Abstract
Here we report the generation of a novel class of supramolecular hydrogelators based on the integration of nucleobase, Arg-Gly-Asp (RGD) peptides, and glucosamine in a single molecule. These novel small molecule hydrogelators self-assemble in water to form stable supramolecular nanofibers/hydrogels and exhibit useful biostability. This approach provides a new opportunity for systematic exploration of the self-assembly of small biomolecules by varying any individual segment to generate a large array of supramolecular hydrogels for biological functions and for biomedical applications.
Introduction
Biological systems have extensively relied on the self-assembly of three classes of biomolecules, nucleic acids, proteins, and polysaccharides for performing biological functions. These biomacromolecules interact with each other through multiple, cooperative non-covalent interactions (e.g., hydrogen bonding, π-π stacking, hydrophobic forces, electrostatics, etc.) to form wide range of complex architectures for carrying out many key activities of living cells. These facts have led to the researches of molecular self-assembly, besides as a ubiquitous process in biology, as an important strategy in soft nanotechnology1 for the preparation of supramolecular nanostructures and nanomaterials by the use of small molecules2, 3 or oligomers.4 The addition of small molecules as possible components of self-assembly offers a unique opportunity to address the usually neglected issue—cost5—in the use of biomacromolecules for self-assembly.
Although it is possible to use biomacromolecules to form assemblies or materials that mimic the structures and (sometimes) the functions of biological systems, it is still relatively prohibitive for using the biomacromolecules in large quantities because of the high cost associated with their production, isolation, and purification. On the other hand, small molecules are much more accessible and less expensive; but how to use the self-assembly of small molecules for mimicking biomacromolecules remains a challenge that has yet to be met through systematic exploration of the self-assembly of small molecules. One of the easiest starting point of such endeavor is to explore the self-assembly of small, biologically important building blocks in water for mimicking the structures and functions of biomacromolecules. Some early works on this direction have underscored the feasibility and promises of this approach. For example, Shinkai et al.6 demonstrated that a uracil-appended cholesterol gelator not only showed excellent ability of self-assembly in most of organic solvents, but also displayed an increased stability of the molecular assembly (in the form of gels) by complementarily binding to oligonucleotide DNA via base-pairing. Shimizu et al.7 reported that nucleotide-appended bolaamphiphiles are capable of gelling water very effectively through the self-assembly that forms a fibrous network, and are able to form supramolecular helical nanofibers in the presence of specific target DNA. Barthelemy et al.8 described that uridine phosphocholine amphiphiles self-assemble into diverse supramolecular structures including vesicles, fibers, hydrogels, and organogels. Inspired by those pioneer works, we have recently demonstrated that the integration of nucleobase, amino acid, and glycoside leads to a new supramolecular system that exhibits diverse structural morphologies in nanoscale (e.g., nanoparticles, nanofibers, or other nanostructures) and has useful biological properties (e.g., biostability, biocompatibility, and the ability to bind and transport DNA into live cells).9 The above-mentioned works not only generated the structural mimic of biological system, but also proved the feasibility of using the self-assembly of small molecules to achieve the functions of biomacromolecules. Moreover, the conjugates of nucleobase, amino acid, and glycoside provide a unique opportunity for a systematic exploration of the self-assembly of small molecules because it is relatively easy to vary any individual segment and to generate a large number of different molecules for self-assembly.
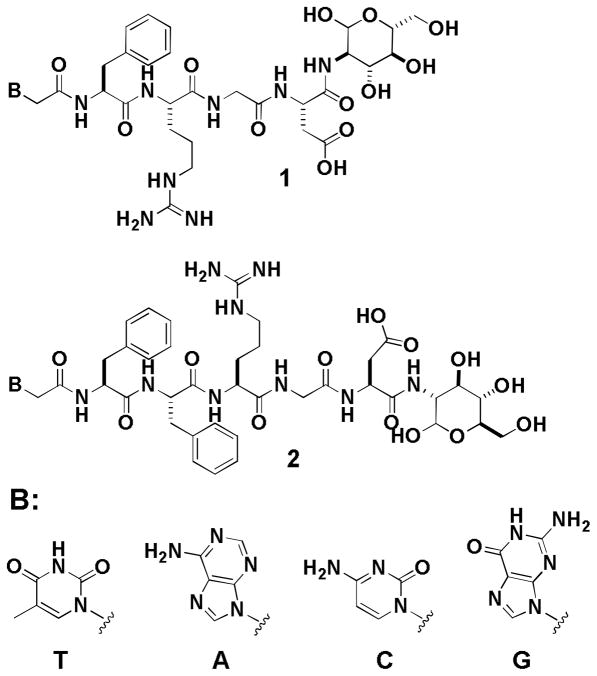
To explore the strategy for expanding the current repertoire of building blocks and generating supramolecular assemblies from biomolecules, and to produce more functional supramolecular hydrogels with biological functions for biomedical applications, we design and synthesize molecules that contain a nucleobase (e.g., adenine, guanine, thymine, or cytosine), a tetrapeptide (e.g., Phe-Arg-Gly-Asp) or a pentapeptide (e.g., Phe-Phe-Arg-Gly-Asp), and a glycoside (e.g., D-glucosamine) via covalent bonds, and successfully obtain a new class of hydrogelators (Scheme 1) that self-assemble in water to afford supramolecular nanofibers and hydrogels. We specifically include Arg-Gly-Asp (RGD) in the peptide segment because RGD, besides as a tripeptide motif responsible for cell adhesion through the selective binding to αvβ3 and αvβ5 integrins,10 is able to help target drugs and diagnostic agents to cancer cells that overexpress integrins.11 Our results show that the designed molecules (1 and 2, Scheme 1) act as hydrogelators that self-assemble in water to form nanofibers and produce hydrogels at the concentration of 3.0 wt% and proper pH. Moreover, the inclusion of glucosamine at the C-terminal of the peptides increases the ability of the hydrogelators to resist enzymatic digestion if the peptide sequences are appropriate. This work illustrates a simple and systematic approach for making a large array of potentially biostable and multifunctional hydrogelators for applications that require long-term biostability.
Scheme 1.
The molecular structures of the hydrogelators 1 and 2 consisting of nucleobase, RGD peptides, and glycoside.
Results and discussion
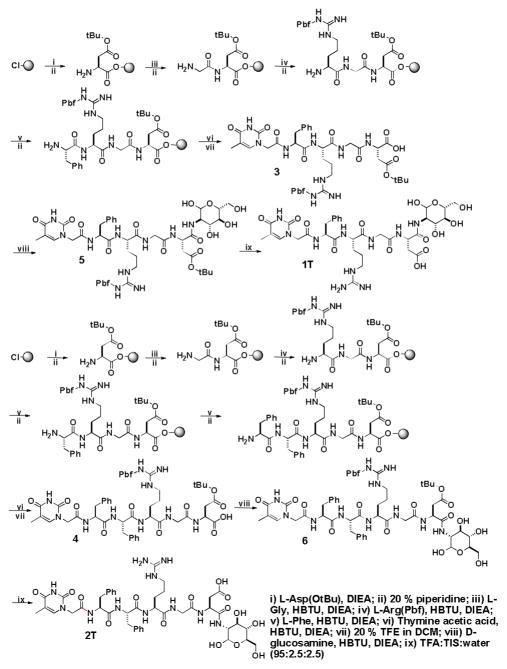
Scheme 2 shows the synthesis of 1T and 2T via the combination of solid phase synthesis and simple coupling reactions in solution phase. Following the protocols reported for making protected peptide fragments,12 we first prepare thymine-Phe-Arg(Pbf)-Gly-Asp(tBu) and thymine-Phe-Phe-Arg(Pbf)-Gly-Asp(tBu) using Fmoc-amino acids and thymine acetic acid. Starting from 2-chlorotrityl resin, the peptide chain is extended from the C-terminal towards the N-terminal through step-by-step peptide chain elongation procedure. After condensations of the peptides with thymine acetic acid, we cleave the protected nucleopeptides fragments from the resin and obtained 3 and 4 in 70–80% yield. After being activated by N-Hydroxysuccinimide (NHS), thymine-Phe-Arg(Pbf)-Gly-Asp(tBu) or thymine-Phe-Phe-Arg(Pbf)-Gly-Asp(tBu) reacts with D-glucosamine, to give the intermediate compounds 5 and 6; the subsequent removal of the protecting groups from them by the addition of trifluoroacetic acid (TFA) gives the hydrogelator 1T or compound 2T in a fair total yield. The syntheses of other hydrogelators (i.e., 1A, 2A, 1C, 2C, 1G and 2G) use other protected nucleobases (i.e., (N6-bis-Boc-adenine-9-yl)-acetic acid, (N4-bis-Boc-cytosine-1-yl)-acetic acid, and (N2-bis-Boc-guanine-9-yl)-acetic acid) (Fig. S1, S2, S3), which are prepared according to the procedures of making nucleobase acetic acids reported by Nieddu.13
Scheme 2.
The synthetic routes of the hydrogelators 1T and 2T.
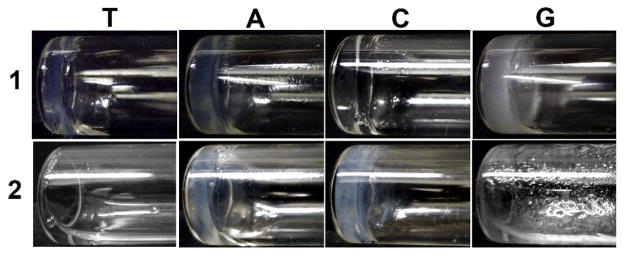
The synthesis of 1 and 2 allows us to test their abilities to form hydrogels in water. All the molecules, except 2T and 2G, behave as small molecule hydrogelators and self-assemble to form hydrogels at the concentration of 3.0 wt% (Table 1). Typically, they have good solubility and give clear solutions in water at a slightly basic condition or raised temperature. The solutions turn into stable hydrogels (Fig. 1) upon the change of their pH to different values. For example, hydrogelator 1T or 1C can form a transparent hydrogel at pH 4.0, and hydrogelator 1A or 1G can afford a semi-transparent hydrogel at pH 7.4. 2C produces a semi-transparent hydrogel at pH=4.0, and hydrogelator 2A self-assembles in water to form a semi-transparent hydrogel at pH around 4.0. The different optical appearances of the hydrogels, as well as the pHs for hydrogelation, between 1T, 1C, 1A, 1G, 2C, and 2A suggest the difference of the self-assembly of these hydrogelators, which probably arises from that purine bases favor the formation of Hoogsteen base pair and strong π-π interaction of purine nucleobases due to two fused five- and six-member heterocyclic rings. Interestingly, 2T and 2G fail to form hydrogels under the condition tested, suggesting that the additional phenylalanine group in 2T and 2G (comparing to 1T and 1G) may disfavor the self-assembly. By following the study of Ulijin et al. on pKa shifts with supramolecular self-assembly14, we calculate the pKa value of carboxylic acid group on the side chain of aspartic acid using SPARC web calculator. The pKa values of the carboxylic groups all are around 3.84, suggesting that hydrogelators exist as carboxylates at the pH higher than 3.84 and also favor ionic association. These results confirm that the covalent connection of nucleobase, RGD peptide, and glycoside offers a valid, simple approach to construct new supramolecular hydrogelators.
Table 1.
Summary of the conditions and properties of the hydrogelators based on nucleobases, Arg-Gly-Asp (RGD) peptides, and glucosamine and corresponding supramolecular nanofibers and hydrogels
| Sample | 1T | 1A | 1C | 1G | 2T | 2A | 2C | 2G |
|---|---|---|---|---|---|---|---|---|
| wt % | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 |
| pH | 4.0 | 7.4 | 4.0 | 7.4 | 4.0a | 4.0 | 4.0 | 4.0a |
| Morphologies of nanostructure | single fibers ribbons, helical fibers | ribbons, bundles | single fibers, double parallel fibers, double helical fibers | fiber with different lengths, bundles | – | fibers, bundle of fibers | fibers, bundles, helical ribbons | single fibers, bundles |
| Width of nanofibers (nm) | 15 | 15 | 10 | 12 | – | 7 | 10 | 6 |
| Critical strain (%) | 0.5 | 0.8 | 0.5 | 0.8 | – | 0.6 | 0.6 | – |
| G′ (kPa) | 35 | 17 | 2.3 | 2.1 | – | 6 | 2.4 | – |
| Biostability (compound remained (%) after 24 hrs) | 45 | 52 | 37 | 50 | 0 | 0 | 0 | 4 |
These compounds fail to result in hydrogels from acidic to basic conditions.
Fig. 1.
Optical images of the hydrogels of 1T (pH=4.0), 1A (pH=7.4), 1C (pH=4.0), 1G (pH=7.4), 2A (pH=4.0), 2C (pH=4.0), and the solution of 2T (pH=4.0) and 2G (pH=4.0). All are at 3.0 wt%.
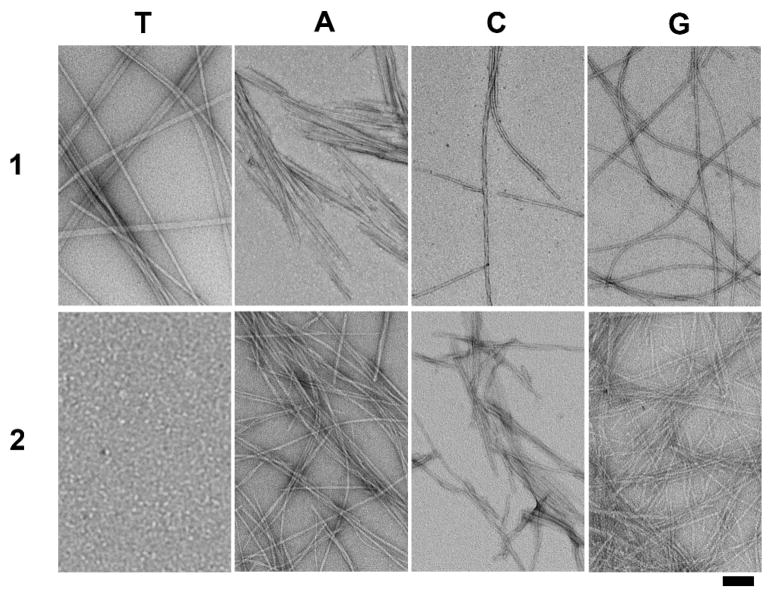
Since supramolecular hydrogelation usually associate with the generation of one-dimensional nanostructures, which physically cross-link and entangle with each other to form a fibrous three-dimensional network as the matrices of the hydrogels, we use transmission electronic microscope (TEM) to image the nanostructures formed by the self-assembly of the hydrogelator 1 or 2 in water. To prepare the TEM samples, we incubate 10 μL of the sample from the hydrogel on a carbon-coated copper grid, followed by removing the excess solution. The grid is negatively stained with 2.0% (w/v) uranyl acetate.15 According to the TEM images shown in Fig. 2, each hydrogelator self-assembles into high-aspect-ratio nanofibers with several micrometers in length and tens of nanometer in width. These well-defined fibrous nanostructures also form bundles to give a hierarchical organization (Fig. S4), indicating that the presence of ionic complementary peptide (RGD segment) may contribute to the formation of the bundles while the difference in the nucleobases may lead to the difference in the morphology and polymorphism of the nanofibers (Table 1). For example, 1T self-assembles to form three different kinds of straight nanofibers, including a large amount of thin single fibers, untwisted nanoribbons formed by parallel association of single fibers, and small quantities of twisted, helical nanofibers. The thin single nanofibers have widths about 15 nm and length over microns. In contrast, the widths of the untwisted nanoribbons range from 25 nm to 50 nm, and the helical nanofibers is twisted along the fiber long axis with a diameter of 23 nm and a helical pitch of 220 nm. Being different from that of 1T, the TEM images from the assembly of 1A show short nanoribbons that aggregate with a width of tens of nanometers to one hundred nanometers and a length of several micrometers. The enlarged images of TEM indicate that the nanoribbons of 1A contain lamellar structures or bundles of parallel nanofibers. TEM images of 1C reveal three self-assembled nanostructures in the hydrogel: individual nanofibers, bundles of two parallel nanofibers, and double helical nanofibers. The single fibers have lengths about several micrometers and uniform widths around 10 nm. The widths of bundles of two parallel nanofibers are about 20 nm, and the double helical nanofibers consist of two single fibers, which twist along the fiber long axis with a diameter of 18 nm and a helical pitch of 85 nm. The hydrogel of 1G contains both short and long fibers with 12 nm in diameter and tens of nanometers to several micrometers in lengths. Two long single nanofibers tend to aggregate together through parallel association to form nanobundles with a width of 16 nm. The easy observation of short nanofibers aggregated from hydrogelator 1A and 1G implies that the presence of purine nucleobase containing two fused five- and six-member heterocyclic rings favors relatively strong π-π interactions and molecular self-assembly, possibly resulting in more nucleating sites, and these nanofibers stop growing when they meet each other rather than continue to grow on top of each other at the junctions of the fibers. The self-assembly of 2A at pH 4.0 affords a large quantity of bundles of nanofibers that have widths of tens of nanometers and a length of hundreds of nanometer to several micrometers, in addition to a small quantity of individual thin nanofibers with a diameter of 7 nm and a length of hundreds of nanometers. The hydrogel of 2C consists of nanofibers, nanobundles and helical nanoribbons. The nanofibers have widths of about 10 nm and lengths of around hundreds of nanometers, the nanobundles are composed of untwisted lamellar structures or aggregates of nanofibers by lateral associations, and the helical nanoribbons self-assemble from the twisted structures in a diameter of 26 nm and a helical pitch of 39 nm. Although 2G fails to form a hydrogel, TEM image of the viscous solution of 2G shows two kinds of nanostructures: individual thin nanofibers which have a width of 6 nm and a length of hundreds of nanometers, and a small amount of nanobundles with 16 nm in diameter and several micrometers in lengths, indicating the self-assembly of 2G in the solution.
Fig. 2.
Transmission electron micrograph (TEM) of the negative stained hydrogels of 1T, 1A, 2A, 1C, 2C, and 1G and the solution of 2T and 2G. Scale bar = 100 nm, and the concentration and pH value for each of them are as same as in Fig. 1.
In order to investigate their viscoelastic properties, one of the essential features of hydrogels, we perform dynamic rheology experiments to study the viscoelasticity of hydrogels 1 and 2. The viscoelastic behavior of a hydrogel gives two key parameters, storage modulus (G′) and loss modulus (G″), to indicate, respectively, the ability of the deformed hydrogel to restore its original geometry and the tendency of a material to flow under exerted stress. For a gel material, G′ is greater than G″, indicating the dominant elastic behavior of the system. In the oscillatory strain sweep experiment, we monitor the variation of storage modulus and loss modulus as a function of strain under a constant frequency 10 rads−1. The hydrogels exhibit typical solid-like rheological behavior with the storage moduli (G′) greater than the loss moduli (G″) within the investigated oscillating strain limit, and the storage moduli show linear viscoelastic responses between 0.1 and 1.0%, providing the elastic properties of hydrogels made of 1T, 1A, 1C, 1G, 2A, and 2C, respectively. Fig. 3 shows the strain sweep profiles of elastic moduli (G′) and loss moduli (G″) at 25 °C. For example, hydrogel 1T exhibits the highest storage modulus of 35.0 kPa among the hydrogels tested in this work, the hydrogels of 1A and 2A possess relatively high storage moduli of 17.0 and 6.0 kPa, respectively, and the hydrogels of 1C, 1G, and 2C show much lower storage moduli of 2.3, 2.1, and 2.4 kPa, respectively (Table 1). The value of critical stain (γc, the strain amplitude at which G′ begins to decrease by 5% from its maximum value) also reflects the stability of hydrogels to strain. The critical strain (γc) is the measurement of the minimum strain amplitude at which storage moduli just begins to decrease by 5% from its maximum value. It derives from the storage-strain profiles of the hydrogel sample when the strain increases from 0.1 to 10% (10 rad/s and 25°C). Over a certain strain, the elastic modulus starts to decrease, and the strain amplitude at the onset of decrease to 5 % of decrease from its maximum value is the critical strain of the hydrogels, which corresponds to the breakdown of the cross-linked network in the hydrogel sample.16 For example, hydrogels of 1A and 1G show the critical strain values of 0.8 %; the hydrogels of 1T, 1C, 2A, and 2C exhibit the relatively low critical strain values at 0.5, 0.5, 0.6, and 0.6 %, respectively (Table 1). The oscillatory frequency sweep experiment help further characterize the strengths of the hydrogels (at 0.1–200 rad/s, a strain of 0.4 %, and within the linear regime). The G′ and G″ values measured in the frequency sweep experiment agree with those obtained from the strain sweep measurement, suggesting the reproducibility of both moduli and the formation of stable hydrogels from hydrogelator 1T, 1A, 1C, 1G, 2A, and 2C, respectively (Fig. S5).
Fig. 3.
(A) Strain dependence of the dynamic storage moduli (G′) and the loss moduli (G″) of the hydrogels of 1T, 1A, 1C, 1G shown in Fig. 1; (B) strain dependence of the dynamic storage moduli (G′) and the loss moduli (G″) of the hydrogels of 2A and 2C shown in Fig. 1.
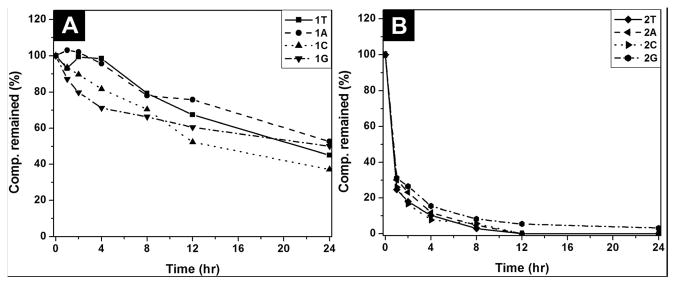
Supramolecular hydrogels, made from small peptides or short L-amino acid sequences, usually are susceptible to biodegradation through hydrolysis catalyzed by proteolytic enzymes.3, 17 Such an inherent susceptibility towards enzymes shortens the in vivo lifetime of these peptide-based hydrogels, thus reducing their efficacy and limiting their scope of applications that require long-term bioavailability.18 Therefore, many efforts have focused on designing and synthesizing peptidomimetic molecules to achieve prolonged or controlled stability and bioavailability. Besides peptidomimics,19, 20 it is also possible to employ glycosylation, a strategy used in biological systems, at the C-terminal of amino acids/peptides for enhancing the stability of the hydrogelators without comprising functions, thus developing biostable and multifunctional hydrogels for applications that require long-term biostability.9, 21 To examine the biostability of 1 and 2, we incubated hydrogelators 1T, 1A, 1C, 1G, 2A, 2C, and compound 2T and 2G with proteinase K, a powerful protease that degrades a wide range of peptidic substrates.20, 22 As shown in Fig. 4A, more than 45 % of 1T, 52% of 1A, 37 % of 1C, and 50% of 1G remain intact after 24 hrs of incubation with proteinase K. Compared with thymine-FRGD, the nucleopeptide without the conjugation of glycoside hydrolyzes completely in 4 hours after the treament with proteinase K (Fig. S6), and these results demonstrates that the incorporation of glycoside renders hydrogelators 1 to be fairly resistance to enzymatic digestion. However, 100% of hydrogelator 2A and 2C, and compound 2T hydrolyzes in 12 hours, and more than 96 % of 2G degrades after 24 hour upon the same treatment (Fig. 4B), indicating the low proteolytic resistance of 2, likely due the longer peptide backbone of 2 and poor tendency to self-assemble to form nanostructrues. We exmanined the biostability of the conjugates in solution state with the concentrations of 0.2 wt%, which are much lower than the gelation concentration (3.0 wt%). This excludes the possibilities that the formation of the supramolecular hydrogels slow down the degradation of the conjugates, thus validating the protective effect conferred by the attachment of glucosamine.
Fig. 4.
(A) The time-dependent course of the digestions of (A) hydrogelators of 1T, 1A, 1C, and 1G; (B) hydrogelators of 2A, 2C, and compound 2T and 2G by proteinase K.
Conclusions
By integrating nucleobase, RGD, and glycoside, we generate a new kind of biostable and biofunctional hydrogels for applications that require long-term biostability. The incorporation of RGD into hydrogelators affords the hydrogels with inherent functionality to bind with live cells through αvβ3 and αvβ5 integrin receptors;23 the attachment of glycoside at the C-terminal not only improves the biostability of the hydrogels, but also allows the self-assembled glycoside to mimic polysaccharides that are the important components of extracellular matrices. For example, hydrogelator 1A or 1G can self-assemble to form stable hydrogels at physiological conditions (pH=7.4), which promise a variety of potential applications, including 3D cell culture, tissue engineering, and drug delivery, which are the subject under continuing exploration. From the systematical evaluation, we find that the conjugation of different nucleobase, FRGD (tetrapeptide), and glucosamine generates stable supramolecular hydrogels with fair biostability to resist proteinase K digestion (Table 1). The integration of FFRGD (pentapeptide) with nucleobases and glucosamine, however, fails to increase biostability. These results underscore the limitation of the predication of the properties of hydrogels from molecular structures, but highlights the importance of the synthesis and experimental evaluation of the supramolecular hydrogels. Nevertheless, this study not only generates new hydrogelators for making gel biomaterials, but also contributes to the rational design of the building blocks for supramolecular nanofibers/hydrogel formation from small bioactive molecules.
Supplementary Material
Acknowledgments
This work was partially supported by a NIH grant (R01CA142746), a HFSP grant (RGP0056/2008), and start-up fund from Brandeis University. Thanks the assistance of Brandeis EM facility.
Footnotes
Electronic Supplementary Information (ESI) available: [Synthesis of hydrogelators 1T, 1A, 1C, 1G, 2A, 2C, and compound 2T and 2G, CD spectra, rheological measurements, cell viability and biostability tests.]. See DOI:10.1039/b000000x/
References
- 1.Whitesides GM, Lipomi DJ. Faraday Discuss. 2009;143:373–384. doi: 10.1039/b917540g. [DOI] [PubMed] [Google Scholar]
- 2.Yang J, Fan EK, Geib SJ, Hamilton AD. J Am Chem Soc. 1993;115:5314–5315. [Google Scholar]; Terech P, Weiss RG. Chem Rev. 1997;97:3133–3159. doi: 10.1021/cr9700282. [DOI] [PubMed] [Google Scholar]; Kolbel M, Menger FM. Chem Commun. 2001:507–507. [Google Scholar]; Lucas LN, van Esch J, Kellogg RM, Feringa BL. Chem Commun. 2001:759–760. [Google Scholar]; Estroff LA, Hamilton AD. Chem Rev. 2004;104:1201–1217. doi: 10.1021/cr0302049. [DOI] [PubMed] [Google Scholar]; Kiyonaka S, Sada K, Yoshimura I, Shinkai S, Kato N, Hamachi I. Nat Mater. 2004;3:58–64. doi: 10.1038/nmat1034. [DOI] [PubMed] [Google Scholar]; Li C, Numata M, Bae AH, Sakurai K, Shinkai S. J Am Chem Soc. 2005;127:4548–4549. doi: 10.1021/ja050168q. [DOI] [PubMed] [Google Scholar]; Sangeetha NM, Maitra U. Chem Soc Rev. 2005;34:821–836. doi: 10.1039/b417081b. [DOI] [PubMed] [Google Scholar]; Iwaura R, Hoeben FJM, Masuda M, Schenning APHJ, Meijer EW, Shimizu T. J Am Chem Soc. 2006;128:13298–13304. doi: 10.1021/ja064560v. [DOI] [PubMed] [Google Scholar]; Jayawarna V, Ali M, Jowitt TA, Miller AE, Saiani A, Gough JE, Ulijn RV. Adv Mater. 2006;18:611–614. [Google Scholar]; Wang G, Sharma V, Cheuk S, Williams K, Dakessian L, Thorton Z. Carbohydr Res. 2006;341:705–716. doi: 10.1016/j.carres.2006.01.023. [DOI] [PubMed] [Google Scholar]; Zhao F, Ma ML, Xu B. Chem Soc Rev. 2009;38:883–891. doi: 10.1039/b806410p. [DOI] [PubMed] [Google Scholar]; Boekhoven J, van Rijn P, Brizard AM, Stuart MCA, van Esch JH. Chem Commun. 2010;46:3490–3492. doi: 10.1039/c000337a. [DOI] [PubMed] [Google Scholar]; Zelzer M, Ulijn RV. Chem Soc Rev. 2010;39:3351–3357. doi: 10.1039/c0cs00035c. [DOI] [PubMed] [Google Scholar]; Banerjee S, Vidya VM, Savyasachi AJ, Maitra U. J Mater Chem. 2011;21:14693–14705. [Google Scholar]; Grahame DAS, Olauson C, Lam RSH, Pedersen T, Borondics F, Abraham S, Weiss RG, Rogers MA. Soft Matter. 2011;7:7359–7365. [Google Scholar]
- 3.Zhang Y, Kuang Y, Gao YA, Xu B. Langmuir. 2011;27:529–537. doi: 10.1021/la1020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vauthey S, Santoso S, Gong HY, Watson N, Zhang SG. Proc Natl Acad Sci USA. 2002;99:5355–5360. doi: 10.1073/pnas.072089599. [DOI] [PMC free article] [PubMed] [Google Scholar]; Rajagopal K, Schneider JP. Curr Opin Struc Biol. 2004;14:480–486. doi: 10.1016/j.sbi.2004.06.006. [DOI] [PubMed] [Google Scholar]; Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 5.Whitesides GM. The Economist - The World in 2012. 2011. vol. Dec-11. [Google Scholar]
- 6.Snip E, Shinkai S, Reinhoudt DN. Tetrahedron Lett. 2001;42:2153–2156. [Google Scholar]; Snip E, Koumoto K, Shinkai S. Tetrahedron. 2002;58:8863–8873. [Google Scholar]; Numata M, Shinkai S. Chem Lett. 2003;32:308–309. [Google Scholar]
- 7.Iwaura R, Yoshida K, Masuda M, Yase K, Shimizu T. Chem Mater. 2002;14:3047–3053. [Google Scholar]; Iwaura R, Yoshida K, Masuda M, Ohnishi-Kameyama M, Yoshida M, Shimizu T. Angew Chem Int Ed. 2003;42:1009. doi: 10.1002/anie.200390257. [DOI] [PubMed] [Google Scholar]; Iwaura R, Ohnishi-Kameyama M, Shimizu T. Chem Commun. 2008:5770–5772. doi: 10.1039/b813592d. [DOI] [PubMed] [Google Scholar]
- 8.Moreau L, Barthelemy P, El Maataoui M, Grinstaff MW. J Am Chem Soc. 2004;126:7533–7539. doi: 10.1021/ja039597j. [DOI] [PubMed] [Google Scholar]; Moreau L, Campins N, Grinstaff MW, Barthelemy P. Tetrahedron Lett. 2006;47:7117–7120. [Google Scholar]
- 9.Li X, Kuang Y, Shi J, Gao Y, Lin HC, Xu B. J Am Chem Soc. 2011;133:17513–17518. doi: 10.1021/ja208456k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruoslahti E. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]; Desgrosellier JS, Cheresh DA. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rust WL, Carper SW, Plopper GE. J Biomed Biotechnol. 2002;2:124–130. doi: 10.1155/S1110724302204015. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cox D, Brennan M, Moran N. Nat Rev Drug Discov. 2010;9:804–820. doi: 10.1038/nrd3266. [DOI] [PubMed] [Google Scholar]; Chen K, Chen X. Theranostics. 2011;1:189–200. doi: 10.7150/thno/v01p0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan WC, White PD. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Oxford University Press; New York: 2004. [Google Scholar]
- 13.Porcheddu A, Giacomelli G, Piredda I, Carta M, Nieddu G. Eur J Org Chem. 2008:5786–5797. [Google Scholar]
- 14.Tang C, Smith AM, Collins RF, Ulijn RV, Saiani A. Langmuir. 2009;25:9447–9453. doi: 10.1021/la900653q. [DOI] [PubMed] [Google Scholar]
- 15.Hayat MA. Principles and Techniques of Electron Microscopy: Biological Applications. 4. Cambridge University Press; 2000. [Google Scholar]
- 16.Song F, Zhang LM. J Phys Chem B. 2008;112:13749–13755. doi: 10.1021/jp8041389. [DOI] [PubMed] [Google Scholar]; Shih WH, Shih WY, Kim SI, Liu J, Aksay IA. Phys Rev A. 1990;42:4772–4779. doi: 10.1103/physreva.42.4772. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Liang G, Xu B. Acc Chem Res. 2008;41:315–326. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]
- 18.Jun HW, Yuwono V, Paramonov SE, Hartgerink JD. Adv Mater. 2005;17:2612–2617. [Google Scholar]
- 19.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. Proc Natl Acad Sci USA. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Umezawa N, Gelman MA, Haigis MC, Raines RT, Gellman SH. J Am Chem Soc. 2002;124:368–369. doi: 10.1021/ja017283v. [DOI] [PubMed] [Google Scholar]; Wender PA, Rothbard JB, Jessop TC, Kreider EL, Wylie BL. J Am Chem Soc. 2002;124:13382–13383. doi: 10.1021/ja0275109. [DOI] [PubMed] [Google Scholar]; Chen F, Zhu NY, Yang D. J Am Chem Soc. 2004;126:15980–15981. doi: 10.1021/ja044493+. [DOI] [PubMed] [Google Scholar]; Farrera-Sinfreu J, Giralt E, Castel S, Albericio F, Royo M. J Am Chem Soc. 2005;127:9459–9468. doi: 10.1021/ja051648k. [DOI] [PubMed] [Google Scholar]; Li X, Yang D. Chem Commun. 2006:3367–3379. doi: 10.1039/b602230h. [DOI] [PubMed] [Google Scholar]; Yang Z, Liang G, Ma M, Gao Y, Xu B. Small. 2007;3:558–562. doi: 10.1002/smll.200700015. [DOI] [PubMed] [Google Scholar]; Yang ZM, Liang GL, Xu B. Chem Commun. 2007:738–740. doi: 10.1039/b516133a. [DOI] [PubMed] [Google Scholar]; Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, Zuckermann RN, Barron AE. Proc Natl Acad Sci USA. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]; Czyzewski AM, Barron AE. Aiche J. 2008;54:2–8. [Google Scholar]; Ma B, Lin G, Yang D. Drug Metab Rev. 2010;42:294–294. [Google Scholar]; Zhang YH, Song K, Zhu NY, Yang D. Chem Eur J. 2010;16:577–587. doi: 10.1002/chem.200901471. [DOI] [PubMed] [Google Scholar]
- 20.Liang GL, Yang ZM, Zhang RJ, Li LH, Fan YJ, Kuang Y, Gao Y, Wang T, Lu WW, Xu B. Langmuir. 2009;25:8419–8422. doi: 10.1021/la804271d. [DOI] [PubMed] [Google Scholar]
- 21.Sola RJ, Griebenow K. J Pharm Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]; Culyba EK, Price JL, Hanson SR, Dhar A, Wong CH, Gruebele M, Powers ET, Kelly JW. Science. 2011;331:571–575. doi: 10.1126/science.1198461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bromme D, Peters K, Fink S, Fittkau S. Arch Biochem Biophys. 1986;244:439–446. doi: 10.1016/0003-9861(86)90611-9. [DOI] [PubMed] [Google Scholar]
- 23.Ruoslahti E. Matrix Biol. 2003;22:459–465. doi: 10.1016/s0945-053x(03)00083-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.