Abstract
Autism spectrum disorders (ASD) are pervasive neurodevelopmental disorders that affect an estimated 1 in 110 individuals. Although there is a strong genetic component associated with these disorders, this review focuses on the multi-factorial nature of ASD and how different genome-wide (genomic) approaches contribute to our understanding of autism. Emphasis is placed on the need to study defined ASD phenotypes as well as to integrate large-scale ‘omics’ data in order to develop a “systems level” perspective of ASD which, in turn, is necessary to allow predictions regarding responses to specific perturbations and interventions.
Autism is among the most enigmatic disorders of child development in that it impacts a broad range of functions and exhibits considerable heterogeneity in its behavioral and physiological manifestations. Defined behaviorally, the 3 core functions affected are centered on social understanding and reciprocity, language acquisition and usage, and restricted interests often coupled with stereotyped, repetitive behaviors (American Psychological Association, 1994; Freitag, Staal, Klauck, Duketis, & Waltes, 2010; Volkmar et al., 1994). Although demonstrated to be highly heritable by twin and family studies (Bailey et al., 1995; Bolton et al., 1994; Folstein & Rutter, 1977; Hallmayer et al., 2002; Ronald et al., 2006), there is currently no gene or group of genes that can be used as biomarkers for diagnostic screening nor have genes been identified that clearly describe the etiology of autism. Thus, autism is generally regarded as a spectrum of disorders, which includes severe, classic autism as described by Kanner (1944), pervasive developmental disorders-not otherwise specified (PDD-NOS), and Asperger’s Syndrome, which is the mildest form described by Asperger (1944) with no cognitive impairment or language delay. Current assumptions are that autism spectrum disorders (ASD) may arise from different etiologies (Geschwind & Levitt, 2007), each of which is likely to be multigenic, or a disorder resulting from cumulative and multigenic risk factors (Jones & Szatmari, 2002).
Due to the expected multigenic nature of ASD, identification of causal genes through classical genetics approaches has been largely unsuccessful, with the exception of genes for syndromic disorders associated with autism (Benvenuto, Manzi, Alessandrelli, Galasso, & Curatolo, 2009), such as Fragile X (Y. Feng et al., 1995), tuberous sclerosis (Smalley, 1998), Retts (Amir & Zoghbi, 2000; Kim & H., 2000; Van den Veyver & Zoghbi, 2001), and Smith-Lemli-Opitz Syndrome (Kelley, 2000). However, the sequencing of the human genome has ushered in a new era of genetic analyses in which the whole genome can be efficiently screened for thousands of variations at once. In addition, similar large-scale genomic approaches (Chen, Jorgenson, & Cheung, 2009; Hawkins, Hon, & Ren, 2010), typically using microarray platforms, have been developed for genome-wide investigations of differences in gene expression (called transcriptomics) between and among individuals with ASD and unaffected individuals as well as regulatory or epigenetic mechanisms that control gene expression (epigenomics). Whereas each type of genome-wide analysis yields a wealth of information, integration of such information is expected to provide a more comprehensive level of understanding of ASD. Finally, given the likely genetic heterogeneity underpinning ASD, it is clear that progress in defining the genetic contributions to the etiologies of ASD as well as the pathobiology underlying different ASD symptoms must rely, at least in part, on the subdivision of the autistic population into more homogeneous subgroups or “phenotypes”, based upon similarity of the behavioral symptoms on which the diagnosis rests or on defined biological parameters, such as a measurable changes in brain structure and neuronal circuitry, which are ultimately responsible for many of the behaviors associated with ASD.
In this review, I describe the large-scale genomics approaches that are required to construct a molecular framework for a “systems level” of understanding of the molecular bases as well as pathological mechanisms of autism. In addition, I emphasize how the integration of genomic approaches, as well as the separation of ASD phenotypes, is necessary to identify and prioritize candidate genes for further study. Finally, I describe the impact of sex hormones on a novel candidate gene for ASD as an example of how gene by environment (G × E) interactions may give rise to the higher levels of testosterone that has been associated with autistic traits (Auyeung, Taylor, Hackett, & Baron-Cohen, 2010) as well as introduce the higher male:female sex bias in autism.
Integrative genomics: Constructing a molecular framework for a “systems level” understanding of autism
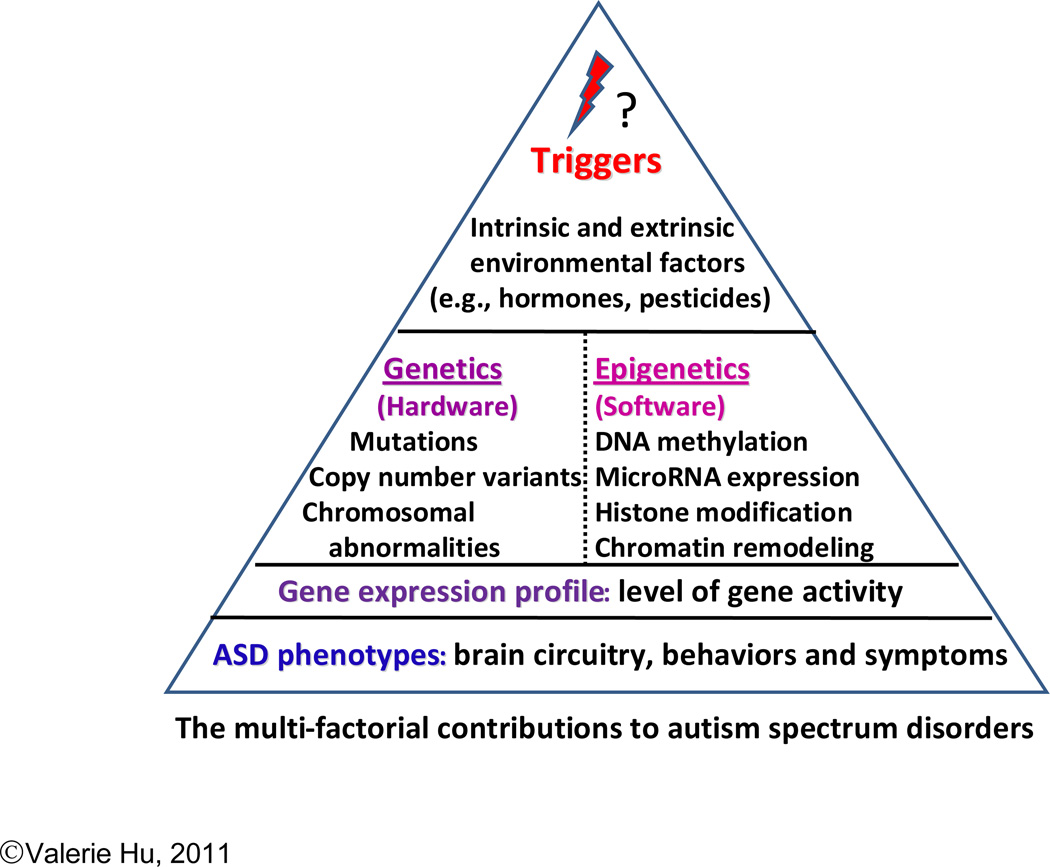
Since the first drafts of the human genome were published in 2001 (Lander et al., 2001; Venter et al., 2001), the term “genomics” has been widely used in the scientific literature. In its broadest definition, “genomics” is the “study of all of the nucleotide sequences, including structural genes, regulatory sequences, and noncoding DNA segments, in the chromosomes of an organism” (The American Heritage Medical Dictionary, 2007). Thus, genomics encompasses not just the study of structural (or sequence) variation in deoxyribonucleic acid (DNA), the heritable genetic material, but also the study of all of the regulatory factors that control the expression of genes, including epigenetic mechanisms and the environment. Figure 1 illustrates these factors in a hierarchical representation in which each level may influence the level or levels below. For example, a particular phenotype of ASD (i.e., that which is observable as a set of behaviors or symptoms reflecting aberrant neural circuitry) is the likely result of gene expression, or ribonucleic acid (RNA) levels, in the brain as well as other tissues in the affected individual. The expression profile, in turn, is a reflection of the aberrant developmental trajectory or end-stage pathology associated with the specific phenotype. Indeed, differentially expressed genes between individuals with and without autism have been demonstrated to affect metabolic and signaling pathways as well as cellular processes, such as cell migration and synapse formation, which are known to be disrupted in autism (as reviewed by Persico and Bourgeron, 2006). However, the pattern of gene expression is dictated by both genetic and epigenetic factors, which also exert reciprocal influences on each other. Whereas genetic factors involve the sequence of nucleotides in DNA which may have a direct impact on protein structure or gene transcription, epigenetic factors, which include DNA methylation, histone modification, chromatin remodeling, and microRNA expression, regulate the extent to which the genes are turned on or off, often in a coordinated manner. Crosstalk between genetic and epigenetic factors includes mutations in DNA which may affect epigenetic modifications, or aberrations in epigenetic mechanisms that result in abnormal chromatin structure leading ultimately to dysregulation of genes.
Fig. 1.
A Hierarchical View of the Multiple Factors That Cause, or Affect Risk for, Autism. In this view, the components of each level can influence those below.
The least understood of all the factors contributing to autism are environmental triggers, which include not just external factors (e.g., drugs, toxins or pesticides), but also intrinsic biological factors, such as hormones or inflammatory mediators, to which a fetus may be exposed in utero, which in turn can interact with susceptibility genes to produce an ASD phenotype. Such factors may exert their effect on gene expression and thus phenotype through either a genetic or epigenetic mechanism or both. A “systems level” of understanding of autism requires the integration of information pertaining to all of the above-mentioned factors contributing to ASD with the ultimate goal of being able to predict: 1) how perturbation in one or more of these factors might result in a particular ASD phenotype, and 2) how pharmacological or other therapeutic intervention might reverse or ameliorate a specific deficit.
The remaining sections of this review first address the rationale for developing a phenotypic approach to studying autism, and then summarize recent findings in each of the fields encompassed by genomics, demonstrating where possible how the integration of different types of ‘omics’ data can enhance our understanding of the pathophysiology of this complex neurological disorder.
Rationale for a phenotypic approach to studies of ASD
There have been many attempts to subdivide autistic individuals according to a variety of different symptoms in order to obtain more homogeneous subgroups of individuals for genetic and other biological analyses. Some studies employ categorical subdivision of affected individuals, for example, separating those who have exhibited regression (e.g., loss of acquired language or social skills), from those who exhibited early onset of autism without regression. Using this approach to subdivide autistic individuals for gene expression analyses of their blood lymphocytes, Gregg et al. (2008) demonstrated that a set of genes involved in natural killer (NK) cell cytotoxicity was shared among the different ASD phenotypes. Inasmuch as ASD is a complex disorder exhibiting a wide variety of different symptoms, other studies employed data from the Autism Diagnostic Interview-Revised (ADI/ADI-R), Autism Diagnostic Observation Schedule (ADOS), as well as other test instruments and clinical observations to identify distinct, phenotypic components within the ASD spectrum (Lord, Leventhal, & Cook Jr., 2001; Nurmi et al., 2003; Rapin, Dunn, Allen, Stevens, & Fein, 2009; Silverman et al., 2001; Stevens et al., 2000; Tadevosyan-Leyfer et al., 2003; Tanguay, Robertson, & Derrick, 1998). One study, which analyzed 98 items on the ADI/ADI-R assessment instrument from 292 individuals, identified six clusters of variables that included spoken language, social intent, compulsions, developmental milestones, savant skills, and sensory aversions (Tadevosyan-Leyfer et al., 2003). By selecting for the "savant skills" phenotype, Nurmi et al. (2003) further demonstrated greatly increased linkage to chromosome 15q11-q13 relative to that obtained with an unsegregated ASD population. Similarly, incorporating language phenotypes into linkage analyses highlighted markers on chromosome regions 7q and 13q (Bradford et al., 2001). However, these loci were not replicated in an independent study focused on language-related endophenotypes (Spence et al., 2006). As an alternative approach to reducing heterogeneity, some studies have stratified individuals with autism dimensionally by quantitating severity of a particular symptom. For example, stratifying the ASD population according to the severity of repetitive behavior allowed quantitative correlations with altered sensitivity of the 5-HT (serotonin) 1d receptor, as manifested by sumatriptan-elicted growth hormone response (Hollander et al., 2000). Dimensional stratification also allows for inclusion of endophenotypes in quantitative trait analyses of genotyped individuals which increases the power of associating a genetic variant with a particular trait. Using this approach, several studies show that quantitative trait loci (QTL) can be clearly associated with social, language-impaired, and nonverbal communication-impaired endophenotypes of ASD (Alarcon, Yonan, Gilliam, Cantor, & Geschwind, 2005; Chen, Kono, Geschwind, & Cantor, 2006; Duvall et al., 2007).
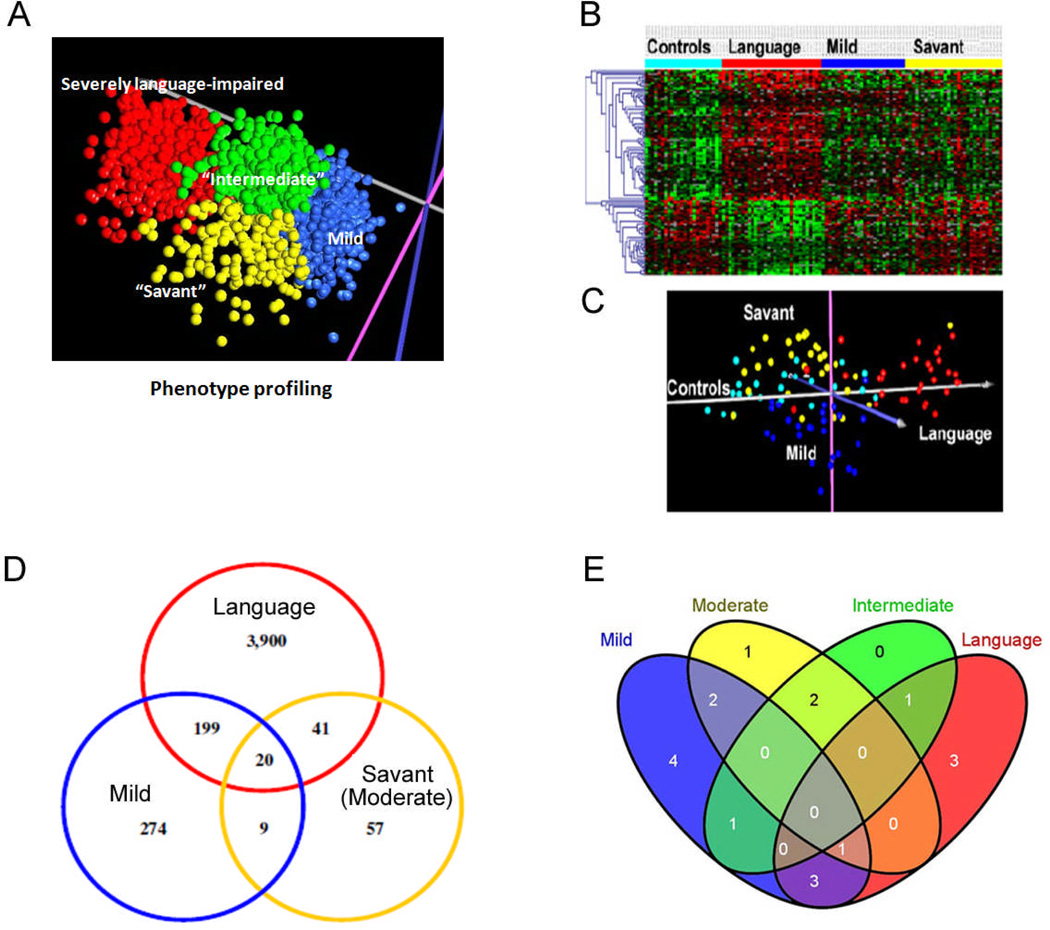
In order to identify phenotypic groups for gene expression profiling, Hu and Steinberg (2009) applied novel multiple clustering algorithms to 123 item scores from the ADI-R diagnostic instrument. This resulted in the identification of 4 ASD subgroups based on severity profiles across the ADI-R items used in this analysis. These subgroups included: 1) individuals with severe language impairment (defined by high severity scores on items related to “spoken language”), 2) those of moderate severity with a notably higher frequency of savant skills (defined by high scores on items related to “savant skills”), 3) those with a mild phenotype (~40% with a clinical diagnosis of Asperger’s Syndrome or PDD-NOS), and 4) those of an intermediate phenotype with neither severe language impairment nor notable savant skills. Figure 2A illustrates the results of phenotype clustering using ADI-R scores, whereas Figures 2B and C show the results of gene expression analyses in lymphoblastoid cell lines (LCL) derived from lymphocytes of individuals selected from different phenotypic clusters (Hu, Sarachana et al., 2009). Aside from demonstrating the distinction of at least 3 of these ASD phenotypes by gene expression analyses (Figure 2D), Hu, Sarachana et al. (2009) discovered that genes involved in circadian rhythm were prominent among those differentially expressed only in LCL from individuals with severe language impairment, demonstrating the value of subtyping by symptomatic profiles. This observation is particularly relevant in that it has been reported that sleep disturbances are prevalent in ASD, particularly among those with more significant language deficits and epilepsy (Malow, 2004). Similar application of phenotyping and quantitative trait analyses to genome-wide genetic (genotype) data also proved to be valuable in identifying ASD subtype-associated single nucleotide polymorphisms (SNPs), some of which were replicated in different subtypes (see Figure 2E) (Hu, Addington, & Hyman, 2011). Thus, a strong argument can be made for subtyping ASD subjects according to phenotype or severity within a specific domain as well as for including related individuals without a clinical diagnosis but expressing the “broad autism phenotype” (Piven, 2001) or a relevant “endophenotype” (trait) (Gottesman & Gould, 2003) in genetic and molecular analyses. By reducing the variability in this heterogeneous population, phenotypic or dimensional approaches are expected to enhance sensitivity with respect to the identification of genes and pathways contributing to, or associated with, the pathobiology of a specific behavioral or symptomatic phenotype of ASD, which may be amenable to therapy.
Fig. 2.
Linking Gene Expression and Genetic Profiles to ASD Phenotypes.
A) Identification of ASD phenotypes by cluster analyses of 123 severity scores from the ADI-R diagnostic instrument described by Hu and Steinberg (2009). The figure shows the results of a principal components analysis of ADI-R scores from 1351 autistic individuals as reported by Hu et al. (2009). Each point on the graph represents an individual, with colors corresponding to the indicated subtypes.
B) Gene expression profiling of lymphoblastoid cell lines (LCL) from the severely language-impaired, mild, and “savant” subtypes identified in (A) and from non-autistic controls (Hu, Sarachana et al., 2009). Note the gradient of gene expression represented by color (red is up-regulated and green is down-regulated in comparison to a standard reference RNA) that relates, either directly or inversely, to severity of ASD.
C) Principal components analysis of the gene expression data in (B) shows separation of the ASD phenotypes, as defined by cluster analyses of ADI-R scores. Each point on the graph represents an individual with colors indicating ASD subtype, as in (A). Turquoise represents controls without autism.
D) A Venn diagram showing the number of differentially expressed genes (RNA transcripts) according to subtype of ASD. There are both overlapping as well as unique genes associated with the 3 different subtypes of ASD that were studied by DNA microarray analyses.
E) Distribution of 18 novel SNPs across 4 subtypes of ASD (Hu et al., 2011). Note that 10 of the 18 SNPs are associated with more than one subtype.
Transcriptomics: Large-scale gene expression profiling
In comparison to genome-wide association or linkage analyses, there have been considerably fewer large-scale or genome-wide analyses of gene expression (RNA levels) in autism. The first such study was conducted with post-mortem brain tissues which revealed a set of 30 differentially expressed genes in the cerebellum of individuals who had been diagnosed with ASD vs. age-matched controls, which included 2 genes of the glutamate excitatory system (Purcell, Jeon, Zimmerman, Blue, & Pevsner, 2001). To explore the possibility that differential gene expression could also be detected in peripherally-derived cells from individuals with or without autism, Hu, Frank, Heine, Lee, and Quackenbush (2006) performed gene expression analyses on LCL from monozygotic twin pairs that were discordant in diagnosis of autism. This study design was used because identical twins who share the same genotype increase the power to identify differentially expressed genes that may be responsible for the discordant phenotype. This study revealed significant differences (≥ 1.5-fold) in gene expression levels between the co-twins, with many of the genes known to be involved in nervous system development and function. These included genes such as ROBO1, EGR2, and CHL1 that are involved in neurite extension and migration as well as genes that are involved in neuronal development and survival (e.g., DAPK1, HOXB2, IL6ST, NTRK2). Aside from demonstrating the diagnostic potential for identifying expressed biomarkers for autism in peripheral (non-neuronal) tissues, these findings also strongly suggested the involvement of epigenetic mechanisms in autism, since the genotypes (DNA sequences) are the same for identical twins.
Other genome-wide expression analyses using either LCL or primary lymphocytes have revealed the involvement of natural killer (NK) activity in autism (Baron, Liu, Hicks, & Gregg, 2006; Gregg et al., 2008) as well as semaphorin 5A (SEMA5A), a gene involved in axon guidance (Melin et al., 2006). The possible involvement of NK activity is important because NK cells are part of the innate immune system which is apparently activated in the brain tissues of some individuals with autism (Vargas, Nascimbene, Krishnan, Zimmerman, & Pardo, 2005). Another study investigated gene expression profiles of LCL from individuals exhibiting autism-related disorders with known genetic etiology, specifically, Fragile X syndrome and 15q11-q13 duplication (dup15) in order to gain insight into genes that are possibly also dysregulated in “idiopathic” (of unknown cause) autism (Nishimura et al., 2007). This study identified 68 shared differentially expressed genes between the two disorders, and further showed that 2 of the genes identified in their study were also differentially expressed in the LCL from male sib pairs who were discordant for idiopathic autism. With respect to expression profiling of idiopathic autism, Hu, Nguyen, Kim, Steinberg, Sarachana, et al. (2009) recently reported in a study of case-control sib pairs that, aside from genes involved in nervous system development and function, genes involved in steroid hormone biosynthesis and immune function were also differentially expressed. Dysregulation of hormone biosynthesis may be responsible for some of the systemic problems (e.g., sleep disturbance, gastrointestinal and immune disorders) associated with autism (Cohly & Panja, 2005; Johnson & Malow, 2008; Jyonouchi, Geng, Ruby, & Zimmerman-Bier, 2005; Pardo, Vargas, & Zimmerman, 2005; Valicenti-McDermott et al., 2006).
Coupling gene expression to ASD phenotype
In a larger study involving unrelated case-controls in which individuals with ASD were divided into subtypes by cluster analyses of ADI-R scores, Hu, Sarachana et al. (2009) demonstrated that, dependent on ASD phenotype, hundreds to thousands of genes were differentially expressed relative to non-autistic controls (see Fig. 2D). Of particular interest were the circadian rhythm genes that were associated only with the subtype with severe language impairment. This finding is of interest because circadian rhythm controls daily physiological activities such as sleep and digestive functions, which are often disturbed in individuals with ASD (Johnson & Malow, 2008). The 20 noncoding transcripts that were found to be differentially expressed among all ASD subtypes relative to controls are particularly intriguing because many of these transcripts were found to be responsive to dihydrotestosterone (a potent metabolite of testosterone), further implicating the involvement of the sex hormones in ASD. As noncoding transcripts do not code for proteins, it is possible that these transcripts may have regulatory functions that are impacted by male hormones. It was also noteworthy that some genes exhibited expression levels that were directly or inversely related to the severity of the phenotype (see Fig. 2C), further suggesting the relevance of these genes to ASD. By associating with severity of autism, transcript abundance may be considered an intermediate quantitative trait connecting genetic variation to behavioral phenotypes of autism. Thus, expression quantitative trait loci (eQTL) analyses may be useful in identifying functionally relevant genetic polymorphisms.
Genetics: The value of quantitative traits and subtyping in the analyses of genome-wide genetic data
Epidemiological studies demonstrating the high heritability of autism (estimated at 60–90% based on concordance rates between monozygotic and dizygotic twins) have long implicated genetics as a major contributing factor towards ASD (Folstein & Rutter, 1977). Recently, genome-wide approaches have been used in an attempt to identify genetic variants or SNPs that are either causal for, or strongly associated with, autism. However, these efforts have been hampered in part by the aforementioned heterogeneity within the autistic population, resulting in the identification of relatively few significant and reproducible SNPs that are associated with ASD. Because genetics and genome-wide association studies (GWAS) of ASD are discussed by Connolly et al. elsewhere in this issue, I will confine my discussion here to a recent study by Hu, Addington, and Hyman (2011) which reported 18 novel and highly significant SNPs that are associated with 4 subtypes of ASD that were identified by cluster analyses of ADI-R scores as described above. This study involved a reanalysis of published GWAS data (Wang et al., 2009) and incorporated both quantitative trait and subtype-dependent association analyses to identify SNPs associated with ASD (Hu et al., 2011). Selected ADI-R item scores from a subset of the individuals genotyped in the original study were first used to derive quantitative traits for deficits in language, nonverbal communication, play skills, and social development, as well as for insistence on sameness. Quantitative trait association analyses were then performed to identify SNPs that associated with these five quantitative traits. These analyses resulted in the identification of 167 unique SNPs out of the 513,312 SNPs on the array that survived quality control tests. These SNPs or quantitative trait loci (QTL) were then used in case-control genetic association analyses in which individuals with ASD were either combined into one group or divided by subtype as described above. Whereas no significant SNPs emerged from the analysis of the combined group of individuals with ASD vs. unaffected individuals, 18 novel SNPs were identified across the 4 ASD subtypes, with 10 of the SNPs associated with two and, in one case three, subtypes (Fig. 2E). The odds ratios for the shared SNPs which are related to the relative risk for ASD were different for the different subtypes, further suggesting genetic heterogeneity among the subtypes. This study thus demonstrates the increased statistical power to identify significant common genetic variants when the heterogeneity of the individuals tested is reduced by subtyping. Interestingly, all of the significant SNPs are located in nonexonic regions of the DNA, which means that they do not affect protein structure and suggests that they may instead play a role in gene regulation.
However, it is important to keep in mind that the candidate genes identified through such analyses are still only “guilty by association.” Gene knockdown or overexpression studies in appropriate animal models are still necessary to prove causation of specific ASD symptoms and behaviors.
Epigenomics of autism: Seeking answers beyond genetics
Epigenomics refers collectively to the epigenetic mechanisms operating on the genome to regulate gene expression and, ultimately, the phenotype of the organism. This includes regulation by DNA methylation, histone modification, chromatin remodeling, and microRNA expression, all of which have been shown to be important to nervous system development and function (Chuang & Jones, 2007; J. Feng & Fan, 2009; Hsieh & Eisch, 2010; Martino, Di Girolamo, Orlacchio, Datti, & Orlacchio, 2009; Mill et al., 2008). So far, there have been relatively few studies on the epigenetic mechanisms involved in autism, although disorders of imprinting (that is, DNA methylation dependent on parent-of-origin) such as Angelman and Praeder-Willi Syndromes which can give rise to some autistic features, implicate methylation anomalies. As mentioned earlier, both behavioral as well as gene expression studies on discordant monozygotic twins are highly suggestive of epigenetic factors in ASD (Bailey et al., 1995; Folstein & Rutter, 1977; Hallmayer et al., 2002; Hu et al., 2006; Le Couteur et al., 1996). However, there are only a handful of studies to date that specifically focus on epigenetics of autism, recently reviewed in Grafodatskaya, Chung, Szatmari, & Weksberg (2010). One such study is that of Nagarajan et al. (2008) which demonstrated increased promoter methylation in the MeCP2 gene that correlated with reduced expression in the frontal cortex of males with autism. Mutations in MeCP2, a methyl-CpG binding protein that is itself a mediator of epigenetic modulation, are causal for Retts Syndrome, which may also be associated with autism (Benvenuto et al., 2009). A more recent study demonstrated methylation differences in the oxytocin receptor gene, OXTR, in both peripheral blood cells and post-mortem temporal cortex that correlated with a diagnosis of ASD (Gregory et al., 2009). OXTR has also been implicated in ASD by family-based association analyses involving the Chinese Han population (Wu et al., 2005) and more recently, by association studies involving Caucasian children and adolescents (Jacob et al., 2007; Yrigollen et al., 2008). With respect to ASD phenotype, the OXTR as well as its ligand, oxytocin (OXT), have been shown to be critical for social cognition, as revealed by OXTR-deficient (Takayanagi et al., 2005) and OXT knockout animal models (Crawley et al., 2007; Winslow & Insel, 2002).
Global methylation analysis reveals a novel ASD candidate gene
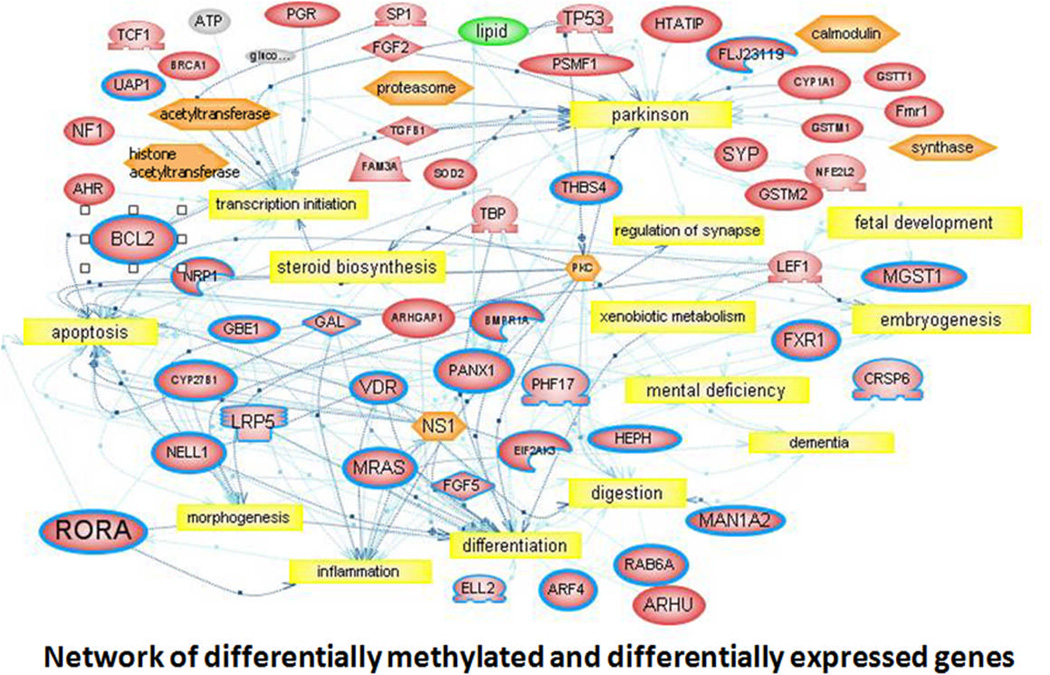
To more fully explore the contributions of DNA methylation to gene expression in autism, Nguyen et al. (2010) conducted a global methylation analysis of LCL from discordant monozygotic twins and sib pairs, and compared the differentially methylated genes with the differentially expressed genes from the same samples (Hu et al., 2006; Hu et al., 2009). This study revealed an overlapping set of genes that exhibited increased methylation and reduced expression. Figure 3 shows a gene interaction network that reveals the inter-relationships between these genes and processes associated with ASD. Further analyses confirmed two of the genes in the network, RORA and BCL2, as being both differentially methylated and expressed. While BCL2 is a gene that is important for cell survival, RORA (retinoic acid-related orphan receptor alpha) is a nuclear transcription factor that plays a critical role in cerebellar development as well as other functions known to be impaired in ASD. It is particularly interesting to note that both RORA and BCL2 proteins were reduced in the post-mortem cerebellum (also frontal cortex for RORA) of individuals with ASD vs. age-matched controls (Nguyen et al., 2010), demonstrating linkage between these molecular alterations in a peripheral tissue and the brain.
Fig. 3.
Network of Differentially Methylated and Differentially Expressed Genes. The network shows interactions among 25 genes (circumscribed in boldface) that were found to be differentially methylated in studies by Nguyen et al (2010) and differentially expressed in separate studies on the same samples by Hu et al. (2006) and by Hu, Sarachana et al. (2009). This interactive gene network was generated using Pathway Studio 5 network prediction software and identified common biological themes, including apoptosis, cellular differentiation, and inflammation. The analysis also revealed neurologically relevant functions and disorders including synaptic regulation, development, and mental deficiency. This information can be used to prioritize ASD candidate genes by function for further testing.
Relevance of RORA to the pathophysiology of autism
Whereas reduction of BCL2 protein has been previously reported in the post-mortem brain of individuals who had been diagnosed with ASD (Fatemi & Halt, 2001; Fatemi, Stary, Halt, & Realmuto, 2001), RORA is a novel ASD candidate gene with important functions related to the pathology observed in autism. RORA was originally identified as a spontaneously mutated gene that is causal for the mouse staggerer phenotype (Dussault, Fawcett, Matthyssen, Bader, & Giguère, 1998; Steinmayr et al., 1998). Using the mouse model, RORA was shown to be critical for Purkinje cell differentiation (Doulazmi et al., 1999; Hadj-Sahraoui et al., 2001) and cerebellar development (Gold et al., 2003; Harding, Atkins, Jaffe, Seo, & Lazar, 1997). It is notable that Purkinje cell deficiency is one of the earliest reported and most consistent neuroanatomical abnormalities in autism (Palmen, van Engeland, Hof, & Schmitz, 2004). RORA also plays a neuroprotective role against oxidative stress (Boukhtouche et al., 2006) and inflammation (Delerive et al., 2001). These functions are significant as there is evidence for both increased oxidative stress (Chauhan & Chauhan, 2006) and inflammation in the brains of at least some individuals with autism (Pardo et al., 2005). RORA is also a regulator of circadian rhythm (Akashi & Takumi, 2005; Sato et al., 2004), the disruption of which has been implicated not only by gene expression studies on ASD phenotypes (Hu, Sarachana et al., 2009), but also by an increasing number of genetic studies (Melke et al., 2008; Nicholas et al., 2007). Although RORA deficiency has been studied primarily with respect to ataxia and hypotonia in the RORA knockout or staggerer mouse models (Gold, Gent, & Hamilton, 2007), it is also associated with restricted behaviors reminiscent of ASD, such as perseverance, limited maze patrolling (Goodall & Gheusi, 1987), reduced exploration (Lalonde, 1987), and anomalous spatial learning (Lalonde & Strazielle, 2003). Thus, the study by Nguyen et al. (2010), which demonstrates reduction of RORA in both brain tissues and LCL derived from peripheral tissues of individuals with ASD, suggests the value of integrating epigenomics with transcriptomics in identifying critical and novel candidate genes with functional relevance to the pathobiology of autism.
MicroRNA expression profiling in tissues and cells from individuals with autism
Another epigenetic mechanism that has been studied to a limited extent with respect to ASD is microRNA (miRNA) expression. MiRNAs are endogenous, single-stranded, non-coding RNA molecules of approximately 22 nucleotides in length that negatively regulate gene expression at both transcriptional and post-transcriptional levels (Nilsen, 2007). There is ample evidence that miRNAs are involved in the development and function of the nervous system (Bicker & Schratt, 2008; Fiore, Siegel, & Schratt, 2008; Kapsimali et al., 2007; Kosik, 2006). Abu-Elneel and colleagues first reported differences in miRNA expression in cerebellar tissues from 13 autistic and 13 age-, gender-, PMI (post-mortem index)- and hemisphere-matched controls (Abu-Elneel et al., 2008). Although not validated, NRXN1 and SHANK3 were postulated to be potential target genes of some of the differentially expressed miRNAs. Talebizadeh, Butler, and Theodoro (2008) also profiled miRNA expression, but in LCL from 6 individuals with ASD and 6 age- and sex-matched individuals. They observed 9 miRNAs to be differentially expressed between the individuals with ASD and controls, with the majority of the miRNAs having predicted gene targets that had been previously identified as candidate genes for ASD by genetic analyses. Interestingly, 4 out the 9 miRNAs overlapped with those identified by the Abu-Elneel study, indicating that differentially expressed brain-related miRNAs can also be differentially expressed in LCL.
In a recent study involving miRNA expression profiling of LCL from discordant monozygotic twins and sib pairs, Sarachana, Zhou, Chen, Manji, and Hu (2010) also observed differentially expressed brain-specific and brain-related miRNAs, in addition to several that are induced by neuronal differentiation. Moreover, by overlaying previously obtained gene expression data from the same samples (Hu et al., 2006; Hu, Sarachana et al., 2009) onto predicted targets of the 49 differentially expressed miRNAs, Sarachana et al. (2010) were able to identify genes that are likely to be regulated by miRNAs in autism. They further validated two of the miRNA target genes (ID3 and PLK2) by miRNA overexpression and knockdown studies, respectively. These genes are involved in circadian rhythm signaling as well as synaptic modulation, a recurrent biological theme in ASD. To summarize, this study once again demonstrates the value of integrating different types of large-scale genomic data in order to obtain a clearer picture of the intricate gene regulatory mechanisms involved in this complex developmental disorder.
Environmental contributions to autism
“Environment” as used here means any factor or agent that is not part of the genome per se. Thus, it includes intrinsic factors, such as hormones, inflammatory mediators, and other biological molecules that may make up the microenvironment around a developing fetus or neonatal brain as well as extrinsic factors such as environmental pollutants, drugs, and food additives that may have an impact on maternal, fetal, or neonatal tissues. Whereas there is much debate regarding the contribution of various environmental factors to autism, there are few rigorous experimental studies that prove or disprove the causal nature of such factors. Much of what we currently know about environmental contributions to ASD comes from epidemiological studies that associate the occurrence of ASD with prenatal exposure or conditions. This information is complemented by a few studies with animal models exposed to suspected environmental agents or conditions. Landrigan (2010) described the environmental contributions to autism in a recent review.
Epidemiological studies
There is cumulative evidence from epidemiological studies that fetal exposure to alcohol or the drugs thalidomide (formerly used to treat morning sickness), valproic acid (VPA, an anticonvulsant), and more recently, misoprostol (a prostaglandin analogue used to treat gastric ulcers) are associated with increased incidence of autism (Landrigan, 2010; Miyazaki, Narita, & Narita, 2005). As early as 1994, autism was diagnosed in 4 out of 100 cases of thalidomide embryopathy in a Swedish study (Stromland, Nordin, Miller, Akerstrom, & Gillberg, 1994). In the same year, fetal valproate syndrome was reported to be associated with a 50% rate of autism in 2 sibling pairs exposed in utero to VPA (Christianson, Chesler, & Kromberg, 1994; Moore et al., 2000; Williams et al., 2001). Over the years, additional cases of autism demonstrated that embryonic exposure to VPA carried a risk factor of ~11% (Landrigan, 2010). While thalidomide is no longer used to treat pregnant women, VPA is still used to treat seizures, bipolar disorder, and sometimes major depression. However, given the elevated risk for craniofacial, skeletal, and limb malformations as well as ASD in the developing fetus (Rodier, Ingram, Tisdale, & Croog, 1997), care must be taken to avoid treating women during pregnancy. Aside from the drugs mentioned above, ASD has also been associated with immune activation (Vargas et al., 2005; Patterson, 2009) or maternal infection with rubella or influenza during pregnancy (Jones & Szatmari, 2002; Landrigan, 2010). These conditions implicate inflammation in the microenvironment of the developing fetus as a risk factor for autism, among other complications. What the epidemiological studies show is that environmental triggers may play a significant role in the etiology of ASD, although little is known about the molecular mechanisms through which these agents increase risk for ASD.
Impact of sex hormones on RORA: An example of an intrinsic environmental trigger
As illustrated in Fig. 1, environmental triggers may impact any of the molecular processes associated with proper expression and function of genes. Because of the reported increase in testosterone levels in some individuals with autism as well as the strong bias towards males, Sarachana et al. (2011) examined the effect of both male and female hormones on RORA, a candidate gene for ASD which Nguyen et al. (2010) identified through a combination of gene expression and methylation analyses. The former study showed that both male and female hormones regulate the expression of RORA, but in opposite directions. The male hormones suppress RORA, while female hormones enhance its expression. Furthermore, the study demonstrated that RORA, a transcription factor, in turn regulates aromatase (CYP19A1), an enzyme that is responsible for the conversion of male to female hormones. Sarachana et al. (2011) therefore postulated that a deficiency in RORA would result in a deficiency in aromatase, leading to the build-up of testosterone which can further inhibit RORA expression. In fact, they observed a strong correlation between the levels of RORA and aromatase proteins in brain tissues of individuals with autism vs. brain tissues from unaffected individuals, with both proteins reduced in the brain tissues from affected individuals (Sarachana et al., 2011). These results which demonstrate a specific gene by environment (G × E) interaction, provide a plausible molecular explanation for the previously unexplained increase in testosterone levels in some individuals with ASD (Auyeung et al., 2010; Ingudomnukul, Baron-Cohen, Wheelwright, & Knickmeyer, 2007). Interestingly, RORA and the estrogen receptor (ER) are transcription factors that share a consensus binding site on DNA (AGGTCA) and, consequently, common target genes. Sarachana et al. (2011) therefore proposed that the existence of shared gene targets may explain why females, with higher levels of estrogens, are less susceptible to autism. That is, estrogens may not only protect females against autism by increasing the level of RORA expression, but also by inducing shared target genes of RORA through ER, thus compensating in part for RORA deficiency. The observation that RORA regulates many genes that function at the glutamatergic synapse (Gold et al., 2003) further suggests that deficiency of RORA will have an impact on synaptic structure and neuronal circuitry.
Connecting genes and pathways to neuronal circuitry
Although much progress has been made in recent years in the identification of genes, pathways, and functions impacted by ASD, there is still a critical need to translate this information into a better understanding of the defective brain circuitry that manifests ultimately as aberrant behaviors. The growing number of ASD candidate genes clearly converges on the synapse as a dysfunctional entity in the brain of individuals with ASD (Geschwind & Levitt, 2007; Persico & Bourgeron, 2006). However, the picture that emerges from the accumulated information on genes is still a static one in that we now know some of the players, but not how they interact dynamically or over the course of development to produce the atypical functional and structural connectivity that has been revealed by numerous neuroimaging studies of individuals with ASD, as described in recent reviews (Anagnostou & Taylor, 2011; Minshew & Keller, 2010). Clearly, there is a pressing need to study the trajectory of genomic changes across both normal and ASD-impacted brain development and to integrate this information with changes in neuronal circuitry and structure that can be studied through a variety of brain imaging methods (Anagnostou & Taylor, 2011).
Summary
This review demonstrates the value of defining phenotypes for genomic analyses of ASD and of integrating the different types of large-scale genomic data to provide a more comprehensive picture of the underlying biological deficits of ASD which are the likely results of an aberrant developmental trajectory or end-state pathology. Identification of these altered pathways and functions in cells and tissues from individuals with ASD is expected to provide a better understanding of the pathobiology of ASD. In turn, this knowledge of the biological underpinnings of autism is important to the identification of novel therapeutic targets and interventions that will hopefully improve the long-term outcomes of the nearly one in 110 individuals affected by this pervasive, yet fascinating, developmental disorder.
Acknowledgements/note
The author is grateful to Dr. Isabelle Rapin (Depts. of Neurology and Pediatrics, Albert Einstein College of Medicine, NY) for thoughtful feedback and suggestions on an earlier version of this manuscript, and to Dr. Anthony-Samuel LaMantia (Dept. of Pharmacology and Physiology, and Institute of Neuroscience, The George Washington University, DC) for helpful discussions. The work that is described from the author’s laboratory was supported by grants from NIMH (R21 MH073393) and Autism Speaks (#2381). The printing cost of this article was in part defrayed by a generous gift from The LIFE Foundation (Aspen, CO).
References
- Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, Kosik KS. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- Akashi M, Takumi T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nature Structural and Molecular Biology. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Alarcon M, Yonan AL, Gilliam TC, Cantor RM, Geschwind DH. Quantitative genome scan and ordered-subsets analysis of autism endophenotypes support language QTLs. Mol Psychiatry. 2005;10:747–757. doi: 10.1038/sj.mp.4001666. [DOI] [PubMed] [Google Scholar]
- The American Heritage Medical Dictionary. Editors of the American Heritage Dictionaries. 3rd ed. Boston, MA: Houghton Mifflin Company; 2007. [Google Scholar]
- American Psychological Association. Diagnostic and Statistical Manual of Mental Disorders. IV ed. Washington, DC: American Psychological Association; 1994. [Google Scholar]
- Amir RE, Zoghbi HY. Rett syndrome: Methyl-CpG-binding protein 2 mutations and phenotype-genotype correlations. Am J Med Genet. 2000;97:147–152. doi: 10.1002/1096-8628(200022)97:2<147::aid-ajmg6>3.0.co;2-o. 2<147::AID-AJMG6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Anagnostou E, Taylor MJ. Review of neuroimaging in autism spectrum disorders: What have we learned and where we go from here. Molecular Autism. 2011 Apr 18;2(4) doi: 10.1186/2040-2392-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asperger H. Die "autistischen psychopathen" im kindesalter. Archiv für Psychiatrie Und Nervenkrankheiten. 1944;117:76–136. [Google Scholar]
- Auyeung B, Taylor K, Hackett G, Baron-Cohen S. Foetal testosterone and autistic traits in 18 to 24-month-old children. Molecular Autism. 2010 Jul 12;1(11) doi: 10.1186/2040-2392-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Baron CA, Liu SY, Hicks C, Gregg JP. Utilization of lymphoblastoid cell lines as a system for the molecular modeling of autism. J Autism Dev Disord. 2006;36:973–982. doi: 10.1007/s10803-006-0134-x. [DOI] [PubMed] [Google Scholar]
- Benvenuto A, Manzi B, Alessandrelli R, Galasso C, Curatolo P. Recent advances in the pathogenesis of syndromic autisms. International Journal of Pediatrics. 2009;2009 doi: 10.1155/2009/198736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker S, Schratt G. microRNAs: Tiny regulators of synapse function in development and disease. Journal of Cellular and Molecular Medicine. 2008;12:1466–1476. doi: 10.1111/j.1582-4934.2008.00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton P, Macdonald H, Pickles A, Rios P, Goode S, Crowson M, Rutter M. A case-control family history study of autism. J Child Psychol Psychiatry. 1994;35:877–900. doi: 10.1111/j.1469-7610.1994.tb02300.x. [DOI] [PubMed] [Google Scholar]
- Boukhtouche F, Vodjdani G, Jarvis CI, Bakouche J, Staels B, Mallet J, Brugg B. Human retinoic acid receptor-related orphan receptor a1 overexpression protects neurones against oxidative stress-induced apoptosis. Journal of Neurochemistry. 2006;96:1778–1789. doi: 10.1111/j.1471-4159.2006.03708.x. [DOI] [PubMed] [Google Scholar]
- Bradford Y, Haines J, Hutcheson H, Gardiner M, Braun T, Sheffield V, Piven J. Incorporating language phenotypes strengthens evidence of linkage to autism. American Journal of Medical Genetics - Neuropsychiatric Genetics. 2001;105:539–547. [PubMed] [Google Scholar]
- Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13(3):171–81. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Chen GK, Kono N, Geschwind DH, Cantor RM. Quantitative trait locus analysis of nonverbal communication in autism spectrum disorder. Molecular Psychiatry. 2006;11:214–220. doi: 10.1038/sj.mp.4001753. [DOI] [PubMed] [Google Scholar]
- Chen X, Jorgenson E, Cheung ST. New tools for functional genomic analysis. Drug Discovery Today. 2009;14:754. doi: 10.1016/j.drudis.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson AL, Chesler N, Kromberg JGR. Fetal valproate syndrome: Clinical and neuro-developmental features in two sibling pairs. Developmental Medicine and Child Neurology. 1994;36:361–369. doi: 10.1111/j.1469-8749.1994.tb11858.x. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- Cohly HH, Panja A. Immunological findings in autism. Int Rev Neurobiol. 2005;71:317–341. doi: 10.1016/s0074-7742(05)71013-8. [DOI] [PubMed] [Google Scholar]
- Crawley JN, Chen T, Puri A, Washburn R, Sullivan TL, Hill JM, Young WS. Social approach behaviors in oxytocin knockout mice: Comparison of two independent lines tested in different laboratory environments. Neuropeptides. 2007;41:145–163. doi: 10.1016/j.npep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Delerive P, Monté D, Dubois G, Trottein F, Fruchart-Najib J, Mariani J, Staels B. The orphan nuclear receptor RORa is a negative regulator of the inflammatory response. EMBO Reports. 2001;2:42–48. doi: 10.1093/embo-reports/kve007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulazmi M, Frédéric F, Lemaigre-Dubreuil Y, Hadj-Sahraoui N, Delhaye-Bouchaud N, Mariani J. Cerebellar Purkinje cell loss during life span of the heterozygous staggerer mouse (Rora+/Rora(sg)) is gender-related. Journal of Comparative Neurology. 1999;411:267–273. 2<267::AID-CNE7>3.0.CO;2–4. [PubMed] [Google Scholar]
- Dussault I, Fawcett D, Matthyssen A, Bader J, Giguére V. Orphan nuclear receptor RORa-deficient mice display the cerebellar defects of staggerer. Mechanisms of Development. 1998;70:147–153. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- Duvall JA, Lu A, Cantor RM, Todd RD, Constantino JN, Geschwind DH. A quantitative trait locus analysis of social responsiveness in multiplex autism families. Am J Psychiatry. 2007;164:656–662. doi: 10.1176/ajp.2007.164.4.656. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Halt AR. Altered levels of Bcl2 and p53 proteins in parietal cortex reflect deranged apoptotic regulation in autism. Synapse. 2001;42:281–284. doi: 10.1002/syn.10002. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Stary JM, Halt AR, Realmuto GR. Dysregulation of reelin and bcl-2 proteins in autistic cerebellum. J Autism Dev Disord. 2001;31:529–535. doi: 10.1023/a:1013234708757. [DOI] [PubMed] [Google Scholar]
- Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int. Rev. Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- Feng Y, Zhang F, Lokey LK, Chastain JL, Lakkis L, Eberhart D, Warren ST. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995;268:731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochimica Et Biophysica Acta - Gene Regulatory Mechanisms. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Infantile autism: A genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- Freitag CM, Staal W, Klauck SM, Duketis E, Waltes R. Genetics of autistic disorders: Review and clinical implications. European Child and Adolescent Psychiatry. 2010;19:169–178. doi: 10.1007/s00787-009-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: Developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Gold DA, Baek SH, Schork NJ, Rose DW, Larsen DD, Sachs BD, Hamilton BA. RORα coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–1131. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold DA, Gent PM, Hamilton BA. RORα in genetic control of cerebellum development: 50 staggering years. Brain Research. 2007;1140:19–25. doi: 10.1016/j.brainres.2005.11.080. [DOI] [PubMed] [Google Scholar]
- Goodall G, Gheusi G. Abnormal patterns of maze patrolling in the mutant mouse staggerer. Behavioral and Neural Biology. 1987;47:307–320. doi: 10.1016/s0163-1047(87)90422-5. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Grafodatskaya D, Chung B, Szatmari P, Weksberg R. Autism spectrum disorders and epigenetics. Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49:794–809. doi: 10.1016/j.jaac.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Gregg JP, Lit L, Baron CA, Hertz-Picciotto I, Walker W, Davis RA, Sharp FR. Gene expression changes in children with autism. Genomics. 2008;91:22–29. doi: 10.1016/j.ygeno.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Gregory SG, Connelly JJ, Towers AJ, Johnson J, Biscocho D, Markunas CA, Pericak-Vance MA. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Medicine. 2009;7:62. doi: 10.1186/1741-7015-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadj-Sahraoui N, Frederic F, Zanjani H, Delhaye-Bouchaud N, Herrup K, Mariani J. Progressive atrophy of cerebellar Purkinje cell dendrites during aging of the heterozygous staggerer mouse (Rora+/sg) Developmental Brain Research. 2001;126:201–209. doi: 10.1016/s0165-3806(01)00095-5. [DOI] [PubMed] [Google Scholar]
- Hallmayer J, Glasson EJ, Bower C, Petterson B, Croen L, Grether J, Risch N. On the twin risk in autism. Am J Hum Genet. 2002;71:941–946. doi: 10.1086/342990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Atkins GB, Jaffe AB, Seo WJ, Lazar MA. Transcriptional activation and repression by RORα, an orphan nuclear receptor required for cerebellar development. Molecular Endocrinology. 1997;11:1737–1746. doi: 10.1210/mend.11.11.0002. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Ren B. Next-generation genomics: An integrative approach. Nature Reviews Genetics. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander E, Novotny S, Allen A, Aronowitz B, Cartwright C, DeCaria C. The relationship between repetitive behaviors and growth hormone response to sumatriptan challenge in adult autistic disorder. Neuropsychopharmacology. 2000;22:163–167. doi: 10.1016/S0893-133X(99)00121-9. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Eisch AJ. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: Unraveling the genome to understand the mind. Neurobiology of Disease. 2010;39:73–84. doi: 10.1016/j.nbd.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Addington A, Hyman A. Novel autism subtype-dependent genetic variants are revealed by quantitative trait and subphenotype association analyses of published GWAS data. PLoS ONE. 2011 Apr 27;6:e19067. doi: 10.1371/journal.pone.0019067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Frank BC, Heine S, Lee NH, Quackenbush J. Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics. 2006;7:118. doi: 10.1186/1471-2164-7-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Nguyen A, Kim KS, Steinberg ME, Sarachana T, Scully MA, Lee NH. Gene expression profiling of lymphoblasts from autistic and nonaffected sib pairs: Altered pathways in neuronal development and steroid biosynthesis. PloS One. 2009;4:e5775. doi: 10.1371/journal.pone.0005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Sarachana T, Kim KS, Nguyen A, Kulkarni S, Steinberg ME, Lee NH. Gene expression profiling differentiates autism case-controls and phenotypic variants of autism spectrum disorders: Evidence for circadian rhythm dysfunction in severe autism. Autism Research : Official Journal of the International Society for Autism Research. 2009;2:78–97. doi: 10.1002/aur.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Steinberg ME. Novel clustering of items from the autism diagnostic interview-revised to define phenotypes within autism spectrum disorders. Autism Research : Official Journal of the International Society for Autism Research. 2009;2:67–77. doi: 10.1002/aur.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingudomnukul E, Baron-Cohen S, Wheelwright S, Knickmeyer R. Elevated rates of testosterone-related disorders in women with autism spectrum conditions. Horm Behav. 2007;51:597–604. doi: 10.1016/j.yhbeh.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Jacob S, Brune CW, Carter CS, Leventhal BL, Lord C, Cook EH., Jr Association of the oxytocin receptor gene (OXTR) in Caucasian children and adolescents with autism. Neuroscience Letters. 2007;417:6–9. doi: 10.1016/j.neulet.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KP, Malow BA. Sleep in children with autism spectrum disorders. Current Neurology and Neuroscience Reports. 2008;8:155–161. doi: 10.1007/s11910-008-0025-y. [DOI] [PubMed] [Google Scholar]
- Jones MB, Szatmari P. A risk-factor model of epistatic interaction, focusing on autism. American Journal of Medical Genetics - Neuropsychiatric Genetics. 2002;114:558–565. doi: 10.1002/ajmg.10513. [DOI] [PubMed] [Google Scholar]
- Jyonouchi H, Geng L, Ruby A, Zimmerman-Bier B. Dysregulated innate immune responses in young children with autism spectrum disorders: Their relationship to gastrointestinal symptoms and dietary intervention. Neuropsychobiology. 2005;51:77–85. doi: 10.1159/000084164. [DOI] [PubMed] [Google Scholar]
- Kanner L. Early infantile autism. The Journal of Pediatrics. 1944;25:211–217. [Google Scholar]
- Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RI. Inborn errors of cholesterol biosynthesis. Adv Pediatr. 2000;47:1–53. [PubMed] [Google Scholar]
- Kim SJ, H CE., Jr Novel de novo nonsense mutation of MECP2 in a patient with rett syndrome. Hum Mutat. 2000;15:382–383. doi: 10.1002/(SICI)1098-1004(200004)15:4<382::AID-HUMU16>3.0.CO;2-8. 4<382::AID-HUMU16>3.0.CO;2–8. [DOI] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7(12):911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Lalonde R. Exploration and spatial learning in staggerer mutant mice. Journal of Neurogenetics. 1987;4:285–292. [PubMed] [Google Scholar]
- Lalonde R, Strazielle C. Motor coordination, exploration, and spatial learning in a natural mouse mutation (nervous) with Purkinje cell degeneration. Behavior Genetics. 2003;33:59–66. doi: 10.1023/a:1021003600900. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Morgan MJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Landrigan PJ. What causes autism? Exploring the environmental contribution. Current Opinion in Pediatrics. 2010;22:219. doi: 10.1097/MOP.0b013e328336eb9a. [DOI] [PubMed] [Google Scholar]
- Le Couteur A, Bailey A, Goode S, Pickles A, Robertson S, Gottesman I, Rutter M. A broader phenotype of autism: The clinical spectrum in twins. J Child Psychol Psychiatry. 1996;37:785–801. doi: 10.1111/j.1469-7610.1996.tb01475.x. [DOI] [PubMed] [Google Scholar]
- Lord C, Leventhal BL, Cook EH., Jr Quantifying the phenotype in autism spectrum disorders. American Journal of Medical Genetics. 2001;105:36–38. 1<36::AID-AJMG1053>3.0.CO;2–4. [PubMed] [Google Scholar]
- Malow BA. Sleep disorders, epilepsy, and autism. Mental Retardation and Developmental Disabilities Research Reviews. 2004;10:122–125. doi: 10.1002/mrdd.20023. [DOI] [PubMed] [Google Scholar]
- Martino S, Di Girolamo I, Orlacchio A, Datti A, Orlacchio A. MicroRNA implications across neurodevelopment and neuropathology. Journal of Biomedicine and Biotechnology. 2009;2009 doi: 10.1155/2009/654346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin M, Carlsson B, Anckarsater H, Rastam M, Betancur C, Isaksson A, Dahl N. Constitutional downregulation of SEMA5A expression in autism. Neuropsychobiology. 2006;54:64–69. doi: 10.1159/000096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melke J, Goubran Botros H, Chaste P, Betancur C, Nygren G, Anckarsäter H, Van Maldergem L. Abnormal melatonin synthesis in autism spectrum disorders. Molecular Psychiatry. 2008;13:90–98. doi: 10.1038/sj.mp.4002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. American Journal of Human Genetics. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshew NJ, Keller TA. The nature of brain dysfunction in autism: Functional brain imaging studies. Current Opinion in Neurology. 2010;23:124–130. doi: 10.1097/WCO.0b013e32833782d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Narita N, Narita M. Maternal administration of thalidomide or valproic acid causes abnormal serotonergic neurons in the offspring: Implication for pathogenesis of autism. Int J Dev Neurosci. 2005;23:287–297. doi: 10.1016/j.ijdevneu.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Moore SJ, Turnpenny P, Quinn A, Glover S, Lloyd DJ, Montgomery T, Dean JCS. A clinical study of 57 children with fetal anticonvulsant syndromes. Journal of Medical Genetics. 2000;37:489–497. doi: 10.1136/jmg.37.7.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan RP, Patzel KA, Martin M, Yasui DH, Swanberg SE, Hertz-Picciotto I, Lasalle JM. MECP2 promoter methylation and X chromosome inactivation in autism. Autism Research. 2008;1:169. doi: 10.1002/aur.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. The FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas B, Rudrasingham V, Nash S, Kirov G, Owen MJ, Wimpory DC. Association of Per1 and Npas2 with autistic disorder: Support for the clock genes/social timing hypothesis. Molecular Psychiatry. 2007;12:581–592. doi: 10.1038/sj.mp.4001953. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends in Genetics. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Martin CL, Vazquez-Lopez A, Spence SJ, Alvarez-Retuerto AI, Sigman M, Geschwind DH. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum Mol Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- Nurmi EL, Dowd M, Tadevosyan-Leyfer O, Haines JL, Folstein SE, Sutcliffe JS. Exploratory subsetting of autism families based on savant skills improves evidence of genetic linkage to 15q11–q13. J Am Acad Child Adolesc Psychiatry. 2003;42:856–863. doi: 10.1097/01.CHI.0000046868.56865.0F. [DOI] [PubMed] [Google Scholar]
- Palmen SJ, van Engeland H, Hof PR, Schmitz C. Neuropathological findings in autism. Brain. 2004;127:2572–2583. doi: 10.1093/brain/awh287. [DOI] [PubMed] [Google Scholar]
- Pardo CA, Vargas DL, Zimmerman AW. Immunity, neuroglia and neuroinflammation in autism. Int Rev Psychiatry. 2005;17:485–495. doi: 10.1080/02646830500381930. [DOI] [PubMed] [Google Scholar]
- Patterson PH. Immune involvement in schizophrenia and autism: Etiology, pathology and animal models. Behavioural Brain Research. 2009;204:313–321. doi: 10.1016/j.bbr.2008.12.016. [DOI] [PubMed] [Google Scholar]
- Persico AM, Bourgeron T. Searching for ways out of the autism maze: Genetic, epigenetic and environmental clues. Trends in Neurosciences. 2006;29:349–358. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Piven J. The broad autism phenotype: A complementary strategy for molecular genetic studies of autism. American Journal of Medical Genetics. 2001;105:34–35. 1<34::AID-AJMG1052>3.0.CO;2-D. [PubMed] [Google Scholar]
- Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57:1618–1628. doi: 10.1212/wnl.57.9.1618. [DOI] [PubMed] [Google Scholar]
- Rapin I, Dunn MA, Allen DA, Stevens MC, Fein D. Subtypes of language disorders in school-age children with autism. Developmental Neuropsychology. 2009;34:66–84. doi: 10.1080/87565640802564648. [DOI] [PubMed] [Google Scholar]
- Rodier PM, Ingram JL, Tisdale B, Croog VJ. Linking etiologies in humans and animal models: Studies of autism. Reproductive Toxicology. 1997;11:417–422. doi: 10.1016/s0890-6238(97)80001-u. [DOI] [PubMed] [Google Scholar]
- Ronald A, Happe F, Bolton P, Butcher LM, Price TS, Wheelwright S, Plomin R. Genetic heterogeneity between the three components of the autism spectrum: A twin study. J Am Acad Child Adolesc Psychiatry. 2006;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- Sarachana T, Xu M, Wu R, Hu VW. Sex hormones in autism: Androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PLoS ONE. 2011;6:e17116. doi: 10.1371/journal.pone.0017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarachana T, Zhou R, Chen G, Manji HK, Hu VW. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Medicine. 2010;2:23. doi: 10.1186/gm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Hogenesch JB. A functional genomics strategy reveals rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Silverman JM, Smith CJ, Schmeidler JM, Buxbaum JD, Lawlor BA, Fitzgerald M. Symptom domains in autism and related conditions: Evidence for familiality. American Journal of Medical Genetics - Neuropsychiatric Genetics. 2001;105:593. doi: 10.1002/ajmg.10048. [DOI] [PubMed] [Google Scholar]
- Smalley SL. Autism and tuberous sclerosis. Journal of Autism and Developmental Disorders. 1998;28:407–414. doi: 10.1023/a:1026052421693. [DOI] [PubMed] [Google Scholar]
- Spence SJ, Cantor RM, Chung L, Kim S, Geschwind DH, Alarcon M. Stratification based on language-related endophenotypes in autism: Attempt to replicate reported linkage. Am J Med Genet B Neuropsychiatr Genet. 2006;141:591–598. doi: 10.1002/ajmg.b.30329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmayr M, André E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, Auclair N, Becker-André M. Staggerer phenotype in retinoid-related orphan receptor α-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3960–3965. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MC, Fein DA, Dunn M, Allen D, Waterhouse LH, Feinstein C, Rapin I. Subgroups of children with autism by cluster analysis: A longitudinal examination. Journal of the American Academy of Child and Adolescent Psychiatry. 2000;39:346–352. doi: 10.1097/00004583-200003000-00017. [DOI] [PubMed] [Google Scholar]
- Stromland K, Nordin V, Miller M, Akerstrom B, Gillberg C. Autism in thalidomide embryopathy: A population study. Dev Med Child Neurol. 1994;36:351. doi: 10.1111/j.1469-8749.1994.tb11856.x. [DOI] [PubMed] [Google Scholar]
- Tadevosyan-Leyfer O, Dowd M, Mankoski R, Winklosky B, Putnam S, McGrath L, Folstein SE. A principal components analysis of the autism diagnostic interview-revised. Journal of the American Academy of Child and Adolescent Psychiatry. 2003;42:864–872. doi: 10.1097/01.CHI.0000046870.56865.90. [DOI] [PubMed] [Google Scholar]
- Takayanagi Y, Yoshida M, Bielsky IF, Ross HE, Kawamata M, Onaka T, Nishimori K. Pervasive social deficits, but normal parturition, in oxytocin receptor-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16096–16101. doi: 10.1073/pnas.0505312102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebizadeh Z, Butler MG, Theodoro MF. Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Research. 2008;1:307. doi: 10.1002/aur.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguay PE, Robertson J, Derrick A. A dimensional classification of autism spectrum disorder by social communication domains. Journal of the American Academy of Child and Adolescent Psychiatry. 1998;37:271–277. doi: 10.1097/00004583-199803000-00011. [DOI] [PubMed] [Google Scholar]
- Valicenti-McDermott M, McVicar K, Rapin I, Wershil BK, Cohen H, Shinnar S. Frequency of gastrointestinal symptoms in children with autistic spectrum disorders and association with family history of autoimmune disease. J Dev Behav Pediatr. 2006;27:S128–S136. doi: 10.1097/00004703-200604002-00011. [DOI] [PubMed] [Google Scholar]
- Van den Veyver IB, Zoghbi HY. Mutations in the gene encoding methyl-CpG-binding protein 2 cause Rett syndrome. Brain Dev. 2001;23(Suppl 1):S147–S151. doi: 10.1016/s0387-7604(01)00376-x. [DOI] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Xiao C. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, Klin A, Siegel B, Szatmari P, Lord C, Campbell M, Kline W. Field trial for autistic disorder in DSM-IV. Am J Psychiatry. 1994;151:1361–1367. doi: 10.1176/ajp.151.9.1361. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Hakonarson H. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G, King J, Cunningham M, Stephan M, Kerr B, Hersh JH. Fetal valproate syndrome and autism: Additional evidence of an association. Developmental Medicine and Child Neurology. 2001;43:202–206. [PubMed] [Google Scholar]
- Winslow JT, Insel TR. The social deficits of the oxytocin knockout mouse. Neuropeptides. 2002;36:221–229. doi: 10.1054/npep.2002.0909. [DOI] [PubMed] [Google Scholar]
- Wu S, Jia M, Ruan Y, Liu J, Guo Y, Shuang M, Zhang D. Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese Han population. Biological Psychiatry. 2005;58:74–77. doi: 10.1016/j.biopsych.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Yrigollen CM, Han SS, Kochetkova A, Babitz T, Chang JT, Volkmar FR, Grigorenko EL. Genes controlling affiliative behavior as candidate genes for autism. Biological Psychiatry. 2008;63:911–916. doi: 10.1016/j.biopsych.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]