Abstract
A major technical challenge in the cost-effective production of cellulosic biofuel is the need to lower the cost of plant cell wall degrading enzymes (PCDE), which is required for the production of sugars from biomass. Several competitive, low-cost technologies have been developed to produce PCDE in different host organisms such as Escherichia coli, Zymomonas mobilis, and plant. Selection of an ideal host organism is very important, because each host organism has its own unique features. Synthetic biology-aided tools enable heterologous expression of PCDE in recombinant E. coli or Z. mobilis and allow successful consolidated bioprocessing (CBP) in these microorganisms. In-planta expression provides an opportunity to simplify the process of enzyme production and plant biomass processing and leads to self-deconstruction of plant cell walls. Although the future of currently available technologies is difficult to predict, a complete and viable platform will most likely be available through the integration of the existing approaches with the development of breakthrough technologies.
1. Introduction
For decades, the biological conversion of plant biomass into biofuels has been a major focus of research [1]. Plant biomass is the most abundant renewable carbon source on Earth, with lignocellulose being its major constituent. Lignocellulose is a highly heterogeneous substrate composed of cellulose, hemicellulose, and lignin. Cellulose is a linear polymer of glucose that is extensively bonded to each other through strong intramolecular hydrogen bonds forming a highly recalcitrant and crystalline structure. Hemicellulose is a complex heteropolymer that comprises a number of polysaccharides such as xylan, galactan, and mannan. Hemicellulose contributes to the heterogeneity, whereas cellulose contributes to the recalcitrance of lignocellulose. Lignin comprises aromatic alcohols and is found to be associated with cellulose and hemicellulose. Lignin protects cellulose from hydrolytic enzymes [2]. Conventional approaches to extract the simple sugars in cellulose involve pretreatment under harsh conditions followed by enzymatic saccharification [3–5]. Simple sugars extracted can then be converted to advanced biofuels that resemble petroleum-based fuels by using recombinant microbes [6].
Biomass hydrolysis remains a unique hurdle and an expensive step in the production of cellulosic fuel. Over the years, emphasis has been placed on the development of inexpensive methodologies to produce hydrolytic enzymes. Cellulolytic microbes isolated from different environmental niches offered various forms of cellulases, and their identification has provided a deeper insight into the mechanism of lignocellulose hydrolysis. Genomic collection has revealed the distribution of a variety of genes encoding hydrolytic enzymes on the chromosome of cellulose-utilizing organisms such as Trichoderma reesei, Acidothermus cellulolyticus, and Clostridium cellulovorans [7–9]. The vast repository of genomic data has confirmed the need to engineer several groups of cellulases (endoglucanase, cellobiohydrolases, exoglucanase, and β-glucosidase) and hemicellulases (xylanase and arabinofuranosidase) with synergistic activity, into a single recombinant host to favor an efficient, single-step hydrolysis of the recalcitrant plant biomass into simple sugars [5]. However, expression of multiple enzymes would impose a huge metabolic burden on the engineered host [4, 10]. Selection of recombinant hosts with a superior ability to produce higher titers of multiple enzymes is a prerequisite for efficient bioprocessing of cellulosic biomass. The advent of recombinant genetic engineering and synthetic biology has enabled manipulation and development of recombinant plants and microbes that favor an efficient, single-step hydrolysis of lignocellulose [4, 11–13]. Many competing technologies have been developed for heterologous expression of plant cell wall degrading enzymes (PCDEs) coupled with simultaneous bioprocessing of cellulosic biomass in various host organisms (e.g., Escherichia coli, fungus, yeast, Zymomonas mobilis, and plant). Selection of an ideal host organism is essential because each host organism not only has its own unique features but also has advantages and disadvantages for a specific application. Thus, good understanding of the unique features of each recombinant host organism is imperative for the heterologous expression of PCDEs. To simplify, the consolidated bioprocessing approach involves the use of recombinant microbes such as E. coli and Z. mobilis, which are capable of hydrolyzing and fermenting plant biomass, whereas an in-planta approach employs plants capable of producing PCDEs, thus allowing autodegradation of plant biomass. In this paper, not only will we provide insight into the expression of PCDEs in three different host organisms, E. coli, Z. mobilis, and plant, but we will also highlight recent advances in the field.
2. Consolidated Bioprocessing
The consolidated bioprocessing (CBP) approach involves engineering cellulolytic ability into major industrially used solventogens such as yeast, E. coli, and Z. mobilis, to facilitate a single-step conversion of plant biomass to fuels. As described above, multiple enzymes are required to depolymerize plant biomass. In addition to the expression of multiple cellulolytic enzymes, engineering a proper protein-secretion system in recombinant microbes is necessary to favor a high titer of extracellular enzyme. Even if an efficient set of cellulases are engineered together with a proficient protein-secretion system, a proper enzyme-enzyme and/or enzyme-microbe synergy should be established to favor continuous hydrolysis of the substrate [14–16]. Without establishing proper synergy between different enzyme groups, the products obtained would begin to accumulate and lead to the inhibition of further substrate hydrolysis [3, 17]. In addition, hydrolysis of plant biomass is highly favored at a higher temperature, but most recombinant hosts are mesophilic, and this presents an additional hurdle in consolidated bioprocessing [8]. It is necessary to coordinate enzyme production, substrate hydrolysis, and fermentation process within a single cell in such a way that one does not lag behind or exceed the other processes. Aided by the powerful tools offered by synthetic biology, systems biology, structural biology, and protein engineering, efforts are underway to eliminate the above-mentioned bottlenecks and develop efficient microbes for consolidated bioprocessing. With the advent of surface-display technology, yeast became an unconquerable host for cellulase expression. However, recombinant hosts such as E. coli and Z. mobilis have recently been considered to have many properties ideal for cellulase expression. In this paper, we will discuss the efforts and hurdles in genetic engineering cellulolytic ability into E. coli and Z. mobilis. Cellulase expression in yeast is reviewed in depth elsewhere [18].
3. Cellulase Expression and Secretion in E. coli
E. coli is one of the most favored industrial microorganisms and has a high potential to become a consolidated bioprocessor owing to the wealth of knowledge available pertaining to this organism that allows for easy genetic manipulation. However, there are some hurdles in the development of E. coli with cellulolytic ability. While most native cellulolytic microbes are extremophiles, living either in conditions of high temperature or low pH, E. coli is a mesophile, and hence, the cellulase system adopted from extremophiles may not function efficiently in E. coli. Some anaerobic mesophiles (Ruminococcus albus and C. cellulovorans) also possess efficient cellulolytic ability [19]. However, unlike the extremophiles, anaerobic mesophiles produce complex cellulases called cellulosomes [20]. Cellulosome consists of a noncellulolytic scaffolding protein to which free cellulases are assembled with their dockerin domain and the entire assembly is displayed on the cell surface. Native proteases of E. coli cleave the dockerin domain of the cellulosomal cellulase, thus, disturbing the assembly process [21]. In addition, the cellulosomal cellulases (e.g., EngB) form inclusion bodies when overexpressed in E. coli. Solubilization of these cellulases was enhanced when their dockerin domain was replaced with the cellulose-binding domain (CBD) from a free cellulase [22]. Maximizing the portion of a soluble protein is a key factor for consolidated bioprocessing.
While solubilization of the expressed cellulases is a potent problem, secretion of the soluble cellulases is another major prerequisite for CBP. E. coli possess a thick outer membrane and very limited number of secretion systems capable of targeting protein to the extracellular space (Figure 1). As such, this thick outer membrane provides an additional hurdle in engineering a secretable cellulolytic system. Overexpression of a cellulolytic system without engineering a new protein-secretion pathway would probably inhibit cell growth due to obstruction of the native transport pathway [5]. Cellulase from Bacillus subtilis was cloned into E. coli and detectable extracellular secretion was achieved without any genetic modification; however, a larger proportion of the enzyme was localized in the periplasmic space [23]. The extracellular secretion may have been caused by the signal peptide of the B. subtilis cellulase that might have specificity toward native protein secretion system in E. coli. However, the localization of a large portion of the protein in the periplasmic space might be because of the differences in the membrane architecture of B. subtilis and E. coli. Being a Gram-positive bacterium, B. subtilis lacks an outer membrane, and, hence, the protein-secretion system is simpler and more efficient, whereas E. coli possesses a thick outer membrane that restricts extracellular transport of periplasmic proteins (Figure 1). E. coli possess an endogenous cellulase that could be secreted when overexpressed [24]. Fusion of the out gene of Erwinia chrysanthemi with a gene encoding for endoglucanase facilitated the secretion of more than 50% of the cellulase produced [25]. Exoglucanase from C. fimi was efficiently secreted from E. coli when fused to ompA sequence and expressed under a weak promoter (PlacUV5) [26]. Recombinant cellulase targeted to the periplasmic region could be secreted into the medium in an lpp-deleted strain of E. coli [27]. Even though cloning and secretion of a few heterologous genes is feasible in E. coli, it is not sufficient for CBP of plant biomass mainly because of the lesser substrate specificity of the readily expressible cellulases of E. coli.
Figure 1.
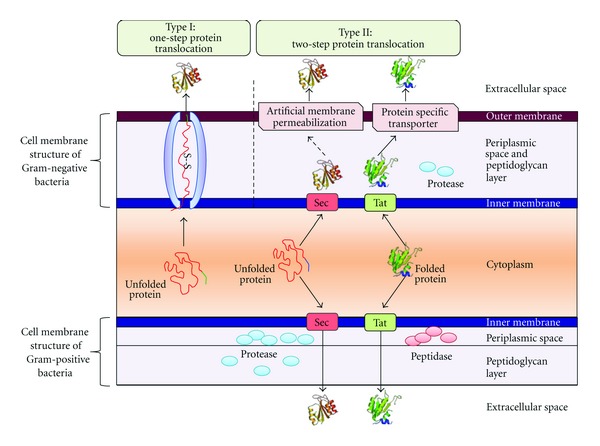
Outline of the major difference between the protein secretion system of Gram-positive and Gram-negative bacteria. Presence of thick outer membrane in Gram-negative bacteria demands for outer membrane permeabilization through the addition of mild detergent, lpp deletion, or by fusing the recombination protein with kill or out gene.
There was an increased search for a new group of cellulase, which is more specific for plant biomass and is readily expressible in E. coli. However, the lack of complete knowledge about the mechanism of cellulase action limits the use of a rational design of cellulase with desired traits for expression in E. coli. Metagenomic libraries are considered powerful tools for the isolation of genes with desired properties. The growing repository of metagenomic information promises to provide a better group of cellulase for expression in E. coli. Novel cellulases were identified in the metagenomic libraries obtained from the insect gut, termite gut, and cow rumen [28, 29]. The cellulases identified in the cow rumen were screened in E. coli and were found to exhibit significant activity against ionic liquid pretreated-plant biomass [30]. The GH5 and GH9 family of cellulases are readily expressible in E. coli. The metagenomic library might help identify GH5 and GH9 family of cellulases for E. coli [31].
Maintenance of the large heterologous DNA segment is rather complex. Despite the availability of several controllable gene expression systems, expression of multiple genes in a soluble form is technically challenging in E. coli. Hence, effort was put forth to identify cellulases with multiple functions. Some hydrolytic enzymes were found to exhibit a significant level of endoglucanase, exoglucanase, and xylanase activity. Cellulase isolated from Caldocellum saccharolyticum was found to have an endoglucanase catalytic domain in the C-terminus and exoglucanase catalytic domain in the N-terminus [32]. Cellulase with a multifunctional catalytic domain (performing both endoglucanase and exoglucanase activity) has been isolated from Bacillus sp. D04 [33]. Cellulase isolated from C. cellulovorans was also capable of functioning as a hemicellulase [34]. A metagenomic repository would be a very useful tool for the isolation of multifunctional cellulase with higher activity at mesophilic conditions.
E. coli is capable of utilizing all monosaccharides that could be obtained from plant biomass [3]. Cellobiose is a dimer of glucose and is the major end-product of cellulose hydrolysis. Accumulated cellobiose inhibits the endo- and exoglucanases, leading to a discontinuity in the overall process. Engineering a cellobiose-uptake system would therefore be the first step toward CBP, because it would help remove the potential inhibitory compound (cellobiose) immediately after it is formed, thus ensuring a continuous hydrolysis process [35]. The cellobiose metabolic pathway of Klebsiella oxytoca was successfully expressed in the ethanol-producing E. coli strain KO11, thus, favoring the growth of this strain on cellobiose [36]. Surface display of β-glucosidase of Thermobifida fusca in E. coli favored direct assimilation of cellooligosaccharides [37]. Cellobiose assimilation was achieved by replacing the promoters of endogenous chitin- (chb-) and salicin- (asc-) utilizing operons of E. coli [35]. Some of the cellobiose-utilizing strains mentioned-above have a superior capability over the glucose-utilizing counterparts to produce significantly higher levels of ethanol when grown on a model sugar substrate [36], while others can cometabolize cellobiose concurrently with sugars such as xylose, galactose, and mannose [38]. Thus, these cellobiose-metabolizing strains could be used as a platform strain in the development of cellulolytic E. coli.
Extensive research has helped strengthen the pool of E. coli biocatalyst with the superior ability to act on a cellulosic substrate; however, the potential of all of these engineered strains to grow on plant biomass directly could not be proved. This is mainly because of the failure of expression of a complete set of hydrolytic enzymes within a single host. Hence, coculture strategies have been proposed to alleviate the metabolic burden of expressing multiple heterologous genes within a single cell. Binary culture has been employed for efficient xylan degradation. Xylan constitutes the major portion of hemicellulose and requires the concomitant action of six enzymes for its hydrolysis to xylose. Cocultivation of two strains of E. coli expressing a different combination of the six major xylanases enhanced ethanol production from the hemicellulosic portion of plant biomass. Coculture of E. coli strains capable of expressing either the cellulolytic or hemicellulolytic enzyme complexes allowed growth on switchgrass pretreated with ionic liquid even without the addition of additional hydrolytic enzymes [33]. These coculture strategies are a promising step toward successful development of cellulolytic E. coli. The minimal genome approach [39] and tightly controllable/switchable gene expression system are well developed for E. coli [40, 41]. Exploitation of these systems to engineer a cellulolytic E. coli would provide precise, spatiotemporal modulation of gene expression of multiple cellulases based on the accumulation of products. A combination of the above-mentioned strategies would help overcome the bottlenecks associated with the development of cellulolytic E. coli.
4. Heterologous Expression and Secretion of Cellulases in Z. mobilis
Zymomonas mobilis is a unique Gram-negative microorganism that can metabolize glucose anaerobically through the Entner-Doudoroff (ED) pathway in contrast to other Gram-negative organisms (e.g., E. coli) that utilize the Embden-Meyerhof-Parnas (EMP) pathway. As an important industrial host organism, Z. mobilis has several advantages for biofuel applications that include higher sugar-uptake ability, a lower cellular biomass yield, and a higher ethanol yield/tolerance. Another advantage is that controlled addition/depletion of oxygen is not required during fermentation, as Z. mobilis can grow microaerobically. However, the limitation of this bacterium is that it can utilize only three sugar substrates: glucose, fructose, and sucrose. For expanding the substrate range, especially for the utilization of plant biomass-derived polymers, several PCDE genes have been cloned and expressed in Z. mobilis. However, similar to other Gram-negative bacteria such as E. coli, the presence of an outer membrane results in inefficient protein secretion, which is a major technical challenge in engineering cellulolytic Z. mobilis (Figure 1).
Previous reports on heterologous expression of PCDE genes in Z. mobilis are summarized (Table 1). Endoglucanase genes such as eglX [48], CMCase [43, 45], and celZ [46] were expressed in Z. mobilis under the control of either the chloramphenicol acyltransferase (cat) promoter [48] or their native promoters [43, 45, 46]. In the case of A. xylinum endoglucanase, approximately 75% of the endoglucanase activity was found in the periplasmic space in recombinant Z. mobilis with no detectable activity in the extracellular fraction. None of the above-described endoglucanases transformed to Z. mobilis could be secreted, mainly because of the protective outer membrane [45, 48]. In contrast, approximately 35% of the endoglucanase celZ from E. chrysanthemi was released into the medium in the absence of detectable cell lysis [46]. This probably occurred because of the correct recognition of the secretion signal of E. chrysanthemi by Z. mobilis. However, whether the secretion observed was due to active secretion or an increased leakiness of the outer membrane at the end of the growth phase was unclear [46]. The β-glucosidase gene from Xanthomonas albilineans or Ruminococcus albus was also expressed in Z. mobilis [50, 51]. Yanase et al. expressed a β-glucosidase gene from R. albus fused with the Tat (twin arginine translocation) signal peptide of a periplasmic enzyme, glucose-fructose oxidoreductase (Gfo), or Sec-dependent secretion signal peptide of gluconolactonase (Gln) [50]. They found that the enzyme thus produced was secreted into both the periplasmic and extracellular space. With the use of Tat and Sec signal peptides, 4.7% and 11.2% of the β-glucosidase activity, respectively, were detected in the extracellular space of the recombinant Z. mobilis [50]. However, these genes could not support the growth of recombinant Z. mobilis on cellobiose as the sole carbon source.
Table 1.
Summary of expression of plant cell wall degrading enzymes in Z. mobilis.
| PCDE | Source organism | Gene Bank ID | M.W (kDa) | Promoter | Secretion signal peptide | Extracellular portion (%) | Periplasmic portion (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Endo-β-1,4-glucanase | A. cellulolyticus | AAA75477.1 | 60.7 | P tac | Z. mobilis PhoC | ~20% | ~30% | [42] |
| A. cellulolyticus | AAA75477.1 | 60.7 | P tac | Z. mobilis ORF ZM 0331 | ~15% | ~25% | [42] | |
| A. cellulolyticus | ABK52392.1 | 41.3 | P tac | Z. mobilis PhoC | 13% | 26% | [42] | |
| A. xylinum | BAA03797.1 | 24 | Native | Native | <1% | 75% | [43] | |
| B. subtilis | NA | NA | Native | Native | NA | NA | [44] | |
| C. uda CB4 | AAA23090.1 | 40.7 | Z. mobilis unidentified gene | Native | NA | NA | [45] | |
| E. chrysanthemi | CAA68604.1 | 46.4 | Native | Native | ~40% | ~80% | [46] | |
| E. cloacae | ABP62583.1 | 40.3 | Native | Native | ~7.6% | ND | [47] | |
| P. cellulosa | NA | NA | ca t*gene on pSUP104 | Native | NA | NA | [48] | |
| X. albilineans | NA | NA | Native | Native | NA | NA | [49] | |
|
| ||||||||
| β-Glucosidase | R. albus | CAA33461.1 | 104.2 | Z. mobilis gfo gene | Z. mobilis Gfo | 4.7% | 61% | [50] |
| R. albus | CAA33461.1 | 104.2 | P tac | Z. mobilis Gln | 11.2% | 34.3% | [50] | |
| X. albilineans | NA | NA | Native | Native | NA | NA | [49, 51] | |
∗Chloramphenicol acyltransferase; NA: not available; ND: not detected.
For the hydrolysis of plant biomass, it is desirable to express multiple PCDE genes in Z. mobilis. Linger et al. heterologously expressed two cellulolytic enzymes, E1 (endo-β-1,4-glucanase) and GH12 (broad substrate range endo-β-1,4-glucanase activity) from an Acidothermus species by using two different secretion signals of Z. mobilis genes: phoC gene and ORFZM0331 [42]. The lack of a secretion signal in their genes resulted in the localization of 96% of GH12 activity within the cytoplasm, whereas the addition of the phoC secretion signal resulted in the localization of approximately 26% of the enzyme activity in the periplasmic space and 13% in the extracellular space. For E1 with the PhoC secretion signal, approximately 20% of the E1 activity was found in the extracellular medium, 30% in periplasmic space, and 50% in cytoplasm [42]. To develop Z. mobilis as a viable platform host organism for cellulosic biofuel production, more studies are needed to engineer Z. mobilis that secretes multiple PCDEs into the extracellular space necessary for the degradation of plant biomass.
5. Plant-Based Expression of Plant Cell Wall Degrading Enzymes
In-planta expression of PCDEs has several advantages over other expression systems (e.g., E. coli, fungus, and yeast) as a means of cost-effective production of cellulosic biofuels. Since plants can serve as both a host organism and a cellulosic biomass substrate for sugar fermentation, expression of PCDEs in plants can lead to self-deconstruction of plant cell walls to generate monomeric sugars. A combination of this approach with synthetic cellulolytic microbes would dramatically improve the economic efficiency of the cellulosic biofuel process (Figure 2). This is because the high production of multiple PCDEs in nonnative cellulolytic and industrial microbes imposes a huge metabolic burden that results in decreased biofuel production, whereas an in-planta approach would favor self-hydrolysable substrate that would reduce the metabolic cost of expression of multiple PCDEs in cellulolytic microbes. Thus, in-planta hydrolysis of plant cells offers a great opportunity to simplify the process of enzyme/biomass production and hydrolysis and minimizes the overall enzyme production cost without requiring the use of expensive bioreactors or complicated purification processes. Moreover, plants are also capable of posttranslational modifications, which may be required for correct protein folding and thus the functionality of specific enzymes [52, 53]. For these reasons, the expression of PCDEs in a plant is gaining increasing popularity. In this paper, we give a brief outline of the new updates on recent publications as well as on many factors to consider in this approach (Table 2). For example, autohydrolysis of plant cells can be avoided by introduction of thermostable cellulase, which has negligible enzyme activity at room temperature. An excellent summary on former studies can be found in previous review papers [53–55].
Figure 2.
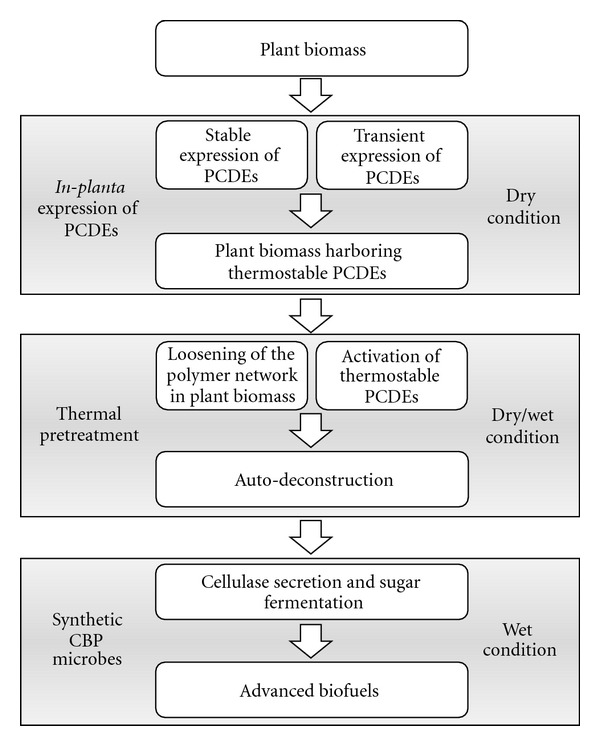
The combination of in-planta expression of PCDEs and synthetic CBP cellulolytic microbes can provide effective production of cellulosic biofuels.
Table 2.
Factors to consider in plant-based expression of plant cell wall degrading enzymes.
| Category | Exemplary factors associated with a common strategy |
|---|---|
| Expression type | Selection between stable transformation in a transgenic plant and transient expression in a wildtype plant |
|
| |
| Host plant | Composition of biomass Ease of DNA cloning Productivity of a plant crop Selection between monocot and dicot plants |
|
| |
| Gene | Development of fusion protein Function and properties of enzyme for example, Thermal stability Subcellular protein localization using transit peptide Synthetic gene design for example, Codon/UTR optimization Use of plant cell wall modifying enzyme |
|
| |
| Expression vector | Development of viral expression system for transient gene expression Easy selection markers for stably transformed plants Optimal promoter RBS optimization for example, Kozak's context sequence for monocot Selection between constitutive and inducible systems |
|
| |
| Functional analysis | Change of enzyme stability with time Cleavage of holoenzyme for example, Truncated enzyme Comparison of plant produced enzyme with E. coli produced enzyme Effect of plant crude extract on enzyme stability Expression of multiple enzymes Microscopic analysis of protein localization Physical properties for example, Activity optima, thermal stability Posttranslational modification for example, Glycosylation Purification process |
|
| |
| Phenotype analysis of transgenic plant | Compositional and structural change in transgenic plant Deleterious phenotype change for example, Fertile, pale leaves Location of protein accumulation for example, High level accumulation in leaf or stem New beneficial traits for example, Resistance to pathogens |
|
| |
| Biomass hydrolysis | Application of enzyme cocktails Composition and property of cellulosic biomass Induction of in-planta hydrolysis Optimal combination of enzymes Resistance of enzymes to pretreatment process |
Heterologous genes can be expressed stably in transgenic plants or transiently in wildtype plants. Most early studies have focused on the generation of stable transformants wherein foreign genes were integrated into plant chromosome or chloroplast genome. Once transgenic plant lines are developed, minimal efforts are needed for the production of the enzymes, since expressed enzymes accumulate in plant organs such as leaves or stems while the plants grow. Alternatively, PCDEs can be transiently produced in plants via Agrobacterium-tumefaciens-mediated transient expression. Compared with stable transformation, the use of a detached wildtype plant as a host for transient expression is advantageous, since it minimizes the environmental impact of genetically modified (GM) crops. In addition, A. tumefaciens-treated plants can produce enzymes for a period of several days [56]. It is assumed that most works reviewed in this paper were performed using transgenic plants unless otherwise specified as there are a very few reports on the transient expression of PCDEs.
For expression of PCDEs, careful selections of host plants, target genes, and expression vectors are essential not only to obtain high-level gene expression but also to reduce the cost of enzyme production, and facilitate pretreatment and hydrolysis of biomass. Along with a few different common dicot plants such as tobacco [57–60], and Arabidopsis [61], a few monocot plants such as sugar cane [62], maize [63, 64], rice [65], and duckweed [66] have been used for the expression of cellobiohydrolase CBH1/CBH2 [62, 63], endoglucanase [57, 60, 62, 63, 67], exoglucanase [60, 67], β-glucosidase [60, 67], xylanase [59, 61, 67], β-mannanase [9], pectate lyases [67], and cutinase [67]. Considering productivity of plant crops in pounds per acre, some popular biofuel monocots, such as Miscanthus and switchgrass, can be a promising host for in-planta expression of cellulases. However, to date, no reports on in-planta expression in biofuel monocotyledons has been published. To avoid self-deconstruction by the expressed PCDEs during growth and to produce highly stable enzymes, thermophilic or hyperthermophilic enzymes are commonly produced in plants [9, 57, 58, 60, 61, 63, 64, 66, 67]. This is perhaps one of the biggest differences when compared with cellulase expression in E. coli or Z. mobilis.
For development of expression vectors, the CaMV (cauliflower mosaic virus) 35S promoter has commonly been used. However, the use of an enhanced 35S promoter with 5′ tobacco etch virus-derived untranslated leader [58] and maize pepC promoter [62] has been reported. To enhance heterologous expression of bacterial or fungal genes in plant, codon optimized synthetic genes might be considered [61, 63]. As an alternative to nuclear integration of genes, overexpression was achieved when an expression cassette was integrated into the chloroplast genome [9, 60, 67].
To increase the level of protein accumulation or maintain high enzyme activity, a commonly adopted approach might be protein subcellular localization by incorporation of a transit peptide into a gene construct. Harrison et al. obtained a higher level of cellobiohydrolase and endoglucanase accumulation when they were fused to a vacuole-sorting determinant rather than an ER retention signal peptide, and the highest level of the endoglucanase was achieved when targeted to chloroplasts [62]. Kim et al. examined three different sources of chloroplast targeting transit peptides such as the light-harvesting chlorophyll a/b-binding protein, Rubisco small subunit (RS), and Rubisco activase. They found that the RS signal peptide favored the highest level of accumulation of cellulase, Cel5A, in transgenic tobacco plants [57]. Jiang et al. directly compared three different subcellular locations of endoglucanase, E2. They achieved the highest enzyme activity when E2 was directed to apoplast by using CLASP (tobacco calreticulin signal peptide) and the lowest enzyme activity from the cytosolic E2 [58]. The study by Hood et al. also supported the idea that protein subcellular localization is a key condition that determines protein accumulation level [63]. The highest endoglucanase E1 accumulation was observed when E1 was targeted to the ER or vacuoles; on the other hand, the activity of CBH I was not detected when it was targeted to vacuole.
Despite successful demonstrations of the subcellular targeting approach, further investigations are needed to determine the best subcellular organs or plant compartments especially when PCDEs are expressed in a senescing plant where proteolytic activity is high and cellular constituents are remobilized. If a large portion of harvested plant biomass is already senesced, high-level accumulation of PCDEs in the senesced plant or even in dead woody parts of plant biomass could be achieved. It is interesting that when the barley α-amylase signal peptide was used to direct xylanase to apoplast, the highest accumulation of xylanase was found in dried stems [61]. As an alternative, Hood et al. expressed thermostable cellulase enzymes in maize seed [63] and Llop-Tous et al. reported a new approach for the production of insoluble aggregates of active xylanase by incorporation of a proline-rich domain of the maize-storage protein into the xylanase-coding sequence [59]. Meanwhile, typical protein accumulation levels of various enzymes in plants ranged between 0.001% and 26% in total soluble protein [53].
While expressing PCDEs, transgenic plants still tended to show a normal phenotype and traits. This might be because of the expression of thermostable enzymes, which are inactive at room temperature or because the enzymes were localized in safe subcellular organs or spaces, and the protein accumulation levels were not sufficient to cause self-deconstruction. Kim et al. reported no changes in phenotype in transgenic tobacco plants expressing Cel5A [57], and Verma et al. observed normal phenotype of Arabidopsis lines expressing CelD, PelB, or PelD [67]. Xylanase-expressing Arabidopsis did not show any deleterious effects on phenotype when compared with wildtype plants [61]. In contrast, transplastomic tobacco plants expressing PCDEs from the chloroplast genome showed a mild phenotypic change in leaf color [9]. Significant phenotypic changes, such as poor plant growth and severe pigment deficiency, were more obvious in homoplasmic plants than in heteroplasmic plants [60]. Interestingly, expression of PCDEs may confer beneficial traits in transgenic plants. For instance, the expression of pectate lyase in tobacco plants improved bacterial resistance to pathogenic E. carotovora probably through induction of protective immune response in plants [67].
With respect to the properties of heterologous enzymes, as plants can process posttranslational modifications, apparent molecular masses of enzymes can be increased probably because of glycosylation and other posttranslational modifications [59, 62]. If a PCDE has multiple domains such as carbohydrate-binding domain (CBD), linker peptide, and catalytic domain, the linker peptide can be cleaved from the holoenzyme depending on the type of enzyme, host plant, and expression method. Harrison et al. reported a decrease in the apparent molecular weights of the expressed CBH I and CBH II in transgenic sugarcane; on the other hand, bacterial EG, which has a monodomain without a CBD, showed the expected molecular mass [62]. Interestingly, PCDEs produced by plants tended to show stability at higher temperature and pH than did those produced by E. coli. Verma et al. directly compared enzyme stability between E. coli- and plant-chloroplast-derived PelB, PelD, CelD, and β-mannanase enzymes at different pH and temperatures. They found that chloroplast-derived enzymes exhibited better stability than E. coli-derived enzymes did [9, 67]. They also showed that when E. coli crude extract containing cell-wall-degrading enzyme was directly loaded in a reaction medium, a high protein concentration reduced enzyme activity; on the other hand, plant extract did not show a reduction in enzyme activity at high protein loading [67].
Since cellulose pretreatment at high temperature allows easy mixing, better substrate solubility, high mass transfer rate, and lowered risk of contamination, the use of plant-expressing thermostable cellulases in cellulosic biofuel production is a promising approach. In addition, due to high temperature and self-deconstruction, in-planta expression helps easy access of externally added catalyst (PCDEs) to inside of plant biomass by loosening cross-linked wall polymers where difficult diffusion has been a significant bottleneck during pretreatment and conversion of lignocellulose to simple sugars. Thus, it may be a good combination between plant-based expression and synthetic CBP for synergistic effect on the production of advanced biofuel (Figure 2).
6. Conclusion
For efficient biofuel production, the expression of plant cell wall degrading enzymes in a host organism requires extensive coupling with bioprocessing of cellulosic biomass. Although diverse strategies depending on the unique feature of each host organism or expression system are available, as previously described, they all aim to achieve the same goal: a high-level production of multiple cellulase enzymes and simultaneous hydrolysis and fermentation of cellulosic biomass. At the present time, the future of currently available competing technologies is difficult to predict; moreover, the most efficient host organism among E. coli, Z. mobilis, fungus, and plant is difficult to determine. However, it is likely that a viable platform will be generated through the integration of multiple existing and future technologies.
Authors' Contributions
S-K. Jung, V. Parisutham and S. H. Jeong are equally contributed to this work.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) through Grants funded by the Ministry of Education, Science and Technology (MEST) (NRF-2009-C1AAA001-2009-0093479, NRF-2009-0076912, NRF-2010-0006436, R32-2008-000-20054-0), by the Intelligent Synthetic Biology Center of Global Frontier Project funded by the MEST (2011-0031948), by a Grant from the Next-Generation BioGreen 21 Program (SSAC, Grant no.: PJ0099058), Rural Development Administration, Republic of Korea, and by UNIST (Ulsan National Institute of Science and Technology) research grant.
References
- 1.Lee SK, Chou H, Ham TS, Lee TS, Keasling JD. Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Current Opinion in Biotechnology. 2008;19(6):556–563. doi: 10.1016/j.copbio.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Sakuragi H, Kuroda K, Ueda M. Molecular breeding of advanced microorganisms for biofuel production. Journal of Biomedicine and Biotechnology. 2011;2011:11 pages. doi: 10.1155/2011/416931.416931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinuselvi P, Park JM, Lee JM, Oh K, Ghim CM, Lee SK. Engineering microorganisms for biofuel production. Biofuels. 2011;2(2):153–166. [Google Scholar]
- 4.Xu Q, Singh A, Himmel ME. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Current Opinion in Biotechnology. 2009;20(3):364–371. doi: 10.1016/j.copbio.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Mazzoli R, Lamberti C, Pessione E. Engineering new metabolic capabilities in bacteria: lessons from recombinant cellulolytic strategies. Trends in Biotechnology. 2012;30(2):111–119. doi: 10.1016/j.tibtech.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Steen EJ, Kang Y, Bokinsky G, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 7.Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454(7206):841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DB. Cellulases and biofuels. Current Opinion in Biotechnology. 2009;20(3):295–299. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal P, Verma D, Daniell H. Expression of Trichoderma reeseiβ-mannanase in tobacco chloroplasts and its utilization in lignocellulosic woody biomass hydrolysis. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0029302.e29302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alper H, Stephanopoulos G. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nature Reviews Microbiology. 2009;7(10):715–723. doi: 10.1038/nrmicro2186. [DOI] [PubMed] [Google Scholar]
- 11.Ghim CM, Kim T, Mitchell RJ, Lee SK. Synthetic biology for biofuels: building designer microbes from the scratch. Biotechnology and Bioprocess Engineering. 2010;15(1):11–21. [Google Scholar]
- 12.Lynd LR, Van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Current Opinion in Biotechnology. 2005;16(5):577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 13.Himmel ME, Ding SY, Johnson DK, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315(5813):804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 14.Maki M, Leung KT, Qin W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. International Journal of Biological Sciences. 2009;5(5):500–516. doi: 10.7150/ijbs.5.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson DB. Three microbial strategies for plant cell wall degradation. Annals of the New York Academy of Sciences. 2008;1125:289–297. doi: 10.1196/annals.1419.026. [DOI] [PubMed] [Google Scholar]
- 16.Wen F, Nair NU, Zhao H. Protein engineering in designing tailored enzymes and microorganisms for biofuels production. Current Opinion in Biotechnology. 2009;20(4):412–419. doi: 10.1016/j.copbio.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.French CE. Synthetic biology and biomass conversion: a match made in heaven? Journal of the Royal Society Interface. 2009;6(4):S547–S558. doi: 10.1098/rsif.2008.0527.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Zyl WH, Lynd LR, Den Haan R, McBride JE. Consolidated bioprocessing for bioethanol production using Saccharomyces cerevisiae. Advances in Biochemical Engineering/Biotechnology. 2007;108:205–235. doi: 10.1007/10_2007_061. [DOI] [PubMed] [Google Scholar]
- 19.Doi RH. Cellulases of mesophilic microorganisms: cellulosome and noncellulosome producers. Annals of the New York Academy of Sciences. 2008;1125:267–279. doi: 10.1196/annals.1419.002. [DOI] [PubMed] [Google Scholar]
- 20.Doi RH, Kosugi A. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nature Reviews Microbiology. 2004;2(7):541–551. doi: 10.1038/nrmicro925. [DOI] [PubMed] [Google Scholar]
- 21.Murashima K, Kosugi A, Doi RH. Synergistic effects on crystalline cellulose degradation between cellulosomal cellulases from Clostridium cellulovorans. Journal of Bacteriology. 2002;184(18):5088–5095. doi: 10.1128/JB.184.18.5088-5095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murasbima K, Kosugi A, Doi RH. Solubilization of cellulosomal cellulases by fusion with cellulose-binding domain of noncellulosomal cellulase EngD from Clostridium cellulovorans. Proteins. 2003;50(4):620–628. doi: 10.1002/prot.10298. [DOI] [PubMed] [Google Scholar]
- 23.Hinchliffe E. Cloning and expression of a Bacillus subtilis endo-1,3-1,4-β- D-glucanase gene in Escherichia coli K12. Journal of General Microbiology. 1984;130(5):1285–1291. doi: 10.1099/00221287-130-5-1285. [DOI] [PubMed] [Google Scholar]
- 24.Park YW, Yun HD. Cloning of the Escherichia coli endo-1,4-D-glucanase gene and identification of its product. Molecular and General Genetics. 1999;261(2):236–241. doi: 10.1007/s004380050962. [DOI] [PubMed] [Google Scholar]
- 25.Zhou S, Yomano LP, Saleh AZ, Davis FC, Aldrich HC, Ingram LO. Enhancement of expression and apparent secretion of Erwinia chrysanthemi endoglucanase (encoded by celZ) in Escherichia coli B. Applied and Environmental Microbiology. 1999;65(6):2439–2445. doi: 10.1128/aem.65.6.2439-2445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam TL, Wong RSC, Wong WKR. Enhancement of extracellular production of a Cellulomonas fimi exoglucanase in Escherichia coli by the reduction of promoter strength. Enzyme and Microbial Technology. 1997;20(7):482–488. doi: 10.1016/s0141-0229(96)00203-7. [DOI] [PubMed] [Google Scholar]
- 27.Shin HD, Chen RR. Extracellular recombinant protein production from an Escherichia coli lpp deletion mutant. Biotechnology and Bioengineering. 2008;101(6):1288–1296. doi: 10.1002/bit.22013. [DOI] [PubMed] [Google Scholar]
- 28.Wenzel M, Schönig I, Berchtold M, Kämpfer P, König H. Aerobic and facultatively anaerobic cellulolytic bacteria from the gut of the termite Zootermopsis angusticollis. Journal of Applied Microbiology. 2002;92(1):32–40. doi: 10.1046/j.1365-2672.2002.01502.x. [DOI] [PubMed] [Google Scholar]
- 29.Fang Z, Fang W, Liu J, et al. Cloning and characterization of a β-glucosidase from marine microbial metagenome with excellent glucose tolerance. Journal of Microbiology and Biotechnology. 2010;20(9):1351–1358. doi: 10.4014/jmb.1003.03011. [DOI] [PubMed] [Google Scholar]
- 30.Hess M, Sczyrba A, Egan R, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331(6016):463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 31.Duan CJ, Feng JX. Mining metagenomes for novel cellulase genes. Biotechnology Letters. 2010;32(12):1765–1775. doi: 10.1007/s10529-010-0356-z. [DOI] [PubMed] [Google Scholar]
- 32.Saul DJ, Williams LC, Grayling RA, Chamley LW, Love DR, Bergquist PL. celB, a gene coding for a bifunctional cellulase from the extreme thermophile “Caldocellum saccharolyticum”. Applied and Environmental Microbiology. 1990;56(10):3117–3124. doi: 10.1128/aem.56.10.3117-3124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bokinsky G, Peralta-Yahya PP, George A, et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(50):19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foong FCF, Doi RH. Characterization and comparison of Clostridium cellulovorans endoglucanases-xylanases EngB and EngD hyperexpressed in Escherichia coli. Journal of Bacteriology. 1992;174(4):1403–1409. doi: 10.1128/jb.174.4.1403-1409.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinuselvi P, Lee SK. Engineering Escherichia coli for efficient cellobiose utilization. Applied Microbiology and Biotechnology. 2011;92(1):125–132. doi: 10.1007/s00253-011-3434-9. [DOI] [PubMed] [Google Scholar]
- 36.Edwards MC, Henriksen ED, Yomano LP, et al. Addition of genes for cellobiase and pectinolytic activity in Escherichia coli for fuel ethanol production from pectin-rich lignocellulosic biomass. Applied and Environmental Microbiology. 2011;77(15):5184–5191. doi: 10.1128/AEM.05700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka T, Kawabata H, Ogino C, Kondo A. Creation of a cellooligosaccharide-assimilating Escherichia coli strain by displaying active beta-glucosidase on the cell surface via a novel anchor protein. Applied and Environmental Microbiology. 2011;77(17):6265–6270. doi: 10.1128/AEM.00459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vinuselvi P, Lee SK. Engineered Escherichia coli capable of co-utilization of cellobiose and xylose. Enzyme and Microbial Technology. 2012;50(1):1–4. doi: 10.1016/j.enzmictec.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Trinh CT, Unrean P, Srienc F. Minimal Eschenchia coli cell for the most efficient production of ethanol from hexoses and pentoses. Applied and Environmental Microbiology. 2008;74(12):3634–3643. doi: 10.1128/AEM.02708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. Journal of Bacteriology. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ham TS, Lee SK, Keasling JD, Arkin AP. A tightly regulated inducible expression system utilizing the fim inversion recombination switch. Biotechnology and Bioengineering. 2006;94(1):1–4. doi: 10.1002/bit.20916. [DOI] [PubMed] [Google Scholar]
- 42.Linger JG, Adney WS, Darzins A. Heterologous expression and extracellular secretion of cellulolytic enzymes by Zymomonas mobilis. Applied and Environmental Microbiology. 2010;76(19):6360–6369. doi: 10.1128/AEM.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamoto T, Yamano S, Ikeaga H, Nakamura K. Cloning of the Acetobacter xylinum cellulase gene and its expression in Escherichia coli and Zymomonas mobilis. Applied Microbiology and Biotechnology. 1994;42(4):563–568. doi: 10.1007/BF00173921. [DOI] [PubMed] [Google Scholar]
- 44.Yoon KH, Park SH, Pack MY. Transfer of Bacillus subtilis endo-β-1,4-glucanase gene into Zymomonas anaerobia. Biotechnology Letters. 1988;10(3):213–216. [Google Scholar]
- 45.Misawa N, Okamoto T, Nakamura K. Expression of a cellulase gene in Zymomonas mobilis. Journal of Biotechnology. 1988;7(3):167–177. [Google Scholar]
- 46.Brestic-Goachet N, Gunasekaran P, Cami B, et al. Transfer and expression of an Erwinia chrysanthemi cellulase gene in Zymomonas mobilis. Journal of General Microbiology. 1989;135(4):893–902. [Google Scholar]
- 47.Thirumalai Vasan P, Sobana Piriya P, Immanual Gilwax Prabhu D, John Vennison S. Cellulosic ethanol production by Zymomonas mobilis harboring an endoglucanase gene from Enterobacter cloacae. Bioresource Technology. 2011;102(3):2585–2589. doi: 10.1016/j.biortech.2010.09.110. [DOI] [PubMed] [Google Scholar]
- 48.Lejeune A, Eveleigh DE, Colson C. Expression of an endoglucanase gene of Pseudomonas fluorescens var. cellulosa in Zymomonas mobilis. FEMS Microbiology Letters. 1988;49(3):363–366. [Google Scholar]
- 49.Su P, Liu CQ, Lucas RJ, Delaney SF, Dunn NW. Simultaneous expression of genes encoding endoglucanase and β-glucosidase in Zymomonas mobilis. Biotechnology Letters. 1993;15(9):979–984. [Google Scholar]
- 50.Yanase H, Nozaki K, Okamoto K. Ethanol production from cellulosic materials by genetically engineered Zymomonas mobilis. Biotechnology Letters. 2005;27(4):259–263. doi: 10.1007/s10529-004-8295-1. [DOI] [PubMed] [Google Scholar]
- 51.Su P, Delaney SF, Rogers PL. Cloning and expression of a β-glucosidase gene from Xanthomonas albilineans in Escherichia coli and Zymomonas mobilis. Journal of Biotechnology. 1989;9(2):139–152. [Google Scholar]
- 52.Gomord V, Faye L. Posttranslational modification of therapeutic proteins in plants. Current Opinion in Plant Biology. 2004;7(2):171–181. doi: 10.1016/j.pbi.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 53.Taylor LE, Dai Z, Decker SR, et al. Heterologous expression of glycosyl hydrolases in planta: a new departure for biofuels. Trends in Biotechnology. 2008;26(8):413–424. doi: 10.1016/j.tibtech.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 54.Sticklen MB. Retraction: plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nature Reviews. 2010;11(4):p. 308. doi: 10.1038/nrg2777. [DOI] [PubMed] [Google Scholar]
- 55.Sainz MB. Commercial cellulosic ethanol: the role of plant-expressed enzymes. In Vitro Cellular and Developmental Biology. 2009;45(3):314–329. [Google Scholar]
- 56.Joh LD, VanderGheynst JS. Agroinfiltration of plant tissues for production of high-value recombinant proteins: an alternative to production in transgenic crops. Journal of the Science of Food and Agriculture. 2006;86(13):2002–2004. [Google Scholar]
- 57.Kim S, Lee DS, Choi IS, Ahn SJ, Kim YH, Bae HJ. Arabidopsis thaliana Rubisco small subunit transit peptide increases the accumulation of Thermotoga maritima endoglucanase Cel5A in chloroplasts of transgenic tobacco plants. Transgenic Research. 2010;19(3):489–497. doi: 10.1007/s11248-009-9330-8. [DOI] [PubMed] [Google Scholar]
- 58.Jiang X, Zhou X, Jiang W, Gao X, Li W. Expressions of thermostable bacterial cellulases in tobacco plant. Biotechnology Letters. 2011;33(9):1797–1803. doi: 10.1007/s10529-011-0642-4. [DOI] [PubMed] [Google Scholar]
- 59.Llop-Tous I, Ortiz M, Torrent M, Ludevid MD. The expression of a xylanase targeted to ER-protein bodies provides a simple strategy to produce active insoluble enzyme polymers in tobacco plants. PLoS ONE. 2011;6(4) doi: 10.1371/journal.pone.0019474.e19474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petersen K, Bock R. High-level expression of a suite of thermostable cell wall-degrading enzymes from the chloroplast genome. Plant Molecular Biology. 2011;76(3-5):311–321. doi: 10.1007/s11103-011-9742-8. [DOI] [PubMed] [Google Scholar]
- 61.Borkhardt B, Harholt J, Ulvskov P, Ahring BK, Jørgensen B, Brinch-Pedersen H. Autohydrolysis of plant xylans by apoplastic expression of thermophilic bacterial endo-xylanases. Plant Biotechnology Journal. 2010;8(3):363–374. doi: 10.1111/j.1467-7652.2010.00506.x. [DOI] [PubMed] [Google Scholar]
- 62.Harrison MD, Geijskes J, Coleman HD, et al. Accumulation of recombinant cellobiohydrolase and endoglucanase in the leaves of mature transgenic sugar cane. Plant Biotechnology Journal. 2011;9(8):884–896. doi: 10.1111/j.1467-7652.2011.00597.x. [DOI] [PubMed] [Google Scholar]
- 63.Hood EE, Love R, Lane J, et al. Subcellular targeting is a key condition for high-level accumulation of cellulase protein in transgenic maize seed. Plant Biotechnology Journal. 2007;5(6):709–719. doi: 10.1111/j.1467-7652.2007.00275.x. [DOI] [PubMed] [Google Scholar]
- 64.Mei C, Park SH, Sabzikar R, Qi C, Ransom C, Sticklen M. Green tissue-specific production of a microbial endo-cellulase in maize (Zea mays L.) endoplasmic-reticulum and mitochondria converts cellulose into fermentable sugars. Journal of Chemical Technology and Biotechnology. 2009;84(5):689–695. [Google Scholar]
- 65.Oraby H, Venkatesh B, Dale B, et al. Enhanced conversion of plant biomass into glucose using transgenic rice-produced endoglucanase for cellulosic ethanol. Transgenic Research. 2007;16(6):739–749. doi: 10.1007/s11248-006-9064-9. [DOI] [PubMed] [Google Scholar]
- 66.Sun Y, Cheng JJ, Himmel ME, et al. Expression and characterization of Acidothermus cellulolyticus E1 endoglucanase in transgenic duckweed Lemna minor 8627. Bioresource Technology. 2007;98(15):2866–2872. doi: 10.1016/j.biortech.2006.09.055. [DOI] [PubMed] [Google Scholar]
- 67.Verma D, Kanagaraj A, Jin S, Singh ND, Kolattukudy PE, Daniell H. Chloroplast-derived enzyme cocktails hydrolyse lignocellulosic biomass and release fermentable sugars. Plant Biotechnology Journal. 2010;8(3):332–350. doi: 10.1111/j.1467-7652.2009.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


