Abstract
The inner cell mass of the mouse pre-implantation blastocyst comprises epiblast progenitor and primitive endoderm cells of which cognate embryonic (mESCs) or extra-embryonic (XEN) stem cell lines can be derived. Importantly, each stem cell type retains the defining properties and lineage restriction of their in vivo tissue of origin. Recently, we demonstrated that XEN-like cells arise within mESC cultures. This raises the possibility that mESCs can generate self-renewing XEN cells without the requirement for gene manipulation. We have developed a novel approach to convert mESCs to XEN cells (cXEN) using growth factors. We confirm that the downregulation of the pluripotency transcription factor Nanog and the expression of primitive endoderm-associated genes Gata6, Gata4, Sox17 and Pdgfra are necessary for cXEN cell derivation. This approach highlights an important function for Fgf4 in cXEN cell derivation. Paracrine FGF signalling compensates for the loss of endogenous Fgf4, which is necessary to exit mESC self-renewal, but not for XEN cell maintenance. Our cXEN protocol also reveals that distinct pluripotent stem cells respond uniquely to differentiation promoting signals. cXEN cells can be derived from mESCs cultured with Erk and Gsk3 inhibitors (2i), and LIF, similar to conventional mESCs. However, we find that epiblast stem cells (EpiSCs) derived from the post-implantation embryo are refractory to cXEN cell establishment, consistent with the hypothesis that EpiSCs represent a pluripotent state distinct from mESCs. In all, these findings suggest that the potential of mESCs includes the capacity to give rise to both extra-embryonic and embryonic lineages.
Keywords: Pluripotent stem cells, Directed differentiation, Extra-embryonic endoderm, FGF, Mouse embryo
INTRODUCTION
Early mammalian embryogenesis is characterized by a gradual restriction in the developmental potential of cells that constitute the embryo. By the blastocyst stage, embryonic and extra-embryonic cells have diverged in their fate and function (Gardner, 1985). The outer cells of the mouse blastocyst form the trophectoderm, which contributes only to the placenta. The inner cell mass (ICM) gives rise to epiblast progenitor cells, which are the source of most embryonic tissues and extra-embryonic mesoderm; and to primitive endoderm (PrE) cells, which form the two extra-embryonic endoderm (ExEn) lineages – visceral endoderm (VE) and parietal endoderm (PE) – of the yolk sac (Gardner and Papaioannou, 1975; Gardner and Rossant, 1979).
Mouse embryonic stem cells (mESCs) are pluripotent self-renewing in vitro cell lines derived from the ICM of blastocysts (Evans and Kaufman, 1981; Martin, 1981) and have overlapping gene expression with epiblast progenitors within the ICM, including the expression of pluripotency-associated transcription factors Oct4, Sox2 and Nanog (Nichols et al., 1998; Avilion et al., 2003; Chambers et al., 2003; Mitsui et al., 2003). In seminal experiments, Beddington and Robertson tested the commitment of mESCs via blastocyst injection and demonstrated that mESCs contribute principally to the epiblast of chimeric embryos (Beddington and Robertson, 1989). Intriguingly, mESCs can also contribute, albeit rarely, to extra-embryonic lineages such as the VE, PE and placental trophoblast (Beddington and Robertson, 1989). Trophoblast stem cells (TS) and extra-embryonic endoderm stem cells (XEN) can also be derived from mouse blastocysts; these are restricted in their developmental potential and express genes specific to trophectoderm or ExEn cells, respectively (Tanaka et al., 1998; Kunath et al., 2005).
In low adherent culture conditions, the aggregation of mESCs promotes the formation of embryoid bodies (EBs) which recapitulate important aspects of early embryogenesis, including both extra-embryonic and embryonic tissue lineage differentiation (Coucouvanis and Martin, 1995). Studies of VE and PE differentiation in EBs have revealed the function of several genes involved in ExEn development, including an essential function for GATA factors Gata4 and Gata6 (Soudais et al., 1995; Morrisey et al., 1998; Capo-Chichi et al., 2005), and the SOX factor Sox17 (Shimoda et al., 2007; Niakan et al., 2010). However, the stochastic nature of EB differentiation complicates the dissection of molecular interactions involved in development. In addition, the ExEn cells formed from EBs cannot be maintained indefinitely in culture as stable cell lines. However, the overexpression of Gata4 or Gata6 is sufficient to drive the establishment of self-renewing XEN cells from mESCs (Fujikura et al., 2002; Shimosato et al., 2007). Nevertheless, it remains unclear whether self-renewing XEN cells can be derived directly from mESCs without requiring transgenic over-expression.
The fibroblast growth factor (FGF) receptor Fgfr2 is enriched in PrE cells, and the ligand Fgf4 is expressed by epiblast progenitor cells within the ICM (Feldman et al., 1995; Arman et al., 1998; Guo et al., 2010). This complementary receptor-ligand expression suggests that epiblast-secreted Fgf4 may be functionally important for PrE development (Rappolee et al., 1994; Goldin and Papaioannou, 2003). It has recently been suggested that PrE formation requires non-cell-autonomous provision of Fgf4 by Nanog-expressing cells (Nichols et al., 2009; Messerschmidt and Kemler, 2010; Yamanaka et al., 2010; Frankenberg et al., 2011). Indeed, Fgf4 or Fgf2, which are not expressed in the early embryo, both function via Fgfr2 and are routinely added during XEN derivation from embryos (Kunath et al., 2005). However, it is unclear whether FGFs are required for XEN cell derivation or whether they function in an autocrine or paracrine manner. FGFs are not required for mESC maintenance and indeed appear to promote differentiation (Kunath et al., 2007; Stavridis et al., 2007). Accordingly, inhibition of FGF or ERK signalling, downstream of the FGF pathway, together with the inhibition of Gsk3 signalling (2i) allows mESCs to be maintained as self-renewing pluripotent stem cells (Ying et al., 2008).
By contrast to their differentiation-promoting effects in mESCs, FGFs contribute to maintenance of pluripotency of stem cells derived from post-implantation mouse epiblast cells (EpiSCs) (Brons et al., 2007; Tesar et al., 2007). EpiSCs thus resemble human embryonic stem cells (hESCs) in their molecular and phenotypic characteristics, including their responsiveness to activin and Nodal as factors that maintain pluripotency (Vallier et al., 2005; Brons et al., 2007; Tesar et al., 2007). Mouse EpiSCs and hESCs both express extra-embryonic lineage-associated genes in response to bone morphogenetic protein (BMP) 4 treatment (Xu et al., 2002; Vallier et al., 2009). Although a capacity for growth factor-induced extra-embryonic differentiation could indicate that EpiSCs and hESCs have broader differentiation potential than mouse ICMs or ESCs, this seems paradoxical, as mouse embryos lose the ability to form trophoblast and ExEn at the expanded blastocyst stage. A recent study suggests that BMP-treated EpiSCs and hESCs instead form a mesodermal subtype that expresses extra-embryonic lineage-associated genes under control of a BRACHYURY-dependent transcriptional network (Bernardo et al., 2011).
In this study, we investigate the capacity of mESCs and EpiSCs to form ExEn. ExEn cells are seen in clonally derived mESC cultures (Evans and Kaufman, 1981; Doetschman et al., 1985; Bradley and Robertson, 1986). Moreover, heterogeneous expression of pluripotency-associated genes Oct4/Pou5f1, Sox2 and Nanog has been noted in mESC cultures (Chambers et al., 2007; Toyooka et al., 2008; Kalmar et al., 2009; Lanner et al., 2010). A small proportion of cells in mESC cultures contain extra-embryonic lineage-associated genes (Sox17- and/or Hex-high-expressing cells) (Canham et al., 2010; Niakan et al., 2010) and chimera experiments in which single Sox17-high-expressing cells were reintroduced into embryos suggested that these cells are committed to an ExEn fate (Niakan et al., 2010). Altogether, these observations confirm those made previously by Beddington and Robertson, suggesting that mESCs maintain an ICM-like capacity for ExEn differentiation, in addition to their ability to form all embryonic lineages (Gardner, 1985; Beddington and Robertson, 1989). We therefore reasoned that stable XEN cells lines could be established directly from mESCs in adherent monolayer culture. We have developed a protocol to convert mESCs into stable self-renewing XEN (cXEN) cell lines without the requirement for gene manipulation. Significantly, we show that mESCs maintained in LIF and serum or 2i can give rise to cXEN cells, unlike EpiSCs, suggesting that distinct pluripotent stem cell states respond uniquely to differentiation-inducing signals. In all, our cXEN protocol uncovers significant and novel molecular mechanisms involved in ExEn development.
MATERIALS AND METHODS
Culture conditions for pluripotent stem cell lines
mESCs were maintained on mouse embryonic fibroblast (MEF) coated pre-gelatinised tissue culture plates (Corning) in serum and LIF: DMEM/F12 media (Invitrogen) supplemented with 20% knockout serum replacement (Invitrogen), 1% L-glutamine (Sigma), 1% penicillin/streptomycin (Sigma), 0.1 mM β-mercaptoethanol (Sigma) and 10 ng/ml of LIF. mESCs were also cultured on pre-gelatinised plates in 2i and LIF: N2B27 media (Stem Cell Science) supplemented with 1 μM of PD0325901 (Tocris), 3 μM of CHIR99021 (Axon) and 10 ng/ml of LIF. EpiSCs were grown on MEF media coated pre-gelatinised plates in CDM/BSA media supplemented with 10 ng/ml recombinant activin A and 12 ng/ml recombinant FGF2 as previously described (Brons et al., 2007). Additional details of the media components can be found in supplementary material Tables S1-S3.
cXEN cell derivation and culture
mESCs were cultured in pluripotency media until they reached 70-80% confluency, then dissociated into single cells with 0.05% Trypsin (Sigma) and seeded at a density of 1 × 104 cells/cm2 in standard mouse XEN cell media: [85% RPMI-1640 (Invitrogen), 13% FBS (Bioserum), 1% Glutamax (Stem Cell Technologies), 1% Penicillin/Streptomycin (Sigma), 0.1 mM β-mercaptoethanol (Sigma)]. Twenty-four hours after initial plating, the media were changed to cXEN cell derivation media: standard XEN media supplemented with 0.01 μM all-trans retinoic acid (Sigma) and 10 ng/ml activin A. Cells were maintained in derivation media for 48 hours. Cells were then dissociated with trypsin and plated at a 1:1 dilution on pre-gelatinized MEF-coated dishes and maintained hereafter in standard XEN media. When the XEN-like cells reached 80-90% confluency they were passaged with trypsin or manually picked and maintained in the absence of MEFs.
For FGF/ERK signalling inhibition, cells were treated with SU5402 (Calbiochem), PD0325901 (Tocris) or PD173074 (Tocris) for up to 6 days at the indicated concentrations. For VE differentiation, XEN cells were cultured in the presence of 50 μg/ml of recombinant BMP4 in standard XEN media.
Cell proliferation and viability assays
mESCs, embryo-derived XEN and cXEN cells were seeded at 0.5 × 104 cells/cm2 on pre-gelatinized 96-well tissue culture plates in mESC or standard XEN media for up to 5 days. An MTT assay (Invitrogen) was performed following inhibitor treatment and absorbance detected using an Envision 2104 Multilabel Plate Reader (Perkin Elmer, Waltham, USA).
Quantitative RT-PCR (qRT-PCR)
RNA was isolated using Trizol Reagent (Invitrogen) with DNaseI treatment. cDNA was synthesized using a First Strand cDNA Synthesis Kit (Fermentas). qRT-PCR was performed using Quantace Sensimix on an Applied Biosystems 7500 machine (Life Technologies Corporation, CA, USA). Primer pairs were designed using Primer3 software or previously published (Molkentin et al., 1997; Fujikura et al., 2002; Niwa et al., 2005; Brown et al., 2010) and are listed in supplementary material Table S4.
Immunohistochemistry and imaging
Samples were fixed in 4% paraformaldehyde at 4°C overnight, permeabilized with 0.5% Tween in 1 × PBS for 20 minutes and blocked with 10% FBS diluted in 0.1% Tween in 1 × PBS for 1 hour. Primary antibodies were diluted at 1:500 in blocking solution and samples incubated at 4°C rotating overnight. Samples were incubated for 1 hour at room temperature in 1:300 dilution of secondary antibody (Molecular Probes), then washed and covered with 0.1% Tween in 1 × PBS containing DAPI Vectashield mounting medium (Vector Lab). A list of the antibodies used can be found in supplementary material Table S5.
Images were taken either on an Olympus 1X71 microscope with Cell^F software (Olympus Corporation, Tokyo, Japan), Zeiss Axiovert 200M microscope with AxioVision Rel 4.7 software (Carl Zeiss, Jena, Germany), or Zeiss LSM 700 confocal microscope and ZEN software. Cell numbers were counted manually using the ImageJ Cell Counter Plugin.
Flow cytometry
Cells were dissociated with 0.05% Trypsin and re-suspended in 500 μl FACS buffer (1 × PBS, 10% FCS) and 7AAD solution (BD Pharmingen, 5 μl/106 cells) to exclude dead cells. Cells were labelled with stage-specific embryonic antigen 1 (SSEA1) primary antibody at a 1:500 dilution in FACS buffer and APC anti-mouse IgM (BD Pharmingen) secondary antibody at a 1:300 dilution, and incubated for 15 minutes on ice. After two washes in FACS buffer, cells were resuspended in 1-2 ml FACS buffer and analyzed on a Beckman Coulter CyAn ADP flow cytometer (Beckman Coulter, High Wycombe, UK). FlowJo software (Becton Dickinson, Oxford, UK) was used to generate dotplots.
Microarray analysis
Total RNA was isolated as above and DNase treated (Ambion). RNA quality was assessed on a Eukaryote Total RNA Nano Series II (Agilent Technologies, Santa Clara, CA, USA) then processed on an Agilent 2100 Bioanalyzer using the RNA electrophoresis program. All RNA samples were amplified using the Total Prep 96 RNA amplification kit (Ambion). Illumina expression microarray MouseWG-6_V2 (Illumina, CA, USA) was used and the data analyzed with Bioconductor packages. Data have been deposited with GEO and will be released six months after publication (Accession Number GSE38477).
RESULTS
A low dose of retinoic acid and activin promotes differentiation of mES to XEN cells
To quantify the proportion of XEN-like cells within mESC cultures in serum and LIF, we used a transgenic reporter cell line in which the gene encoding a green fluorescent protein has been introduced into the endogenous Sox17 locus (Sox17GFP/+) (Kim et al., 2007). We performed flow cytometry to capture the GFPhigh population and compared the expression of ExEn versus pluripotency-associated genes (supplementary material Fig. S1). The expression of the cell surface antigen SSEA1 (Solter and Knowles, 1978) was used to distinguish the pluripotent cell population. The majority of Sox17GFP/+ mESCs have high to moderate expression of SSEA1 and were GFPlow (92.3%), whereas a small proportion (1.9%) of cells have moderate SSEA1 expression and were GFPhigh. We then isolated and compared the gene expression of the GFPhigh versus GFPlow cells by qRT-PCR (supplementary material Fig. S1). The GFPhigh cells upregulate the expression of ExEn-associated genes Gata6, Pdgfra, Sox7, Sox17, Lama1, Sparc and Dab2 (Brown et al., 2010) compared with GFPlow cells, which either lack the expression of these genes, or express them at very low levels. Conversely, we find that the GFPlow cells express Pou5f1, Nanog and Sox2 at a higher level compared with the GFPhigh population (supplementary material Fig. S1). Altogether, this suggests that XEN-like cells exist within mESC cultures and may be coaxed to proliferate in XEN-promoting conditions.
Retinoic acid (RA) is a general differentiation-promoting agent that has previously been used to drive ExEn differentiation from F9 embryonal carcinoma cell lines (Strickland et al., 1980) and mESCs (Capo-Chichi et al., 2005; Soprano et al., 2007; Artus et al., 2010), suggesting that RA may be used to promote XEN derivation. Additionally, the TGFβ family member Nodal is expressed in PrE (Mesnard et al., 2006), suggesting that components of this pathway may also promote XEN derivation. However, a high dose of RA or activin, which acts via the Nodal-signalling pathway, can promote the differentiation of mESCs to neurons or definitive endoderm, respectively (Bibel et al., 2004; Kubo et al., 2004). We therefore reasoned that treatment of mESCs with a low-dose of RA and activin for a short duration could be used to promote XEN cell conversion.
We used Sox17GFP/+ mESCs to test RA and activin concentrations up to 100 μM and 20 ng/ml, respectively, in standard XEN media. We performed flow cytometry to quantify GFP expression 4 days after the start of the cXEN protocol reflecting active cell proliferation without compromising viability due to over confluency. We observed the highest proportion of GFP-expressing cells from 0.01-10 μM of RA together with 10 ng/ml activin (supplementary material Table S6) and have subsequently used the lowest effective concentration of 0.01 μM RA (Fig. 1A,B). Altogether, this suggests that RA and activin promote the conversion of mESCs to XEN (cXEN) cells.
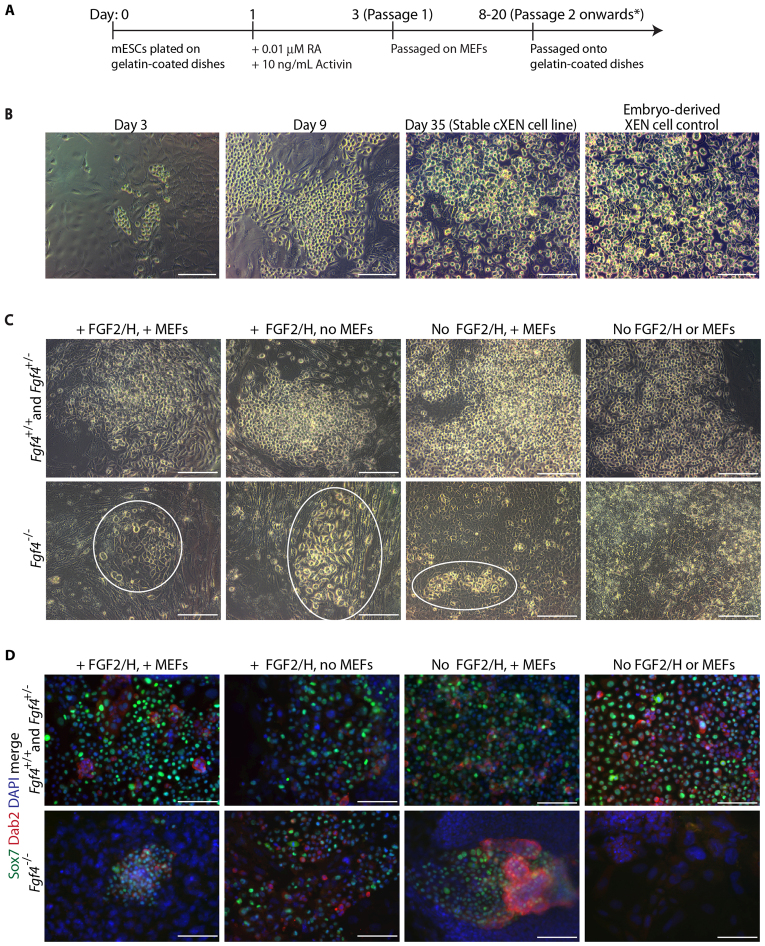
Fig. 1.
A growth factor-based method to convert mESCs into stable cXEN cell lines. (A) cXEN derivation protocol in standard XEN media. Cells were passaged onto MEFs at passage 1 and maintained on gelatin thereafter. *cXEN cells were passaged when they reached 70-80% confluency. (B) Representative phase-contrast images of wild-type cells at the defined time points during cXEN derivation (day 3 prior to the first passage, day 8 after the second passage and day 35 stable cXEN cell lines) compared with embryo derived XEN cell control. Scale bars: 100 μm. (C) Representative phase-contrast images of Fgf4+/+, Fgf4+/– and Fgf4–/– mESCs at day 13 following initiation of the cXEN protocol in the conditions indicated. XEN-like cells are circled in the Fgf4–/– conditions. Scale bars: 100 μm. (D) Immunofluorescence analysis for Sox7 (green) and Dab2 (red) expression with DAPI merge (blue) of Fgf4+/+, Fgf4+/– and Fgf4–/– in cXEN cell conditions at day 18. Scale bars: 100 μm.
Endogenous FGF is sufficient for cXEN cell derivation
FGF2 or FGF4 are routinely added during XEN cell derivation from pre-implantation embryos (Kunath et al., 2005). However, it is unclear whether FGF is required for XEN derivation and, if so, whether it functions in an autocrine or paracrine fashion. To test the effect of FGF signalling, we treated mESCs with cXEN derivation media supplemented with 24 ng/ml FGF2 and 1 μg/ml heparin, which facilitates FGF-receptor binding (Yayon et al., 1991). Surprisingly, we observed a greater proportion of Sox17GFP/+-expressing cells in the absence of exogenous FGF2 and heparin (supplementary material Table S6).
To further test the requirement for FGF signalling we derived Fgf4 mutant mESCs from blastocyst stage embryos (Feldman et al., 1995). We plated Fgf4+/+, Fgf4+/– and Fgf4–/– mESCs at the same density in cXEN derivation media with or without FGF2 and heparin (Fig. 1C). Cells were subsequently grown in either the presence or absence of MEFs, which provide a source of FGF (Wang et al., 2005). Consistently, both Fgf4+/+ and Fgf4+/– cells gave rise to cXEN cells in the absence of exogenous FGF. By contrast, substantial cell death was observed for Fgf4–/– cells grown in the absence of exogenous FGF following RA and activin treatment, and none of the surviving cells became cXEN cells (Fig. 1C).
We tested the efficiency of cXEN differentiation by immunofluorescence analysis for Nanog and Sox7 (supplementary material Table S7). In the absence of exogenous FGF, Sox7-positive cells were present in Fgf4+/+ and Fgf4+/– but not Fgf4–/– cells. Conversely, the highest proportion of Nanog-positive cells was observed in Fgf4–/– cells grown in the absence of exogenous FGF. Further immunofluorescence analysis for Sox7 and Dab2 confirmed that Fgf4–/– cells grown in the absence of exogenous FGF are indeed unable to give rise to cXEN cells (Fig. 1D). However, exogenous FGF can compensate for the endogenous deficiency, thereby allowing cXEN derivation from Fgf4–/– mESCs. This defect in commitment is reminiscent of the inability of Fgf4–/– mESCs to differentiate into other lineages, including neurons (Kunath et al., 2007; Stavridis et al., 2007), suggesting that FGF signalling is required to exit mESC self-renewal.
We then asked whether cXEN cells could be derived from mESCs in the presence of the FGF-receptor inhibitors PD173074 or SU5402 (Mohammadi et al., 1997; Mohammadi et al., 1998) or the ERK signalling inhibitor PD0325901 (Bain et al., 2007). We tested the toxic effect of the inhibitors on mESCs and determined that 10 μM SU5402, 5 μM PD0325901 or 1 μM PD173074 maintained mESC self-renewal, consistent with previous observations (Ying et al., 2008; Nichols et al., 2009) (supplementary material Fig. S2). Wild-type mESCs were subsequently plated in XEN derivation media supplemented with these inhibitors in the absence of exogenous FGF. To determine the proportion of XEN- or mESC-like cells, we performed immunofluorescence analysis for Gata6 and Sox7 compared with Nanog and Oct4. PD0325901 and PD173074 prevent cXEN derivation by restricting Gata6 and Sox7 expression and a larger proportion of cells express Nanog and Oct4 (supplementary material Table S8). Surprisingly, SU5402 did not restrict Gata6 and Sox7 expression to the same extent even at 10 or 100 μM concentrations (data not shown) suggesting that SU5402 may not be equivalent to PD173074 or PD0325901. Altogether, this suggests that endogenous Fgf4 functions via FGF/ERK signalling during cXEN derivation, rather than via a non-canonical mechanism.
We next asked whether FGF/ERK signalling is required to maintain cXEN cells after they are established. We plated embryo-derived XEN and cXEN cells in standard XEN media treated with SU5402, PD173074 or PD0325901 and determined the rate of cell proliferation by performing an MTT assay for up to 5 days following inhibitor treatment (supplementary material Fig. S2). FGF/ERK signalling inhibition had no effect on cell proliferation or cell identity, as determined by immunofluorescence analysis (supplementary material Fig. S2), confirming that it is not required to maintain XEN cells.
In all, this protocol has allowed us to derive stable cXEN cells from 16 mESC lines from a variety of backgrounds (supplementary material Table S9), demonstrating the robustness and reproducibility of this approach. In cases where we failed to derive cXEN cells, a crucial requirement for gene function within the PrE lineage has been suggested. We have maintained cXEN cells for over 4 months with no appreciable change in morphology.
cXEN cells are indistinguishable from embryo-derived XEN cells
Based on their morphological similarities, we next asked whether cXEN cells are molecularly identical to embryo-derived XEN cells. We performed immunofluorescence analysis for ExEn-associated cell surface or extracellular matrix proteins laminin, Dab2 and Sparc, and key endoderm transcription factors, including Gata4, Gata6, Sox7 and Sox17 (Fig. 2A), which are robustly expressed in cXEN cells, similar to embryo-derived XEN cells. Importantly, cXEN cells lacked expression of the pluripotency-associated transcription factor Oct4.
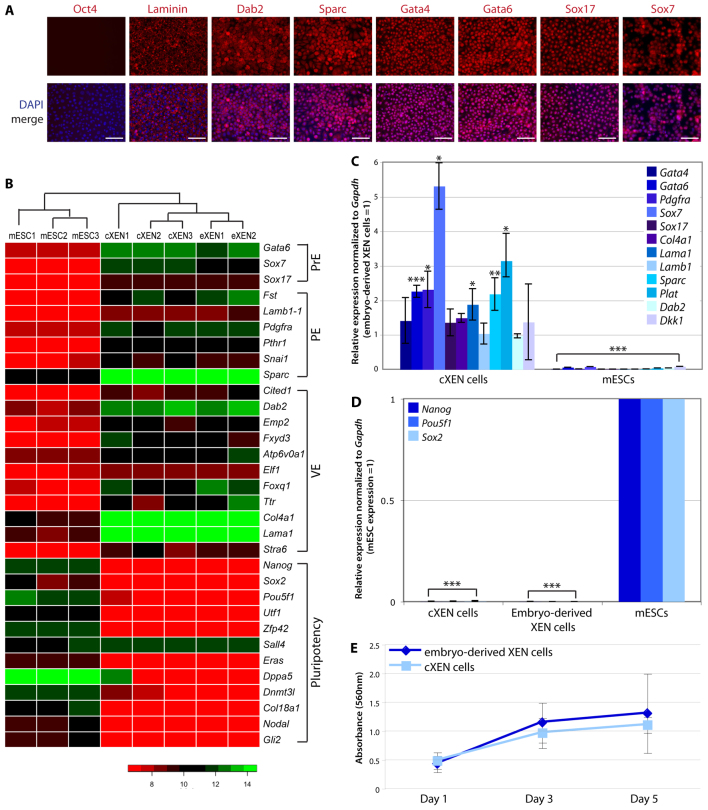
Fig. 2.
cXEN cells are identical to embryo-derived XEN cells. (A) Immunofluorescence analysis of stable cXEN cells (day 35) for Oct4, laminin, Dab2, Sparc, Gata4, Gata6, Sox17 and Sox7 expression (red) and DAPI (blue) merge (representative of three cXEN cell biological replicates). Scale bars: 100 μm. (B) Heat map of microarray data for pluripotency, primitive endoderm (PrE), visceral endoderm (VE), parietal endoderm (PE) or additional XEN cell-associated genes. Normalized gene expression values are represented by a colour heat map spectrum from high expression (green) to low expression (red). Data shown are biological replicates of cXEN cells, embryo-derived XEN cells (eXEN) and mESCs. (C,D) qRT-PCR analysis of cXEN cells (day 35) and mESCs for the expression of ExEn lineage-associated genes (C) (Gata4, Gata6, Pdgfra, Sox7, Sox17, Col4a1, Lama1, Lamb1, Sparc, Plat, Dab2 and Dkk1) or pluripotency associated genes (D) (Nanog, Pou5f1 and Sox2). Relative expression reflected as a fold difference compared with embryo-derived XEN cells or mESCs (=1), respectively. Data are mean±s.e.m. Three biological replicates. *P<0.05; **P<0.01; ***P<0.001. (E) MTT assay of the rate of proliferation of cXEN cells (day 35) compared with embryo-derived XEN cells. Cells were plated at the same cell density and analyzed after 1, 3 or 5 days of culture in standard XEN media. Data are the mean±s.e.m. Three biological and three technical replicates.
We next asked whether gene expression of cXEN cells is globally similar to embryo-derived XEN cells using cDNA microarray analysis (Fig. 2B). We performed hierarchical cluster analysis of absolute gene expression values after quantile normalization (Bolstad et al., 2003). A dendrogram of the clustering analysis illustrates the similarity between cXEN and embryo-derived XEN cells, which cluster together based on their gene expression patterns (Fig. 2B). mESCs have a distinct identity, clustering neither with cXEN nor with embryo-derived XEN cells (Fig. 2B). mESCs express the pluripotency-associated genes Nanog, Pou5f1, Sox2, Utf1, Zfp42, Eras, Dppa5, Dnmt3l, Col18a1, Nodal and Gli2, which is in contrast to cXEN and embryo-derived XEN cells (Mitsui et al., 2003). Our results confirm previous observations that Sall4 is expressed in both mESCs and XEN cell lines (Lim et al., 2008). cXEN and embryo-derived XEN cells similarly express Gata6, Sox7 and Sox17 (Fig. 2B). Moreover, cXEN and embryo-derived XEN cells similarly express genes associated with parietal endoderm (Fst, Snai1, Pth1r, Pdgfra, Lamb1-1 and Sparc) and visceral endoderm (Cited1, Dab2, Emp2, Fxyd3, Atp6v0a1, Elf1, Foxq1, Col4a1, Lama1, Ttr and Stra6) (Kunath et al., 2005; Brown et al., 2010). Pearson correlation analysis confirms that cXEN and embryo-derived XEN cells have stronger correlation (R2 value=0.97±0.00) than the correlation between either mESCs and cXEN or embryo-derived XEN cells (R2 value=0.90±0.00) (supplementary material Fig. S3).
We further validated the microarray data by performing qRT-PCR analysis on candidate genes and comparing the mRNA expression of cXEN to embryo-derived XEN cells and mESCs (Fig. 2C,D). We confirm that cXEN cells robustly express Gata4, Gata6, Sox7 and Sox17 as well as genes encoding ExEn-associated cell surface proteins or basement membrane components Pdgfra, Col4a1, Lama1, Lamb1, Sparc, Plat, Dkk1 and Dab2, which is in contrast to mESCs that express these genes at a low level or lack expression altogether (Fig. 2C). Furthermore, we confirm that cXEN and embryo-derived XEN cells lack the expression of Nanog, Pou5f1 and Sox2 (Fig. 2D).
To determine whether cXEN cells are functionally equivalent to embryo-derived XEN cells, we established cXEN cells from mESCs that have a constitutively active promoter driving a gene encoding a red fluorescent protein. Single UbiC::dTomato cXEN cells were plated with wild-type mESCs to generate embryoid bodies (EBs) (supplementary material Fig. S4, Table S10). By comparison, we also plated single UbiC::dTomato mESCs or CAG::GFP embryo-derived XEN cells as a negative or positive control, respectively. We performed confocal imaging to determine the contribution of cXEN, XEN or mES cells. As expected, XEN and cXEN cells were restricted to the periphery of EBs, consistent with their commitment to an ExEn lineage. By contrast, mESCs contributed primarily to cells within the centre of EBs and were also observed peripherally. Finally, to determine whether cXEN cells proliferate at the same rate as embryo-derived XEN cells we performed an MTT assay for up to 5 days in standard XEN media (Fig. 2E) and show that cXEN cells do indeed proliferate equivalently to embryo-derived XEN cells. In all, this demonstrates that cXEN cells are functionally and molecularly equivalent to embryo-derived XEN cells and are distinct from the mESCs from which they were derived.
cXEN derivation requires expression of Gata6, Gata4, Sox17 and Pdgfra and downregulation of Nanog
In PrE development, Gata6 is downstream of Grb2, an effector of the FGF-signalling pathway, and upstream of Gata4, Pdgfra and Sox17 (Chazaud et al., 2006; Artus et al., 2011; Plusa et al., 2008; Artus et al., 2010; Niakan et al., 2010). Although PrE cells are established in Gata6 mutant embryos, their further ExEn differentiation is compromised (Morrisey et al., 1998; Koutsourakis et al., 1999). Gata6–/– mESCs lack expression of Gata4, Pdgfra and Sox17, and fail to establish VE or PE cells in EB assays (Morrisey et al., 2000; Capo-Chichi et al., 2005). We find that Gata6–/– mESCs can undergo differentiation in cXEN derivation conditions, suggesting that they are not blocked from exiting mESC self-renewal (Fig. 3A). We performed immunofluorescence analysis for Sox7 and Nanog to quantify the proportion of XEN- or mESC-like cells, respectively (Fig. 3B). No Sox7-expressing XEN-like cells emerged from the Gata6–/– mESCs and we observed few Nanog-expressing cells after prolonged culture in XEN media (Fig. 3B). These results confirm that Gata6 is functionally required for cXEN establishment, thereby recapitulating the role of this gene in ExEn development.
Fig. 3.
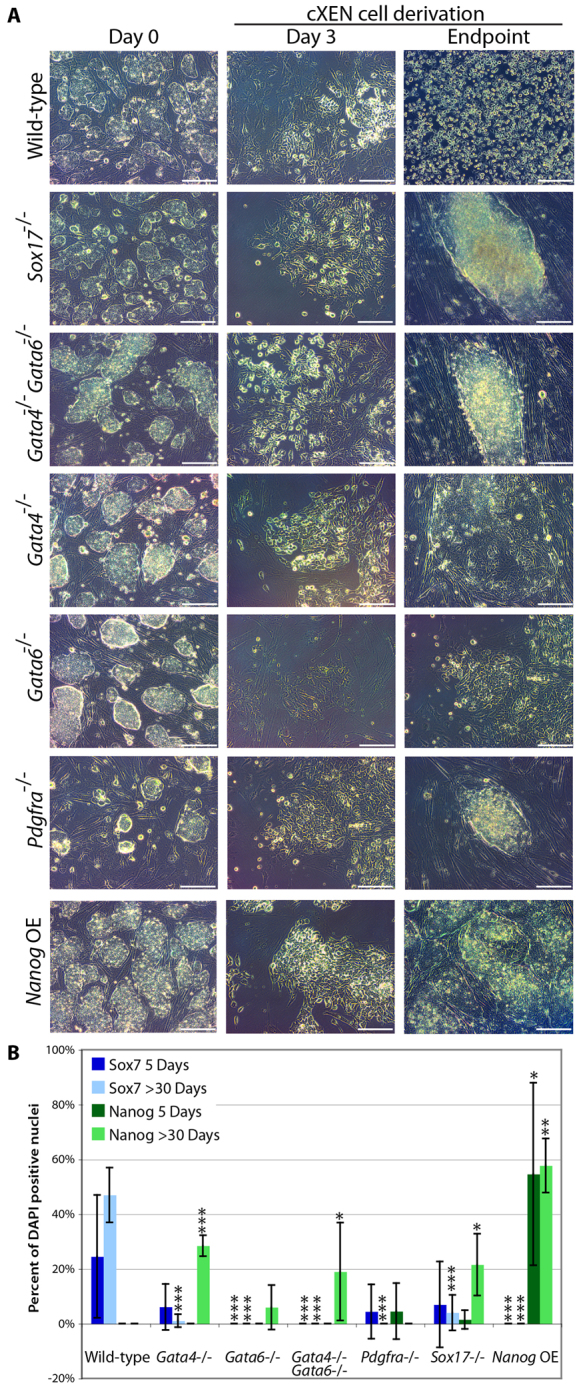
Gata6, Gata4, Sox17 and Pdgfra are required for cXEN establishment. (A) mESCs mutant for Sox17, Gata4, Gata6, Pdgfra or Gata4 and Gata6 double-mutant cells were subjected to the cXEN protocol along with wild-type or Nanog overexpressing (OE) mESCs. Representative phase-contrast images were taken at defined time points during the course of derivation [day 0 in pluripotency conditions, day 3 prior to the first passage and the endpoint: after prolonged culture (day 30)]. Scale bars: 100 μm. (B) Quantification of Sox7 and Nanog expression following immunofluorescence analysis at 5 days and 30 days after cXEN derivation. Data are mean±s.e.m. of five separate counts. *P<0.05; **P<0.01; ***P<0.001.
In mouse embryos, mutation of either Gata4, Pdgfra or Sox17 has no effect on PrE initiation, which probably reflects their in vivo genetic redundancy in this cell fate decision (Kuo et al., 1997; Narita et al., 1997; Kanai-Azuma et al., 2002; Shimoda et al., 2007; Artus et al., 2011; Artus et al., 2010). However, in embryos undergoing delayed implantation, knockout of Pdgfra–/– or Sox17–/– reduces PrE cell numbers (Artus et al., 2011; Artus et al., 2010) and blocks XEN cell derivation (Artus et al., 2010; Niakan et al., 2010). Moreover, Gata4–/– mESCs cannot spontaneously form VE or PE cells in EBs (Soudais et al., 1995), and the mutation of either Pdgfra or Sox17 leads to mis-expression of ExEn-associated genes and causes defects in XEN maintenance (Shimoda et al., 2007; Artus et al., 2010; Niakan et al., 2010).
A small proportion of Sox7-expressing XEN-like cells emerged from the Gata4–/–, Pdgfra–/– and Sox17–/– mESCs during the cXEN protocol; however, these cells could not be expanded or maintained, confirming previous observations that these genes are required for XEN derivation (Fig. 3A,B). This suggests that there may be compensatory mechanisms accounting for the ability of XEN cells to emerge, albeit rarely, from these mutant backgrounds. Intriguingly, we observed Nanog expression in the mutant cell lines after prolonged culture in the absence in LIF, suggesting that Gata4, Pdgfra and Sox17 expression antagonizes the expression of a key pluripotency-associated gene.
Nanog overexpression has been demonstrated to block differentiation of mESCs and can maintain stem cell self-renewal in the absence of LIF (Chambers et al., 2003). As expected, the overexpression of Nanog alone is sufficient to block Sox7 expression and cXEN derivation from mESCs, demonstrating that the downregulation of Nanog is required for XEN derivation (Fig. 3B). In all, these results suggest that cXEN cells require the same molecular mechanism for their derivation as embryo-derived XEN cells.
cXEN cells undergo VE differentiation in response to BMP treatment
Recently, a subtype of VE cells has been differentiated from embryo-derived XEN cells treated with BMP4, demonstrating the plasticity of XEN cells (Paca et al., 2012; Artus et al., 2012). We therefore asked whether cXEN cells could also differentiate into VE cells in response to BMP4. We established cXEN cells from an α-fetoprotein (Afp) Afp::GFPTg/+ transgenic reporter cell line, as the expression of this gene distinguishes VE cells (Kwon et al., 2006) (supplementary material Fig. S5). We found small patches of Afp::GFP-high-expressing cells in the course of cXEN cell establishment, but once stable cXEN cell lines were derived there was no appreciable expression of the transgene. Treatment with 50 ng/ml of BMP4 in standard XEN media resulted in the upregulation of Afp::GFP-high expression in the cells which grew as epithelialized colonies (supplementary material Fig. S6). To enrich for the Afp-high population, we manually picked colonies for further expansion followed by qRT-PCR analysis for the expression of VE-associated genes: Afp, ApoE, Cited1 and Ihh, which are upregulated 1.9-fold or more in the BMP4-treated cXEN cells (supplementary material Fig. S6). The expression of endoderm transcription factors is not upregulated compared with untreated controls, with the exception of Gata4, which is over twofold enriched, confirming previous observations (Artus et al., 2012). Taken together, this demonstrates that cXEN cells, like embryo-derived XEN cells, have the capacity to differentiate into VE cells in response to BMP4 treatment.
mESCs from 2i and LIF conditions can be converted to cXEN cells
We have previously demonstrated that mESC cultures maintained in serum and LIF spontaneously undergo differentiation at a low frequency (1.9%) into XEN-like cells (Niakan et al., 2010) (supplementary material Fig. S1). We next asked whether XEN cells also arise from pluripotent cells grown in 2i and LIF (Ying et al., 2008). To address this, we cultured mESCs in 2i and LIF conditions for eight passages (supplementary material Fig. S7). As expected, mESCs in 2i and LIF grew as uniform dome-shaped colonies on gelatin-coated dishes and were positive for alkaline phosphatase activity. Using qRT-PCR, we confirmed that these cells express Nanog, Oct4 and Sox2 (supplementary material Fig. S7).
We performed immunofluorescence analysis for Oct4 as well as for Sox17, Gata4, laminin and Dab2 to distinguish whether XEN-like cells emerge under distinct pluripotency conditions (Fig. 4). As previously observed (Niakan et al., 2010), cells maintained in serum and LIF have Sox17-high expression within a subset of cells (Fig. 4). Sox17-high cells within serum and LIF lacked the expression of Oct4 and had nuclear colocalized Gata4 and Sox17, as well as cell surface-localized laminin and Dab2 expression. mESCs in 2i and LIF robustly express Oct4 in all of the colonies, confirming that in these defined culture conditions, pluripotency transcription factors are more homogeneously expressed (Ying et al., 2008). However, mESCs maintained in 2i and LIF, lacked the expression of Sox17, Gata4 and Dab2, suggesting that XEN-like cells are either absent in these conditions or below the detection of our analysis. To further test whether XEN-like cells emerge in 2i and LIF, we established Sox17GFP/+ mESCs in this condition and performed flow cytometry after staining for SSEA1 (supplementary material Fig. S1). We find that, compared with mESCs in serum and LIF, cells cultured in 2i and LIF express SSEA1 in a slightly greater proportion (94.5% compared with 92.3%) of cells. Importantly, in 2i and LIF conditions, no GFPhigh-expressing cells were observed.
Fig. 4.
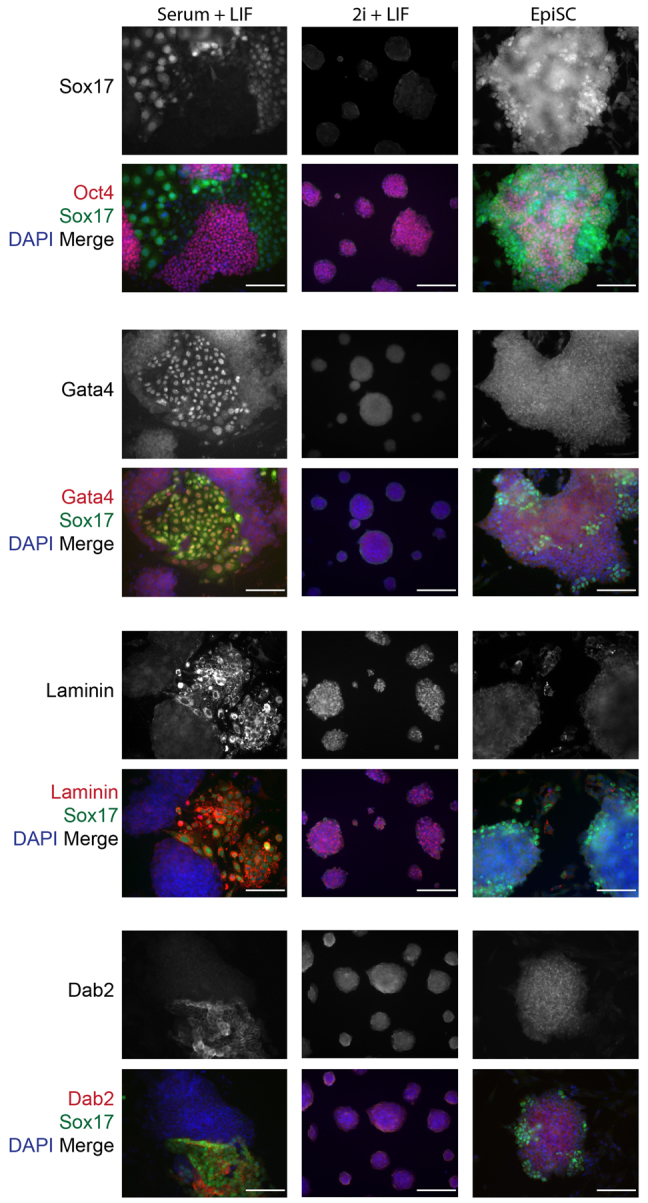
XEN-like cells are present in mESC cultures in serum and LIF. Immunofluorescence analysis of Sox17 expression: in mESCs maintained in serum and LIF; in mESC maintained in Erk and Gsk3 inhibitors, and LIF (2i+LIF); and in EpiSC maintained in the presence of Fgf2 and activin. Sox17-high-expressing cells were analyzed for colocalization with Oct4, Gata4, laminin or Dab2 (red) expression along with DAPI (blue) merge. Scale bars: 100 μm.
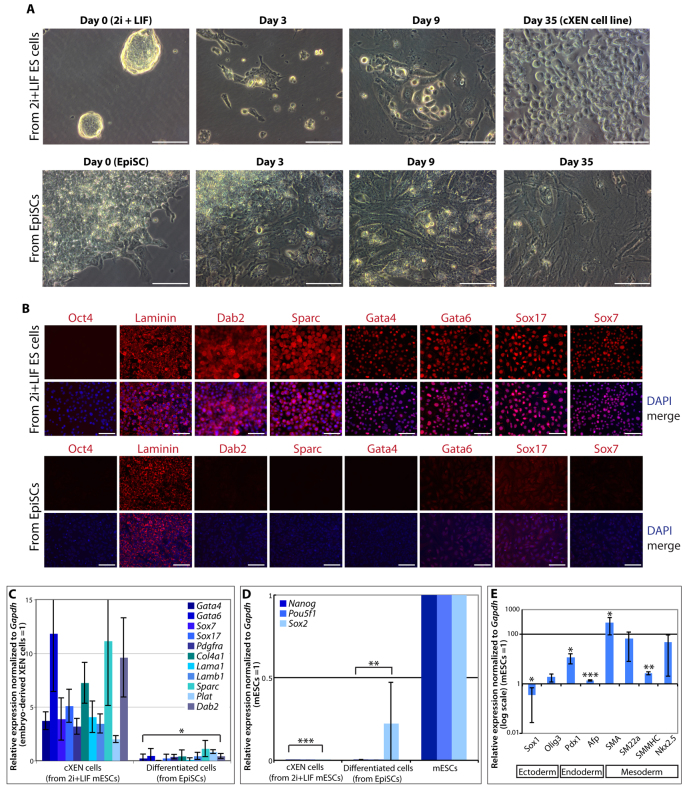
We next asked whether mESCs grown in 2i and LIF can give rise to cXEN cells. We repeated the cXEN protocol and found that mESCs from 2i and LIF conditions can give rise to cXEN cells, similar to mESCs from serum and LIF (Fig. 5A,B). Furthermore, cXEN cells derived from mESCs in 2i and LIF lacked the expression of Oct4 and robustly expressed laminin, Dab2, Sparc, Gata4, Gata6, Sox17 and Sox7 by immunofluorescence analysis (Fig. 5C). We confirmed by qRT-PCR analysis that the cXEN cells derived from mESCs in 2i and LIF also expressed Gata4, Gata6, Pdgfra, Sox7, Sox17, Col4a1, Lama1, Lamb1, Sparc, Plat and Dab2 at a comparable level with embryo-derived XEN cells (Fig. 5D). Importantly, these cXEN cells lacked the expression of Pou5f1, Nanog and Sox2 (Fig. 5E). In all, this demonstrates that mESCs from either serum or 2i and LIF are equivalently responsive to XEN-promoting signals.
Fig. 5.
mESCs from 2i and LIF conditions can give rise to cXEN cells, unlike EpiSCs. (A) Representative phase-contrast images of mESCs from 2i and LIF conditions or EpiSCs at defined time-points during the cXEN protocol (day 0 in pluripotency conditions, day 3 prior to the first passage, day 9 after the second passage and day 35) (representative image of three biological replicates of stem cell lines from 2i and LIF conditions or two biological and six technical replicates of EpiSCs). Scale bars: 50 μm. (B) Immunofluorescence analysis of cXEN cells from 2i and LIF or differentiated cells from EpiSCs at day 35 of the protocol. Cells were immunostained and analyzed for the expression of Oct4, laminin, Dab2, Sparc, Gata4, Gata6, Sox17 and Sox7 expression (red), and DAPI (blue) merge. Scale bars: 100 μm. (C,D) qRT-PCR analysis of cXEN cells from 2i and LIF conditions or differentiated cells from EpiSCs for the expression of XEN-associated genes (C) or pluripotency-associated genes (D). Relative expression reflected as a fold difference compared with embryo-derived XEN or mESCs (=1), respectively. Data are the mean±s.e.m. Three biological replicates (from 2i and LIF) or two biological and three technical replicates (from EpiSCs). (E) qRT-PCR analysis of differentiated cells from EpiSCs. Relative expression reflected as a fold difference (log scale) compared with mESCs (=1) for germ cell lineage-associated genes: Sox1 and Olig3 (ectoderm); Pdx1 and Afp (endoderm); Sma, Sm22a, Smmhc and Nkx2.5 (mesoderm). Data are the mean±s.e.m. Two biological and three technical replicates. *P<0.05; **P<0.01; ***P<0.001.
Different pluripotent stem cell states have distinct ExEn differentiation capacities
We next asked whether EpiSCs are equivalently responsive to our cXEN protocol. We cultured EpiSCs as previously described; cells grew as monolayer colonies and, characteristically, lacked the expression of alkaline phosphatase (Brons et al., 2007) (supplementary material Fig. S7). qRT-PCR analysis confirmed that EpiSCs uniquely express Fgf5 and have lower expression of Nanog compared with mESCs in serum or 2i and LIF (supplementary material Fig. S7).
EpiSCs give rise to Sox7- and Cdx2-expressing cells (Brons et al., 2007), which have been suggested to reflect extra-embryonic XEN or trophoblast stem-like cells and a possible reversion of developmental commitment. We next asked whether XEN-like cells exist within EpiSC cultures. Like mESCs, EpiSC cultures contained Sox17-high-expressing cells that colocalized with the expression of Gata4 (Fig. 4). Sox17-high-expressing cells lacked the expression of Oct4, suggesting that they are either poised or committed to differentiate. However, unlike mESCs, the Sox17-high-expressing cells within the EpiSC cultures do not overlap with the expression of Sox7, Dab2 and laminin. Altogether, this suggests that the heterogeneity within EpiSC cultures is distinct and that the cells within EpiSC cultures may be more reminiscent of germ layer lineages such as definitive endoderm (Sox17, Gata4, Gata6) or mesoderm (Sox7) (Gandillet et al., 2009), which would be consistent with the epiblast origin from which EpiSCs are derived.
Intriguingly, the cXEN protocol results in the differentiation of EpiSCs into cells that lacked the morphology of XEN cells (Fig. 5A,B). By qRT-PCR analysis we confirm that the EpiSC differentiated cells downregulated the expression of Pou5f1, Nanog and Sox2 (Fig. 5E). Significantly, with the exception of laminin, these cells lacked the expression of ExEn-associated proteins (Fig. 5C). Moreover, cells differentiated from EpiSCs lacked the expression of Gata4, Sox7, Sox17, Pdgfra, Lama1, Lamb1 and Dab2, and have low to moderate expression of Gata6, Col4a1, Sparc and Plat (Fig. 5D). We next performed qRT-PCR analysis for genes associated with later stages of germ cell lineages (ectoderm, endoderm or mesoderm), as the EpiSCs we used for this analysis had been maintained in cXEN derivation conditions for 1 month. We used the Sox factor Sox1 (Wood and Episkopou, 1999) and the oligodendrocyte factor Olig3 (Muller et al., 2005) as markers of ectoderm neural differentiation; the pancreatic and duodenal homeobox 1 gene Pdx1 (Jonsson et al., 1994) and Afp (Kuhlmann, 1979) as markers of endoderm pancreatic or liver differentiation; smooth muscle actin (Sma) (Shimizu et al., 1995), smooth muscle 22 alpha (Sm22a) (Li et al., 1996) and smooth muscle myosin heavy chain (Smmhc) (Watanabe et al., 1996) as markers of mesoderm smooth muscle differentiation; and the homeobox-containing gene Nkx2.5 (Lints et al., 1993) as a marker of cardiac mesoderm (Fig. 5F). The cells differentiated from EpiSCs in cXEN conditions robustly express Sma (287.9±192.5-fold upregulation), Nkx2.5 (49.0±47-fold upregulation) and Sm22a (67.7±59.6-fold upregulation), and have low-moderate Smmhc expression (2.7±0.4-fold upregulation). The expression of Pdx1 (11.4±5.0-fold upregulation) suggests that there is some heterogeneity within these differentiation conditions and the large standard deviation for gene expression points to the variability in differentiation efficiency between biological replicates of EpiSC lines (Fig. 5F). As the cells differentiated from EpiSCs lack the expression of Sox1, Olig3 and Afp, this suggests that neuroectoderm and liver differentiation may be blocked under these conditions (Fig. 5F). Altogether, these results demonstrate that EpiSCs, in contrast to mESCs, are blocked from cXEN differentiation and may be refractory to extra-embryonic differentiation.
DISCUSSION
mESC contribution towards extra-embryonic lineages in chimera experiments has been noted (Beddington and Robertson, 1989) and, in conjunction with a growing body of work on mESC heterogeneity, suggests that mESCs can give rise to both epiblast progenitor and ExEn-like cells. We have previously demonstrated that Sox17-high expression in cells within mESC cultures distinguishes XEN-like cells (Niakan et al., 2010). However, it was unclear whether self-renewing XEN cell lines could be established directly from mESCs without genetic manipulation. Here, we describe an approach for directing the differentiation of mESCs to cXEN cells, which are identical to XEN cells derived from the PrE of pre-implantation embryos. Interestingly, mESCs may also generate trophoblast-like cells without genetic manipulation (Schenke-Layland et al., 2007; He et al., 2008). Altogether, our work further suggests that mESCs have a greater differentiation capacity than has been previously appreciated.
The absence of XEN-like cells in 2i and LIF culture conditions suggests that our cXEN protocol probably reflects both a differentiation and selection process in which a low dose of RA and activin limits the range of differentiated cells types that can emerge from mESCs. One of these differentiated cell types are XEN cells that can be enriched and expanded in conditions that favour their proliferative advantage. Based on our observations, we present a model (Fig. 6) in which we suggest that mESCs exit pluripotency and transit through a pre-XEN state where key pluripotency genes are downregulated and ExEn-associated genes are upregulated. Eventually, in conditions that favour XEN expansion, we select and grow stable XEN stem cell lines. By contrast, EpiSCs represent a distinct pluripotent stem cell state unable to undergo XEN conversion, suggesting that these cells do not reverse their developmental commitment. The hypothesis that EpiSCs are limited in their differentiation potential to embryonic germ layer lineages is supported by recent observations that EpiSCs, like hESCs, undergo extra-embryonic mesoderm rather than trophoblast differentiation in the presence of BMP4 (Bernardo et al., 2011). It will be interesting to determine whether, like EpiSCs, hESCs are refractory to XEN differentiation.
Fig. 6.
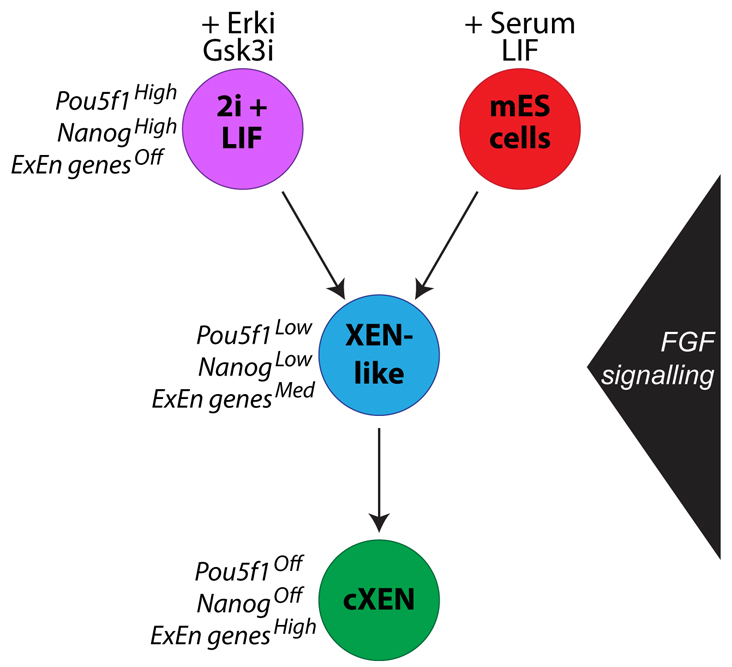
A schematic model of cXEN cell derivation from mESCs. mESCs maintained in LIF and either 2i (purple) or serum (red) can give rise to cXEN cells. Our data suggest that, during cXEN derivation, cells transit through a XEN-like cell state (blue), whereby pluripotency genes are downregulated and extra-embryonic endoderm (ExEn) genes are upregulated. FGF signalling is required to exit mESC self-renewal but not for cXEN maintenance (green).
Our FGF/ERK signalling results are consistent with previous studies that have demonstrated that VE and PE differentiation in EBs is blocked in the presence of FGFR inhibitors or in mESCs that have a dominant-negative mutation of FGFR2 (Chen et al., 2000; Li et al., 2001; Li et al., 2004; Hamazaki et al., 2006). Furthermore, our results confirm previous studies suggesting that FGF signalling is required to exit mESC self-renewal (Kunath et al., 2007; Stavridis et al., 2007). In all, this suggests that FGF/ERK signalling is also required for mESC differentiation to cXEN cells. Intriguingly, cXEN derivation demonstrates that exogenous FGF is not required unless endogenous Fgf4 is compromised. In conclusion, a notable advantage to the cXEN protocol is its potential application to a wide selection of mutant mESCs, representing a novel approach to the genetic study of ExEn development.
Supplementary Material
Acknowledgments
We thank Stephen Duncan for Gata4–/–, Gata6–/–, and Gata4 and Gata6 double-mutant ESCs; Sean Morrison for Sox17GFP/+ reporter ESCs; Austin Smith for Nanog overexpressing ESCs; Janet Rossant for embryo-derived XEN cell lines. We thank Marko Hyvonen for recombinant LIF, activin A, BMP4 and FGF2 proteins. We thank Developmental Studies Hybridoma Bank for the SSEA1 antibody. We thank the Cambridge Genomic Service for technical assistance.
Footnotes
Funding
This work is supported by the Human Frontier Science Program, National Institutes of Health (NIH) [RO1-HD052115 and RO1-DK084391 to A.K.H.]; the New York State Department of Health [NYSTEM IDEA grant N08G-175 to A.K.H.); and the March of Dimes Foundation (K.K.N. and R.A.P.). L.T.Y.C. is supported by a MRC Capacity-Building studentship. K.K.N. is supported by a Centre for Trophoblast Research Next Generation Fellowship. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.078519/-/DC1
References
- Arman E., Haffner-Krausz R., Chen Y., Heath J. K., Lonai P. (1998). Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl. Acad. Sci. USA 95, 5082–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J., Panthier J. J., Hadjantonakis A. K. (2010). A role for PDGF signaling in expansion of the extra-embryonic endoderm lineage of the mouse blastocyst. Development 137, 3361–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J., Piliszek A., Hadjantonakis A. K. (2011). The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev. Biol. 350, 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artus J., Douvaras P., Piliszek A., Isern J., Baron M. H., Hadjantonakis A. K. (2012). BMP4 signaling directs primitive endoderm-derived XEN cells to an extraembryonic visceral endoderm identity. Dev. Biol. 361, 245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003). Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007). The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beddington R. S., Robertson E. J. (1989). An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 105, 733–737 [DOI] [PubMed] [Google Scholar]
- Bernardo A. S., Faial T., Gardner L., Niakan K. K., Ortmann D., Senner C. E., Callery E. M., Trotter M. W., Hemberger M., Smith J. C., et al. (2011). BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibel M., Richter J., Schrenk K., Tucker K. L., Staiger V., Korte M., Goetz M., Barde Y. A. (2004). Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat. Neurosci. 7, 1003–1009 [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- Bradley A., Robertson E. (1986). Embryo-derived stem cells: a tool for elucidating the developmental genetics of the mouse. Curr. Top. Dev. Biol. 20, 357–371 [DOI] [PubMed] [Google Scholar]
- Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- Brown K., Doss M. X., Legros S., Artus J., Hadjantonakis A. K., Foley A. C. (2010). eXtraembryonic ENdoderm (XEN) stem cells produce factors that activate heart formation. PLoS ONE 5, e13446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canham M. A., Sharov A. A., Ko M. S., Brickman J. M. (2010). Functional heterogeneity of embryonic stem cells revealed through translational amplification of an early endodermal transcript. PLoS Biol. 8, e1000379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi C. D., Rula M. E., Smedberg J. L., Vanderveer L., Parmacek M. S., Morrisey E. E., Godwin A. K., Xu X. X. (2005). Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev. Biol. 286, 574–586 [DOI] [PubMed] [Google Scholar]
- Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003). Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 [DOI] [PubMed] [Google Scholar]
- Chazaud C., Yamanaka Y., Pawson T., Rossant J. (2006). Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 10, 615–624 [DOI] [PubMed] [Google Scholar]
- Chen Y., Li X., Eswarakumar V. P., Seger R., Lonai P. (2000). Fibroblast growth factor (FGF) signaling through PI 3-kinase and Akt/PKB is required for embryoid body differentiation. Oncogene 19, 3750–3756 [DOI] [PubMed] [Google Scholar]
- Coucouvanis E., Martin G. R. (1995). Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell 83, 279–287 [DOI] [PubMed] [Google Scholar]
- Doetschman T. C., Eistetter H., Katz M., Schmidt W., Kemler R. (1985). The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 87, 27–45 [PubMed] [Google Scholar]
- Evans M. J., Kaufman M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- Feldman B., Poueymirou W., Papaioannou V. E., DeChiara T. M., Goldfarb M. (1995). Requirement of FGF-4 for postimplantation mouse development. Science 267, 246–249 [DOI] [PubMed] [Google Scholar]
- Frankenberg S., Gerbe F., Bessonnard S., Belville C., Pouchin P., Bardot O., Chazaud C. (2011). Primitive endoderm differentiates via a three-step mechanism involving Nanog and RTK signaling. Dev. Cell 21, 1005–1013 [DOI] [PubMed] [Google Scholar]
- Fujikura J., Yamato E., Yonemura S., Hosoda K., Masui S., Nakao K., Miyazaki J.-i., Niwa H. (2002). Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 16, 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandillet A., Serrano A. G., Pearson S., Lie A. L. M., Lacaud G., Kouskoff V. (2009). Sox7-sustained expression alters the balance between proliferation and differentiation of hematopoietic progenitors at the onset of blood specification. Blood 114, 4813–4822 [DOI] [PubMed] [Google Scholar]
- Gardner R. L. (1985). Clonal analysis of early mammalian development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 312, 163–178 [DOI] [PubMed] [Google Scholar]
- Gardner R. L., Papaioannou V. E. (1975). Differentiation in the trophectoderm and the inner cell mass. In The Early Development of Mammals: (ed. Balls M., Wild A. E.), pp. 107–132 Cambridge: Cambridge University Press; [Google Scholar]
- Gardner R. L., Rossant J. (1979). Investigation of the fate of 4-5 day post-coitum mouse inner cell mass cells by blastocyst injection. J. Embryol. Exp. Morphol. 52, 141–152 [PubMed] [Google Scholar]
- Goldin S. N., Papaioannou V. E. (2003). Paracrine action of FGF4 during periimplantation development maintains trophectoderm and primitive endoderm. Genesis 36, 40–47 [DOI] [PubMed] [Google Scholar]
- Guo G., Huss M., Tong G. Q., Wang C., Li Sun L., Clarke N. D., Robson P. (2010). Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685 [DOI] [PubMed] [Google Scholar]
- Hamazaki T., Kehoe S. M., Nakano T., Terada N. (2006). The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol. Cell. Biol. 26, 7539–7549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Pant D., Schiffmacher A., Meece A., Keefer C. L. (2008). Lymphoid enhancer factor 1-mediated Wnt signaling promotes the initiation of trophoblast lineage differentiation in mouse embryonic stem cells. Stem Cells 26, 842–849 [DOI] [PubMed] [Google Scholar]
- Jonsson J., Carlsson L., Edlund T., Edlund H. (1994). Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371, 606–609 [DOI] [PubMed] [Google Scholar]
- Kalmar T., Lim C., Hayward P., Munoz-Descalzo S., Nichols J., Garcia-Ojalvo J., Martinez Arias A. (2009). Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 7, e1000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai-Azuma M., Kanai Y., Gad J. M., Tajima Y., Taya C., Kurohmaru M., Sanai Y., Yonekawa H., Yazaki K., Tam P. P., et al. (2002). Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129, 2367–2379 [DOI] [PubMed] [Google Scholar]
- Kim I., Saunders T. L., Morrison S. J. (2007). Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell 130, 470–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsourakis M., Langeveld A., Patient R., Beddington R., Grosveld F. (1999). The transcription factor GATA6 is essential for early extraembryonic development. Development 126, 723–732 [PubMed] [Google Scholar]
- Kubo A., Shinozaki K., Shannon J. M., Kouskoff V., Kennedy M., Woo S., Fehling H. J., Keller G. (2004). Development of definitive endoderm from embryonic stem cells in culture. Development 131, 1651–1662 [DOI] [PubMed] [Google Scholar]
- Kuhlmann W. D. (1979). Immunoperoxidase labelling of alpha 1-fetoprotein (AFP) in normal and regenerating livers of a low and a high AFP producing mouse strain. Histochemistry 64, 67–75 [DOI] [PubMed] [Google Scholar]
- Kunath T., Arnaud D., Uy G. D., Okamoto I., Chureau C., Yamanaka Y., Heard E., Gardner R. L., Avner P., Rossant J. (2005). Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development 132, 1649–1661 [DOI] [PubMed] [Google Scholar]
- Kunath T., Saba-El-Leil M. K., Almousailleakh M., Wray J., Meloche S., Smith A. (2007). FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134, 2895–2902 [DOI] [PubMed] [Google Scholar]
- Kuo C. T., Morrisey E. E., Anandappa R., Sigrist K., Lu M. M., Parmacek M. S., Soudais C., Leiden J. M. (1997). GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11, 1048–1160 [DOI] [PubMed] [Google Scholar]
- Kwon G. S., Fraser S. T., Eakin G. S., Mangano M., Isern J., Sahr K. E., Hadjantonakis A. K., Baron M. H. (2006). Tg(Afp-GFP) expression marks primitive and definitive endoderm lineages during mouse development. Dev. Dyn. 235, 2549–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanner F., Lee K. L., Sohl M., Holmborn K., Yang H., Wilbertz J., Poellinger L., Rossant J., Farnebo F. (2010). Heparan sulfation-dependent fibroblast growth factor signaling maintains embryonic stem cells primed for differentiation in a heterogeneous state. Stem Cells 28, 191–200 [DOI] [PubMed] [Google Scholar]
- Li L., Miano J. M., Cserjesi P., Olson E. N. (1996). SM22 alpha, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ. Res. 78, 188–195 [DOI] [PubMed] [Google Scholar]
- Li L., Arman E., Ekblom P., Edgar D., Murray P., Lonai P. (2004). Distinct GATA6- and laminin-dependent mechanisms regulate endodermal and ectodermal embryonic stem cell fates. Development 131, 5277–5286 [DOI] [PubMed] [Google Scholar]
- Li X., Chen Y., Scheele S., Arman E., Haffner-Krausz R., Ekblom P., Lonai P. (2001). Fibroblast growth factor signaling and basement membrane assembly are connected during epithelial morphogenesis of the embryoid body. J. Cell Biol. 153, 811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C. Y., Tam W. L., Zhang J., Ang H. S., Jia H., Lipovich L., Ng H. H., Wei C. L., Sung W. K., Robson P., et al. (2008). Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell 3, 543–554 [DOI] [PubMed] [Google Scholar]
- Lints T. J., Parsons L. M., Hartley L., Lyons I., Harvey R. P. (1993). Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119, 419–431 [DOI] [PubMed] [Google Scholar]
- Martin G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnard D., Guzman-Ayala M., Constam D. B. (2006). Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development 133, 2497–2505 [DOI] [PubMed] [Google Scholar]
- Messerschmidt D. M., Kemler R. (2010). Nanog is required for primitive endoderm formation through a non-cell autonomous mechanism. Dev. Biol. 344, 129–137 [DOI] [PubMed] [Google Scholar]
- Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003). The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- Mohammadi M., McMahon G., Sun L., Tang C., Hirth P., Yeh B. K., Hubbard S. R., Schlessinger J. (1997). Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955–960 [DOI] [PubMed] [Google Scholar]
- Mohammadi M., Froum S., Hamby J. M., Schroeder M. C., Panek R. L., Lu G. H., Eliseenkova A. V., Green D, Schlessinger J., Hubbard S. R. (1998). Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 17, 5896–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J. D., Lin Q., Duncan S. A., Olson E. N. (1997). Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11, 1061–1072 [DOI] [PubMed] [Google Scholar]
- Morrisey E. E., Tang Z., Sigrist K., Lu M. M., Jiang F., Ip H. S., Parmacek M. S. (1998). GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 12, 3579–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey E. E., Musco S., Chen M. Y., Lu M. M., Leiden J. M., Parmacek M. S. (2000). The gene encoding the mitogen-responsive phosphoprotein Dab2 is differentially regulated by GATA-6 and GATA-4 in the visceral endoderm. J. Biol. Chem. 275, 19949–19954 [DOI] [PubMed] [Google Scholar]
- Muller T., Anlag K., Wildner H., Britsch S., Treier M., Birchmeier C. (2005). The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes Dev. 19, 733–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita N., Bielinska M., Wilson D. B. (1997). Wild-type endoderm abrogates the ventral developmental defects associated with GATA-4 deficiency in the mouse. Dev. Biol. 189, 270–274 [DOI] [PubMed] [Google Scholar]
- Niakan K. K., Ji H., Maehr R., Vokes S. A., Rodolfa K. T., Sherwood R. I., Yamaki M., Dimos J. T., Chen A. E., Melton D. A., et al. (2010). Sox17 promotes differentiation in mouse embryonic stem cells by directly regulating extraembryonic gene expression and indirectly antagonizing self-renewal. Genes Dev. 24, 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A. (1998). Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. (2009). Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136, 3215–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. (2005). Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929 [DOI] [PubMed] [Google Scholar]
- Paca A., Seguin C. A., Clements M., Ryczko M., Rossant J., Rodriguez T. A., Kunath T. (2012). BMP signaling induces visceral endoderm differentiation of XEN cells and parietal endoderm. Dev. Biol. 361, 90–102 [DOI] [PubMed] [Google Scholar]
- Plusa B., Piliszek A., Frankenberg S., Artus J., Hadjantonakis A. K. (2008). Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development 135, 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappolee D. A., Basilico C., Patel Y., Werb Z. (1994). Expression and function of FGF-4 in peri-implantation development in mouse embryos. Development 120, 2259–2269 [DOI] [PubMed] [Google Scholar]
- Schenke-Layland K., Angelis E., Rhodes K. E., Heydarkhan-Hagvall S., Mikkola H. K., Maclellan W. R. (2007). Collagen IV induces trophoectoderm differentiation of mouse embryonic stem cells. Stem Cells 25, 1529–1538 [DOI] [PubMed] [Google Scholar]
- Shimizu R. T., Blank R. S., Jervis R., Lawrenz-Smith S. C., Owens G. K. (1995). The smooth muscle alpha-actin gene promoter is differentially regulated in smooth muscle versus non-smooth muscle cells. J. Biol. Chem. 270, 7631–7643 [DOI] [PubMed] [Google Scholar]
- Shimoda M., Kanai-Azuma M., Hara K., Miyazaki S., Kanai Y., Monden M., Miyazaki J. (2007). Sox17 plays a substantial role in late-stage differentiation of the extraembryonic endoderm in vitro. J. Cell Sci. 120, 3859–3869 [DOI] [PubMed] [Google Scholar]
- Shimosato D., Shiki M., Niwa H. (2007). Extra-embryonic endoderm cells derived from ES cells induced by GATA factors acquire the character of XEN cells. BMC Dev. Biol. 7, 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solter D., Knowles B. B. (1978). Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1). Proc. Natl. Acad. Sci. USA 75, 5565–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano D. R., Teets B. W., Soprano K. J. (2007). Role of retinoic acid in the differentiation of embryonal carcinoma and embryonic stem cells. Vitam. Horm. 75, 69–95 [DOI] [PubMed] [Google Scholar]
- Soudais C., Bielinska M., Heikinheimo M., MacArthur C. A., Narita N., Saffitz J. E., Simon M. C., Leiden J. M., Wilson D. B. (1995). Targeted mutagenesis of the transcription factor GATA-4 gene in mouse embryonic stem cells disrupts visceral endoderm differentiation in vitro. Development 121, 3877–3888 [DOI] [PubMed] [Google Scholar]
- Stavridis M. P., Lunn J. S., Collins B. J., Storey K. G. (2007). A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development 134, 2889–2894 [DOI] [PubMed] [Google Scholar]
- Strickland S., Smith K. K., Marotti K. R. (1980). Hormonal induction of differentiation in teratocarcinoma stem cells: generation of parietal endoderm by retinoic acid and dibutyryl cAMP. Cell 21, 347–355 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Kunath T., Hadjantonakis A. K., Nagy A., Rossant J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 [DOI] [PubMed] [Google Scholar]
- Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., McKay R. D. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 [DOI] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. (2008). Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135, 909–918 [DOI] [PubMed] [Google Scholar]
- Vallier L., Alexander M., Pedersen R. A. (2005). Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118, 4495–4509 [DOI] [PubMed] [Google Scholar]
- Vallier L., Mendjan S., Brown S., Chng Z., Teo A., Smithers L. E., Trotter M. W., Cho C. H., Martinez A., Rugg-Gunn P., et al. (2009). Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development 136, 1339–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Zhang H., Zhao Y., Li J., Cai J., Wang P., Meng S., Feng J., Miao C., Ding M., et al. (2005). Noggin and bFGF cooperate to maintain the pluripotency of human embryonic stem cells in the absence of feeder layers. Biochem. Biophys. Res. Commun. 330, 934–942 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Sakomura Y., Kurabayashi M., Manabe I., Aikawa M., Kuro-o M., Suzuki T., Yazaki Y., Nagai R. (1996). Structure and characterization of the 5 ′-flanking region of the mouse smooth muscle myosin heavy chain (SM1/2) gene. Circ. Res. 78, 978–989 [DOI] [PubMed] [Google Scholar]
- Wood H. B., Episkopou V. (1999). Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 86, 197–201 [DOI] [PubMed] [Google Scholar]
- Xu R. H., Chen X., Li D. S., Li R., Addicks G. C., Glennon C., Zwaka T. P., Thomson J. A. (2002). BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264 [DOI] [PubMed] [Google Scholar]
- Yamanaka Y., Lanner F., Rossant J. (2010). FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 137, 715–724 [DOI] [PubMed] [Google Scholar]
- Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991). Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64, 841–848 [DOI] [PubMed] [Google Scholar]
- Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.