chr 11 near the QTL peak detected.
Material and methods
Genotyping
From an initial panel of 458 F2 mice (11) genomic DNAs from 140 mice representing DF phenotype extremes were selected for additional SNP genotyping. Among these 140 mice 75 were from the low DF group and 65 were from the high DF group. 112 SNPs representing Chrs 2 and 11 were selected and genotyped utilizing the Genetic Analysis service at The Jackson Laboratory (Bar Harbor, ME USA). The 112 SNPs are a subset of the 1638 SNPs described by Petkov and colleagues for inbred strain identification (12). These SNP assays selected detected polymorphisms between A/J and 129P3/J and increased the number of informative SNPs from 29 to 108 for Chr 2 and from 14 to 47 for Chr 11. The SNPs were genotyped using the KASPar SNP Genotyping system developed at KBiosciences (Hoddesdon, Hertfordshire, UK) (http://www.kbioscience.co.uk/chemistry/chemistry_Kasp_intro.html).
Interval mapping
Interval mapping was performed in R with R/qtl package (13). The DF phenotypes from 140 F2 mice were dichotomized by pooling the low DF mice with modified TF scores of 1 and 2; and pooling the high DF mice with modified TF scores of 3 and 4 (11). The binary phenotype was analyzed by the scanone function with the expectation maximization (EM) algorithm. Logarithm of the odds (to the base 10)(LOD) scores were calculated at each 1 cM (centimorgan) position on the genome. Sex averaged cM were converted to National Center for Biotechnology Information (NCBI) Build 37 base pair values.
Two strain haplotype analysis
SNP datasets of the A/J and 129P3/J inbred strains from the Imputed Diversity Array, Build 37 were used with the Mouse Strain Comparison Tool (The Center for Genome Dynamics at the The Jackson Laboratory, Bar Harbor, ME USA)(http://cgd.jax.org/straincomparison/)(14) to determine genetic intervals on Chr 2 and Chr 11 where 129P3/J = A/J. Genetic intervals were considered equivalent between 129P3/J and A/J where there are at least 10 consecutive SNP calls that match. Every other part of the genome on Chrs 2 and 11 were then considered nonequivalent.
Animals
Mice homozygous for targeting of the Accn1 (ASIC2, BNaC1a, BNC1, and Mdeg) gene were obtained as gifts from Dr. Michael Welsh (Howard Hughes Medical Institute, Department of Internal Medicine, University of Iowa College of Medicine, Iowa City USA). Accn1 encodes an amiloride-sensitive cation channel 1, neuronal (degenerin) also named acid-sensing ion channel 2 (ASIC2) which plays a key role as a component of a mechanosensory complex and when joined to ASIC1 form the acid-sensing ion channel to produce H+-gated currents activated by acidic extracellular solutions.(15–17). A neomycin cassette was inserted into the Accn1 gene to disrupt two exons encoding the second transmembrane domain, which is common to both splice variants of the gene, as well as a channel pore (15). Male and female Accn1tm1Wsh/Accn1tm1Wsh (Accn1−/−) animals were used to generate a cohort of animals for fluoride treatment. The C57BL6/J strain (The Jackson Laboratory, Bar Harbor, ME USA) was used as wildtype controls since the Accn1 −/− allele has been backcrossed on the C57BL6/J strain. All animal studies were performed in accordance with the University of North Carolina at Chapel Hill Animal Care and Use Committee guidelines.
Fluoride treatment
Accn1−/− and C57BL6/J mice at 3–5 weeks of age were treated with 0 ppm [F-] or 50 ppm [F-] as NaF in the drinking water for 60 days. Mice were fed a constant nutrition diet containing 16 ppm fluoride, 1.00% Ca+2, 0.75% P04−3 and 2.4 IU/gm vitamin D3. Mice were allowed food and water ad libitum. DF affecting the lower mandibular incisors was assessed using the modified TF scale and quantitative fluorescence as previously described (1, 5, 11). One-way ANOVA (PASW Statistics version 18.0.0, SPSS Inc., Chicago, IL, USA) followed by Bonferroni post hoc testing was used for comparisons between genotypes and treatment groups. P ≤ 0.05 was considered significant.
Results and Discussion
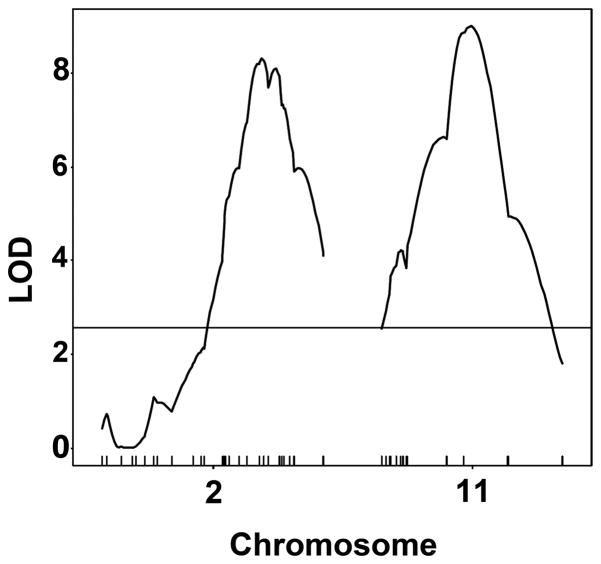
Initial detection of the Dfs1 and Dfs2 QTLs identified intervals of 77.62 Mb and 74.29 Mb, respectively on a cohort of 102 phenotypic extreme F2 animals (11). There are various strategies to refine QTL intervals which include but are not limited to: increasing the number of F2 mice (in order to increase the number of informative recombinations); combining multiple QTL data for the same trait (same or different species); expression QTL mapping; haplotype association mapping; or increasing marker densities. We have chosen to perform the later which has had some success in other studies (18–21). Interval mapping using increased marker densities on Chrs 2 and 11 resulted in a modest narrowing of QTL intervals and an improved estimation of maximum LOD scores with peak LOD = 8.252 located at 68 cM on Chr 2 and peak LOD = 8.845 located at 35 cM on Chr 11 (Fig. 1). LOD scores were calculated along genetic distances of 95 cM for Chr 2 and 76 cM for Chr 11 and include 43 anchor SNPs where chromosome positions in bp for select LOD scores could be obtained (Tables 1 and 2). For Chr 2 at LOD ≥ 6.0 the interval was 57–84 cM (~51 Mb), LOD ≥ 7.0 the interval was 62–79 cM (~32 Mb), and LOD ≥ 8.0 the interval was 65–74 cM (~17 Mb). For Chr11 at LOD ≥ 6.0 the interval was 18–51 cM (~53 Mb), LOD ≥ 7.0 the interval was 28–48 cM (~34 Mb), and LOD ≥ 8.0 the interval as 31–45 cM (~22 Mb). This incremental improvement in QTL interval lengths may have been limited to some degree by the number of informative recombinations carried by the subset of F2 animals studied.
Fig. 1.
Interval mapping output from Rqtl for Chrs 2 and 11. LOD scores are plotted along the genetic positions (cM). The tick marks along the x-axis are the anchor SNP marker locations described in Tables 1 and 2.
Table 1.
Interval mapping on chromosome 2
| Marker | Genetic Distance (cM) | LOD | Chromosome Position of Marker (bp) * |
|---|---|---|---|
| rs3680350 | 0.000 | 0.414 | 3,880,017 |
| rs6331390 | 1.682 | 0.742 | 5,895,118 |
| rs3719255 | 7.836 | 0.030 | 10,677,314 |
| rs13476365 | 12.877 | 0.011 | 19,202,258 |
| rs6165425 | 14.066 | 0.042 | 20,680,270 |
| mCV23574676 | 17.772 | 0.254 | 26,733,409 |
| rs13459062 | 22.004 | 1.090 | 31,027,542 |
| mCV25370550 | 23.437 | 0.974 | 33,798,339 |
| rs6265423 | 29.296 | 0.786 | 47,020,132 |
| rs13476535 | 38.715 | 1.798 | 61,594,843 |
| rs13476547 | 41.905 | 2.050 | 65,384,303 |
| rs4136610 | 43.338 | 2.126 | 69,989,588 |
| rs13476589 | 50.919 | 3.989 | 77,938,379 |
| 02-081-544 | 51.493 | 4.444 | 80,194,100 |
| rs13476610 | 51.831 | 4.824 | 83,283,198 |
| mCV25115393 | 52.301 | 5.170 | 85,788,243 |
| rs6312617 | 53.977 | 5.361 | 93,528,209 |
| rs3694656 | 53.977 | 5.361 | 97,038,674 |
| rs13476687 | 58.192 | 5.974 | 106,567,750 |
| rs13476719 | 61.623 | 6.939 | 116,028,112 |
| rs13476765 | 66.914 | 8.190 | 128,300,336 |
| rs4223510 | 68.583 | 8.252 | 131,679,416 |
| rs13476785 | 70.753 | 7.673 | 134,586,818 |
| rs13476811 | 75.243 | 7.917 | 142,540,462 |
| rs6363071 | 76.432 | 7.301 | 145,858,075 |
| rs6209325 | 77.379 | 7.235 | 147,881,059 |
| rs13476846 | 79.787 | 6.644 | 152,857,172 |
| UT_2_156.443943 | 81.693 | 5.896 | 155,644,451 |
| rs3714936 | 94.258 | 4.106 | 168,912,439 |
MGSCv37 Build 37.1
Table 2.
Interval mapping on chromosome 11
| Marker | Genetic Distance (cM) | LOD | Chromosome Position of Marker (bp) * |
|---|---|---|---|
| gnf11.006.472 | 0.000 | 2.545 | 8,934,218 |
| rs13480871 | 1.672 | 2.941 | 11,457,493 |
| rs13480881 | 3.348 | 3.321 | 14,278,768 |
| rs13480889 | 3.582 | 3.660 | 17,364,752 |
| rs3708339 | 6.003 | 3.891 | 21,757,367 |
| rs6398304 | 7.925 | 4.193 | 24,653,078 |
| rs3718803 | 8.633 | 4.205 | 28,192,020 |
| rs4228647 | 10.309 | 3.838 | 32,130,040 |
| rs13480955 | 10.779 | 4.264 | 34,315,729 |
| 11-059-257 | 27.507 | 6.592 | 58,438,767 |
| rs3694522 | 34.772 | 8.845 | 70,520,265 |
| rs3656982 | 53.606 | 5.144 | 93,517,065 |
| rs6161623 | 53.826 | 4.949 | 95,851,719 |
| rs13481245 | 76.667 | 1.812 | 115,470,957 |
MGSCv37 Build 37.1
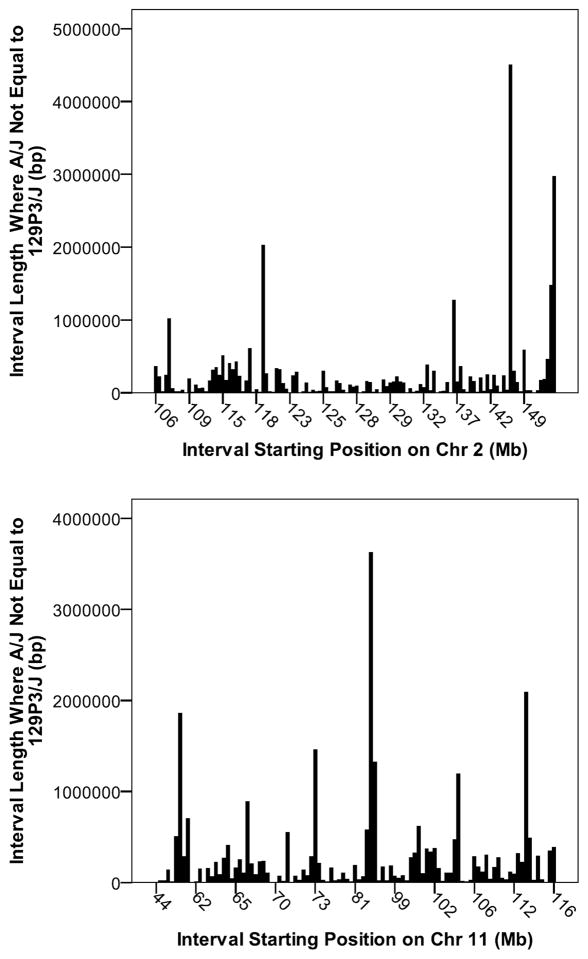
Two strain haplotype analysis was performed to compare genomes between parental A/J (DF susceptible) and 129P3/J (DF resistant) inbred strains on Chrs 2 and 11 (Fig. 2A and 2B). This has been made efficient through the recent development of analytical tools at the Center for Genome Dynamics, The Jackson Laboratory (14). Across the entire Chr 2 there are 416 segments totaling 80.09 Mb of nonequivalence and comprising 44.1% of the chromosome length. Similarly across Chr 11 there are 197 segments totaling 49.06 Mb of nonequivalence and comprising 40.26% of the chromosome length. For the QTL intervals on Chr 2 and Chr 11 where LOD ≥ 6.0 we have identified segments where nonequivalence in SNP genotypes exist (Fig. 3). We hypothesize that genes residing within these segments of nonequivalence are likely to be associated with DF susceptibility/resistance. Conversely segments where A/J and 129P3/J are identical would contain genes that would not likely to be associated with DF susceptibility/resistance. Within the Chr 2 QTL (LOD ≥ 6.0) there are 121 intervals ranging in length from 468 to 4,502,998 bp. This represents ~27.9 Mb and contains approximately 507 genes. For the QTL interval (LOD ≥ 6.0) on Chr 11 there are 103 segments/intervals ranging in length from 1284 to 2,090,072 bp and totaling 27.26 Mb and contains approximately 829 genes. By narrowing the two QTL intervals we have reduced the number genes to interrogate by almost half. This number can be further refined by segregating pseudogenes and non-annotated genes from early consideration.
Fig. 2.
Haplotype analysis across Chrs 2 and 11 for A/J and 129P3/J. Black bars identify segments of Chr 2 genome (A) and Chr 11 genome (B) where A/J is not equal to 129P3/J. Segment of nonequivalence on Chr 11 near the peak LOD score (C). Distances are in Mb. The figure is a modification of an on screen output from http://cgd.jax.org/straincomparison/.
Fig. 3.
Haplotype analysis within QTLs (LOD ≥ 6.0) on Chrs 2 and 11 for A/J and 129P3/J. Bars represent intervals in basepair (bp) where A/J and 129P3/J are not equivalent. For each starting interval the corresponding positions in sex-averaged cM are provided in parentheses and were converted from NCBI 37 Build Base pair value using Mouse Map Converter (http://cgd.jax.org/mousemapconverter/)(25). Chr 2: 106 (55.54), 109 (56.55), 115 (58.01), 118 (59.37), 123 (60.69). 125 (61.25), 128 (62.29), 129 (62.77), 132 (64.15), 137 (67.78), 142 (70.41), and 149 (73.80); Chr 11: 44 (25.94), 62 (37.96), 65 (40.59), 70 (42.99), 73 (45.25), 81 (48.80). 99 (62.33), 102 (65.48), 106 (68.89), 112 (75.82), and 116 (80.92).
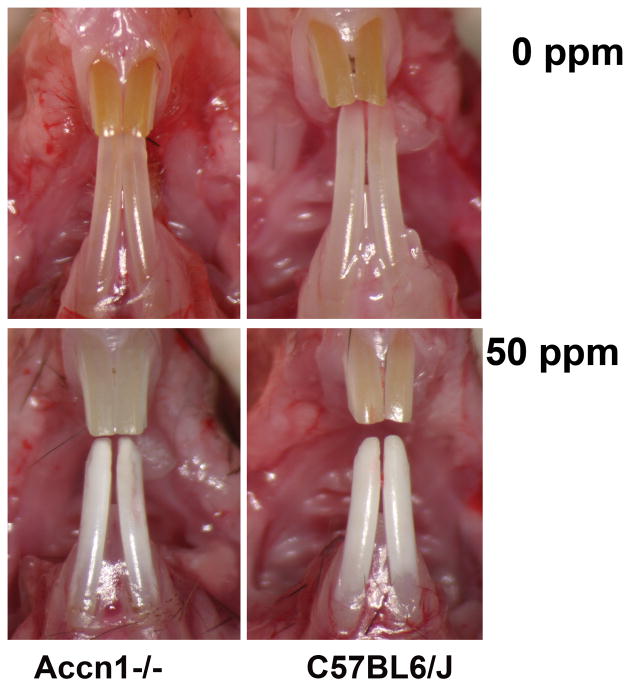
Among the genes on Chr 11 within LOD ≥ 8.0 QTL interval resides Accn1 (Asic2, BNaC1a, BNC1, or Mdeg). Accn1 residing at 80,693,671–81,781,959 bp on Chr 11 was selected for further investigation for the following reasons: a large number of SNPs within 3′ end of this gene are informative between the A/J and 129P3/J strains (Fig. 3C). ACCN1 is member of the H+-gated subgroup of the degenerin/epithelial Na+ channel family and as an acid-sensing ion channel (ASIC) has some distant homology with the flr-1gene. The flr1 gene which encodes an ion channel belonging to the degenerin/epithelial sodium channel superfamily was mapped as a recessive fluoride resistant gene in C. elegans (22–24). Accn1 null mice, which have been backcrossed to the C57BL/6J strain, have no overt enamel pathology (data not shown) and appear indistinguishable from the wildtype C57BL/6J strain. The C57BL/6J strain is DF susceptible, but not to the same degree as A/J (9). Following F treatment Accn1 null mice developed DF that was not statistically different from the DF in similarly treated C57BL/6J mice (Fig. 4)(Table 3). These data suggest that loss of function of Accn1 alone in the C57BL/6J strain is either insufficient (not a major effect gene) to contribute to DF susceptibility, that Accn1 plays no direct role in DF susceptibility, or there is epistasis between other C57BL/6J alleles. In conclusion the intervals of previously detected DF QTLs Dfs1 and Dfs2 have been reduced and contain approximately 1336 genes. The narrowing the DF QTL intervals will facilitate additional candidate gene selections and interrogation.
Fig. 4.
DF development in Accn1 null mice compared to wildtype C57BL6/J mice treated for 60 days with 0 ppm F− or 50 ppm F− as NaF in the drinking water.
Table 3.
Dental fluorosis assessment
| Strain | Fluoride treatment
|
|||
|---|---|---|---|---|
| 0 ppm F− | 50 ppm F− | |||
| Clinical* | QF** | Clinical | QF | |
|
| ||||
| Male | ||||
| Accn1 KO (n = 6) | 0.0 ± 0.0 | 13.95 ± 1.21 | 3.19 ± 0.9 | 24.04 ± 5.23 |
| Wildtype $(n = 9) | 0.6 ± 0.6 | 15.44 ± 2.25 | 3.28 ± 0.4 | 23.70 ± 4.66 |
| p = 0.029 | p = 0.164 | p = 0.790 | p = 0.888 | |
| Female | ||||
| Accn1 KO (n = 5) | 0.2 ± 0.4 | 13.60 ± 1.16 | 3.00 ± 0.9 | 25.12 ± 4.67 |
| Wildtype (n = 8) | 0.1 ± 0.3 | 14.88 ± 4.25 | 3.13 ± 0.5 | 21.07 ± 5.15 |
| p = 0.841 | p = 0.491 | p = 0.760 | p = 0.181 | |
| Combined | ||||
| Accn1 KO (n = 11) | 0.1 ± 0.3 | 13.77 ± 1.14 | 3.1 ± 0.9 | 24.45 ± 4.85 |
| Wildtype (n = 17) | 0.4 ± 0.6 | 15.17 ± 3.24 | 3.2 ± 0.5 | 22.46 ± 4.93 |
| p = 0.095 | p = 0.164 | p = 0.717 | p = 0.278 | |
Modified TF scale
Quantitative fluorescence
C57BL/6J inbred strain
Acknowledgments
The authors thank Dr. Michael Welsh at the Howard Hughes Medical Institute, Department of Internal Medicine, University of Iowa College of Medicine, Iowa City USA for the gift of Accn1−/− mice. Supported by NIH DE014853
References
- 1.Everett ET. Fluoride’s Effects on the Formation of Teeth and Bones, and the Influence of Genetics. J Dent Res. 2011;90:552–560. doi: 10.1177/0022034510384626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronckers AL, Lyaruu DM, DenBesten PK. The impact of fluoride on ameloblasts and the mechanisms of enamel fluorosis. J Dent Res. 2009;88:877–893. doi: 10.1177/0022034509343280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fejerskov O, Manji F, Baelum V. The nature and mechanisms of dental fluorosis in man. J Dent Res. 1990;69(Spec No):692–700. doi: 10.1177/00220345900690S135. discussion 721. [DOI] [PubMed] [Google Scholar]
- 4.Beltran-Aguilar ED, Barker LK, Canto MT, Dye BA, Gooch BF, Griffin SO, Hyman J, Jaramillo F, Kingman A, Nowjack-Raymer R, Selwitz RH, Wu T. Surveillance for dental caries, dental sealants, tooth retention, edentulism, and enamel fluorosis--United States, 1988–1994 and 1999–2002. MMWR Surveill Summ. 2005;54:1–43. [PubMed] [Google Scholar]
- 5.Everett ET, McHenry MA, Reynolds N, Eggertsson H, Sullivan J, Kantmann C, Martinez-Mier EA, Warrick JM, Stookey GK. Dental fluorosis: variability among different inbred mouse strains. J Dent Res. 2002;81:794–798. doi: 10.1177/0810794. [DOI] [PubMed] [Google Scholar]
- 6.Mousny M, Banse X, Wise L, Everett ET, Hancock R, Vieth R, Devogelaer JP, Grynpas MD. The genetic influence on bone susceptibility to fluoride. Bone. 2006;39:1283–1289. doi: 10.1016/j.bone.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Mousny M, Omelon S, Wise L, Everett ET, Dumitriu M, Holmyard DP, Banse X, Devogelaer JP, Grynpas MD. Fluoride effects on bone formation and mineralization are influenced by genetics. Bone. 2008;43:1067–1074. doi: 10.1016/j.bone.2008.07.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan D, Gurumurthy A, Wright M, Pfeiler TW, Loboa EG, Everett ET. Genetic background influences fluoride’s effects on osteoclastogenesis. Bone. 2007;41:1036–1044. doi: 10.1016/j.bone.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan D, Willett TL, Gu X, Martinez-Mier EA, Sardone L, McShane L, Grynpas M, Everett ET. Phenotypic variation of fluoride responses between inbred strains of mice. Cells Tissues Organs. 2011;194:261–267. doi: 10.1159/000324224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vieira AP, Hanocock R, Eggertsson H, Everett ET, Grynpas MD. Tooth quality in dental fluorosis genetic and environmental factors. Calcif Tissue Int. 2005;76:17–25. doi: 10.1007/s00223-004-0075-3. [DOI] [PubMed] [Google Scholar]
- 11.Everett ET, Yan D, Weaver M, Liu L, Foroud T, Martinez-Mier EA. Detection of dental fluorosis-associated quantitative trait Loci on mouse chromosomes 2 and 11. Cells Tissues Organs. 2009;189:212–218. doi: 10.1159/000151383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, Tucker P, Boursot P, McMillan L, Churchill GA, de Villena FP. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price MP, Lewin GR, McIlwrath SL, Cheng C, Xie J, Heppenstall PA, Stucky CL, Mannsfeldt AG, Brennan TJ, Drummond HA, Qiao J, Benson CJ, Tarr DE, Hrstka RF, Yang B, Williamson RA, Welsh MJ. The mammalian sodium channel BNC1 is required for normal touch sensation. Nature. 2000;407:1007–1011. doi: 10.1038/35039512. [DOI] [PubMed] [Google Scholar]
- 16.Askwith CC, Wemmie JA, Price MP, Rokhlina T, Welsh MJ. Acid-sensing ion channel 2 (ASIC2) modulates ASIC1 H+-activated currents in hippocampal neurons. J Biol Chem. 2004;279:18296–18305. doi: 10.1074/jbc.M312145200. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Ma X, Sabharwal R, Snitsarev V, Morgan D, Rahmouni K, Drummond HA, Whiteis CA, Costa V, Price M, Benson C, Welsh MJ, Chapleau MW, Abboud FM. The ion channel ASIC2 is required for baroreceptor and autonomic control of the circulation. Neuron. 2009;64:885–897. doi: 10.1016/j.neuron.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyers SN, Rodriguez-Zas SL, Beever JE. Fine-mapping of a QTL influencing pork tenderness on porcine chromosome 2. BMC Genet. 2007;8:69. doi: 10.1186/1471-2156-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riquet J, Coppieters W, Cambisano N, Arranz JJ, Berzi P, Davis SK, Grisart B, Farnir F, Karim L, Mni M, Simon P, Taylor JF, Vanmanshoven P, Wagenaar D, Womack JE, Georges M. Fine-mapping of quantitative trait loci by identity by descent in outbred populations: application to milk production in dairy cattle. Proc Natl Acad Sci U S A. 1999;96:9252–9257. doi: 10.1073/pnas.96.16.9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Zhang H, Li H, Li N, Zhang Y, Zhang Q, Wang S, Wang Q, Wang H. Fine-mapping quantitative trait loci for body weight and abdominal fat traits: effects of marker density and sample size. Poult Sci. 2008;87:1314–1319. doi: 10.3382/ps.2007-00512. [DOI] [PubMed] [Google Scholar]
- 21.Yu H, Edderkaoui B, Cortez A, Davidson HM, Wergedal JE, Baylink DJ, Mohan S. Mapping of the chromosome 17 BMD QTL in the F(2) male mice of MRL/MpJ x SJL/J. Genetica. 2009;135:59–66. doi: 10.1007/s10709-008-9258-6. [DOI] [PubMed] [Google Scholar]
- 22.Katsura I. In search of new mutants in cell-signaling systems of the nematode Caenorhabditis elegans. Review Genetica. 1993;88:137–146. doi: 10.1007/BF02424470. [DOI] [PubMed] [Google Scholar]
- 23.Katsura I, Kondo K, Amano T, Ishihara T, Kawakami M. Isolation, characterization and epistasis of fluoride-resistant mutants of Caenorhabditis elegans. Genetics. 1994;136:145–154. doi: 10.1093/genetics/136.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Take-Uchi M, Kawakami M, Ishihara T, Amano T, Kondo K, Katsura I. An ion channel of the degenerin/epithelial sodium channel superfamily controls the defecation rhythm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:11775–11780. doi: 10.1073/pnas.95.20.11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA, Wergedal JE, Bult C, Paigen B, Flint J, Tsaih SW, Churchill GA, Broman KW. A new standard genetic map for the laboratory mouse. Genetics. 2009;182:1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]