Glycosylphosphatidylinositol (GPI) membrane-anchored proteins have attracted considerable attention as markers of detergent-insoluble cholesterol/sphingolipid-rich “lipid rafts” (1). GPI-anchored proteins are ubiquitous among the eukaryotes and are particularly abundant, in terms of surface density, in a number of protozoan organisms. The term GPI was first introduced in 1985 for the membrane anchor of the variant surface glycoprotein (VSG) of Trypanosoma brucei and the first complete GPI structure (2) and descriptions of GPI biosynthesis (3, 4) were for T. brucei VSG.
The African trypanosomes are protozoan parasites that divide in the blood of the mammalian host and cause human sleeping sickness and Nagana in cattle. These diseases are fatal if untreated and the current drugs are highly toxic and difficult to administer. With an alarming upward trend and currently at least 350,000 new cases of sleeping sickness each year, uninterrupted vector control and new therapeutic agents are urgently required to stem this disease. The trypanosomes undergo a complex life cycle between the tsetse fly vector and the mammalian host. The best-studied life cycle stages are the bloodstream forms and the procyclic forms, which divide in the mid-gut of the tsetse fly. The plasma membranes of both forms are covered with GPI-anchored proteins (Fig. 1). In a previous issue of PNAS, Nagamune et al. (5) provide clear-cut and compelling evidence that GPI biosynthesis is essential to bloodstream form T. brucei parasites. These results validate the long-held belief that the GPI pathway is a viable target for the development of new drugs against sleeping sickness and Nagana. Surprisingly, the same report shows that GPI biosynthesis in the insect-dwelling procyclic form of the parasite is nonessential, even though it is rich in GPI-anchored procyclin glycoproteins.
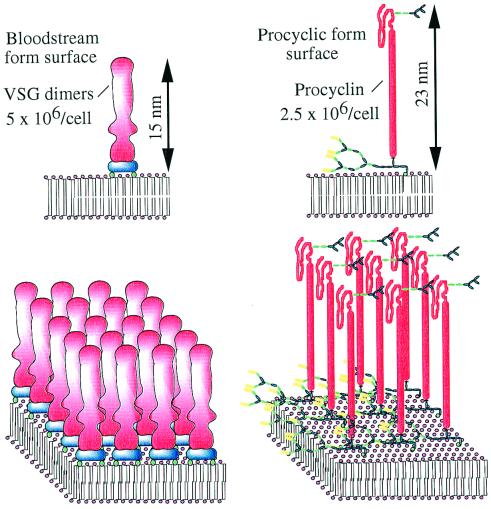
Figure 1.
The major surface molecules of T. brucei bloodstream and procyclic forms. The cartoons represent 20 nm × 20 nm portions of plasma membrane. The blue components of the VSGs represent the two GPI anchors that attach the VSG dimers to the membrane. The mature GPI anchors of the procyclins have very complex side chains and some procyclins contain small N-linked oligosaccharides beyond the polyanionic rod domain, as shown here.
The bloodstream forms of T. brucei are covered with a surface coat of 107 VSG molecules (6, 7). The coat acts as a diffusion barrier, allowing access of small nutrient molecules but preventing the approach of macromolecules, like components of the innate immune system, to the plasma membrane. The parasite contains several hundred VSG genes and their sequential expression enables the parasite population to evade specific immune attack through antigenic variation (8). The VSGs exist as homodimers and, despite very low sequence identity, adopt almost identical tertiary structures, which explains how antigenically distinct VSGs assemble into functionally identical coat arrays (9). Antigenic variation makes VSG vaccine development a nonstarter and other invariant surface molecules appear to be unavailable for immune surveillance. On the other hand, generic processes involved in the production of a stable VSG coat are considered potential therapeutic targets. This includes the biosynthesis of GPI anchors and their addition to VSG (Fig. 2) and to the novel GPI-anchored trypanosome transferrin receptor (10).
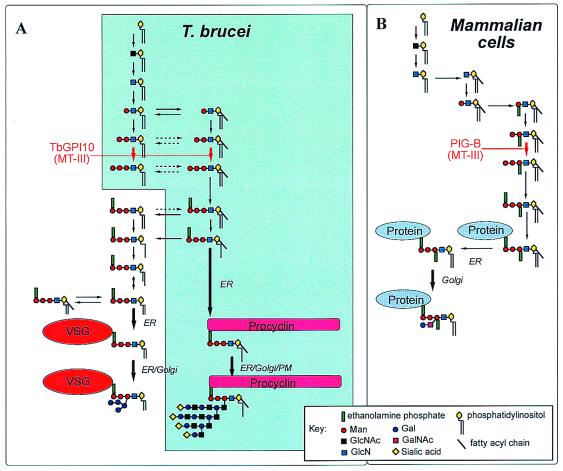
Figure 2.
The GPI biosynthetic pathways of T. brucei and mammalian cells. This is a consensus view based on the work of many groups, reviewed in refs. 11 and 23. (A) The shaded area represents the pathway in T. brucei procyclic cells and the nonshaded area represents the additional fatty acid remodeling steps and attachment to VSG unique to T. brucei bloodstream forms. (A and B) The location of the MT-III enzyme, encoded by the TbGPI10 and PIG-B genes in the parasite (A) and mammalian (B) cells, respectively, is indicated in red.
Genes associated with GPI biosynthesis have been cloned by complementation of GPI-deficient mammalian cell lines and temperature-sensitive yeast GPI mutants. Kinoshita and colleagues (11–13) have played a major role in this area and have cloned and characterized the lion's share of the 20 genes known to be associated with GPI biosynthesis in mammalian cells. Furthermore, they have made seminal contributions to our understanding of the organization of the endoplasmic reticulum-resident components of the GPI pathway, the role of GPI mutations in the disease paroxysmal nocturnal hemoglobinurea, and the role of GPIs in mammalian tissue development (11–15). The cloning of the PIG-B gene, which encodes the third mannosyltransferase (MT-III) of mammalian GPI biosynthesis, led to the identification of the yeast orthologue GPI10 (16) and, through a fragment in the T. brucei genome project database, the parasite gene TbGPI10. In the present study, TbGPI10 was shown to restore GPI-protein expression in a PIG-B deficient T lymphoma cell line and to rescue GPI10-disrupted Saccharomyces cerevisiae, demonstrating that it is the orthologue of PIG-B and GPI10. Attempts to knock out both alleles of TbGPI10 in the bloodstream form of T. brucei by homologous recombination resulted in gene amplification, suggesting that the gene is essential. This finding was confirmed by introduction of an episomal copy, which allowed both chromosomal copies to be deleted and was retained without drug selection in the TbGPI10−/− double knockout clones but lost from single-knockout TbGPI10+/− clones.
Why is TbGPI10 essential for bloodstream form T. brucei growth in vitro? Disruption of GPI biosynthesis by deletion of MT-III will prevent the assembly of the VSG coat. Although this would be expected to render the parasites noninfectious to animals, because of the host's innate and specific immune systems, it is not obvious why this might be lethal in vitro. However, proteolytic removal of the N-terminal domain (70% of the VSG polypeptide) from the cell surface causes the cells to lose their characteristic shape (17), suggesting that the coat may be essential for maintaining cell morphology and viability. Alternatively, the lethal phenotype may be caused by the loss of the GPI-anchored trypanosome transferrin receptor, expressed only in the bloodstream stage of the parasite where transferrin uptake from host serum appears to be essential (18). One or both of these effects probably explains the essential nature of the TbGPI10 gene in bloodstream form T. brucei. The formal possibility that there is a toxic build up of early GPI intermediates in the absence of MT-III seems less likely because the parasites are capable of catabolizing excess GPIs (19).
Regardless of the reason(s) for the lethality of MT-III disruption, the results provide encouragement for the development of drug leads based on inhibitors of this and other enzymes of the GPI pathway, with the proviso that parasite-specific inhibitors can be developed. Although the T. brucei and mammalian GPI biosynthetic pathways have much in common, giving rise to the same GPI core structure of EtN-P-6Manα1–2Manα1–6Manα1–4GlcNα1–6myo-inositol-1-P-lipid, there are significant differences (Fig. 2). Thus, mammalian (and yeast) MT-IIIs are more stringent with respect to their acceptor substrate than that of T. brucei; the former require the presence of a side-chain ethanolamine phosphate on the first mannose residue of the acceptor (16) whereas the parasite enzyme does not. This bodes well for the development of a parasite-specific MT-III inhibitor. A natural-product yeast/mammalian-specific inhibitor that prevents addition of the side-chain EtN-P group, and consequently prevents the action of MT-III, already has been described (20). Other exploitable differences include the T. brucei-specific fatty acid remodeling reactions (21) and earlier steps in the pathway. For example, in vitro parasite-specific inhibitors of GlcN-PI mannosylation and inositol acylation already have been synthesized (22) and differences in the host and parasite GlcNAc-PI de-N-acetylases have been described.
Taken together, the gene knockout study described in a previous issue of PNAS (5) and the existence of first-generation parasite-specific GPI pathway inhibitors provides a proof-of-concept that therapeutic intervention via inhibition of the GPI pathway is feasible. The GPI pathways of other protozoan pathogens are also likely to be good targets for the development of novel therapeutics (23). For example, the surface membranes of Plasmodium (malaria), Toxoplasma (toxoplamosis), and Trypanosoma cruzi (Chagas' disease) are rich in GPI-anchored proteins, and the Leishmania parasites are rich in essential GPI-related free glycolipids (24). In addition, GPI anchors are essential for cell-wall biogenesis in yeast and probably also in fungal pathogens like Candida and Aspergillus. Sadly, a major stumbling block in the development of antiparasite therapeutics is the general lack of interest in tropical diseases shown by the pharmaceutical industry. Hopefully the new political will to “roll back” major infectious diseases such as tuberculosis and malaria (which together kill over 6 million people a year) may provide some impetus to screen compound libraries against validated pathogen targets, including the parasite GPI pathways. The antiprotozoal pharmacopoeia is extremely limited and drug-resistance is a major problem. The need to follow up on promising targets with high-throughput screening is acute.
Finally, Nagamune et al. (5) show that they can delete both alleles of TbGPI10 in the procyclic form of T. brucei and that these GPI-deficient cells successfully colonized tsetse fly mid-guts, albeit at lower efficiency than wild type. This is an extraordinary result considering our current view of the procyclic cell surface (Fig. 1). The double knockout procyclic forms are completely devoid of cell surface procyclins and can only be cultured in nonadherent culture ware. This latter point may explain why Roditi and colleagues (25) obtained slightly different results when deleting individual genes of the procyclin family. In that case, all but one gene could be deleted before producing nonviable cells. How can procyclics survive without any of their GPI-anchored procyclin repertoire? This would be easier to answer if we had a clear idea of their function in vivo. Thus far, it has been argued that (i) their polyanionic nature, extended conformation, and large carbohydrate side chains provide the parasites with protection in the hydrolytic environment of the fly mid-gut, and (ii) a family of genes encoding structurally distinct procyclins is needed to satisfy specific events in the maturation processes in the fly (26). With respect to i, some protective role can still be proposed to explain the reduced efficiency of fly infection by the double knockouts. This finding may be crucial in the wild where the rates of tsetse fly infection are extremely low. With respect to ii, the results provide no clues for specific roles for procyclins in the tsetse fly. On the other hand, the tsetse fly studies were limited, for technical reasons, to the analysis of mid-gut infections and did not include analysis of the transformations needed to complete the life cycle and prepare the parasite for reentry into a mammalian host. Thus, there is still much to learn about the function of procyclins, and the possibility that other molecules may replace procyclin in their absence requires investigation. These are challenging issues in trypanosome cell biology and the work published recently in PNAS, and similar gene knockout studies in Leishmania (27–29), are stimulating and demand reassessment of several aspects of parasite–vector and host–parasite interactions.
Footnotes
See companion article on page 10336 in issue 19 of volume 97.
References
- 1.Simons K, Ikonen E. Nature (London) 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson M A J, Homans S W, Dwek R A, Rademacher T W. Science. 1988;239:753–759. doi: 10.1126/science.3340856. [DOI] [PubMed] [Google Scholar]
- 3.Masterson W J, Doering T L, Hart G W, Englund P T. Cell. 1989;56:793–800. doi: 10.1016/0092-8674(89)90684-3. [DOI] [PubMed] [Google Scholar]
- 4.Menon A K, Schwarz R T, Mayor S, Cross G A M. J Biol Chem. 1990;265:9033–9042. [PubMed] [Google Scholar]
- 5.Nagamune K, Nozaki T, Maeda Y, Ohishi K, Fukuma T, Hara T, Schwarz R T, Sütterlin C, Brun R, Riezman H, Kinoshita T. Proc Natl Acad Sci USA. 2000;97:10336–10341. doi: 10.1073/pnas.180230697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vickerman K, Luckins A G. Nature (London) 1969;224:1125–1127. doi: 10.1038/2241125a0. [DOI] [PubMed] [Google Scholar]
- 7.Cross G A M. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- 8.Cross G A M. BioEssays. 1996;18:283–291. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- 9.Blum M L, Down J A, Gurnett A M, Carrington M, Turner M J, Wiley D C. Nature (London) 1993;362:603–609. doi: 10.1038/362603a0. [DOI] [PubMed] [Google Scholar]
- 10.Steverding D, Stierhof Y D, Fuchs H, Tauber R, Overath P. J Cell Biol. 1995;131:1173–1182. doi: 10.1083/jcb.131.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinoshita T, Ohishi K, Takeda J. J Biochem. 1997;122:251–257. doi: 10.1093/oxfordjournals.jbchem.a021746. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe R, Murakami Y, Marmor M D, Inoue N, Maeda Y, Hino J, Kangawa K, Julius M, Kinoshita T. EMBO J. 2000;19:4402–4411. doi: 10.1093/emboj/19.16.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohishi K, Inoue N, Maeda Y, Takeda J, Riezman H, Kinoshita T. Mol Biol Cell. 2000;11:1523–1533. doi: 10.1091/mbc.11.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nozaki M, Ohishi K, Yamada N, Kinoshita T, Nagy A, Takeda J. Lab Invest. 1999;79:293–299. [PubMed] [Google Scholar]
- 15.Takahashi M, Inoue N, Ohishi K, Maeda Y, Nakamura N, Endo Y, Fujita T, Takeda J, Kinoshita T. EMBO J. 1996;15:4254–4261. [PMC free article] [PubMed] [Google Scholar]
- 16.Sütterlin C, Escribano M, Gerold P, Maeda Y, Mazon M, Kinoshita T, Schwarz R, Riezman H. Biochem J. 1998;332:153–159. doi: 10.1042/bj3320153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cross G A M, Johnson J G. In: Biochemistry of Parasites and Host–Parasite Relationships. Van den Bossche H, editor. Amsterdam: North–Holland; 1976. pp. 413–420. [Google Scholar]
- 18.Steverding D. Parasitol Int. 2000;48:191–198. doi: 10.1016/s1383-5769(99)00018-5. [DOI] [PubMed] [Google Scholar]
- 19.Güther M L S, Masterson W J, Ferguson M A J. J Biol Chem. 1994;269:18694–18701. [PubMed] [Google Scholar]
- 20.Sütterlin C, Horvath A, Gerold P, Schwarz R, Wang Y, Dreyfuss M, Riezman H. EMBO J. 1997;16:6374–6383. doi: 10.1093/emboj/16.21.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morita Y S, Kimberly S P, Englund P T. Science. 2000;288:140–143. doi: 10.1126/science.288.5463.140. [DOI] [PubMed] [Google Scholar]
- 22.Smith T, Sharma D K, Crossman A, Brimacombe J, Ferguson M A J. EMBO J. 1999;18:5922–5930. doi: 10.1093/emboj/18.21.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferguson M A J. J Cell Sci. 1999;112:2799–2809. doi: 10.1242/jcs.112.17.2799. [DOI] [PubMed] [Google Scholar]
- 24.Ilgoutz S C, Zawadzki J L, Ralton J E, McConville M J. EMBO J. 1999;8:2746–2755. doi: 10.1093/emboj/18.10.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruepp S, Furger A, Kurath U, Kunz-Renggli C, Hemphill A, Brun R, Roditi I. J Cell Biol. 1997;137:1369–1379. doi: 10.1083/jcb.137.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vassella E, Abbeele J V D, Bütikoffer P, Renggli C K, Furger A, Brun R, Roditi I. Genes Dev. 2000;14:615–626. [PMC free article] [PubMed] [Google Scholar]
- 27.Hilley J D, Zawadzki J L, McConville M J, Coombs G H, Mottram J C. Mol Biol Cell. 2000;11:1183–1195. doi: 10.1091/mbc.11.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ilg T. EMBO J. 2000;19:1953–1962. doi: 10.1093/emboj/19.9.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Späth G F, Epstein L, Leader B, Singer S M, Avila H A, Turco S J, Beverley S M. Proc Natl Acad Sci USA. 2000;97:9258–9263. doi: 10.1073/pnas.160257897. [DOI] [PMC free article] [PubMed] [Google Scholar]