Box C/D small (nucleolar) ribonucleoproteins [s(no)RNPs] catalyze RNA-guided 2′-O-ribose methylation in two of the three domains of life. Recent structural studies have led to a controversy over whether box C/D sRNPs functionally assemble as monomeric or dimeric macromolecules. To examine box C/D sRNP architecture more closely, the authors investigate the role of the omnipresent sRNA loop as a structural determinant of sRNP assembly. They show through sRNA mutagenesis, native gel electrophoresis, and single-particle EM that a di-sRNP is the near exclusive architecture obtained when reconstituting box C/D sRNPs with natural or artificial sRNAs containing an internal loop.
Keywords: Archaea, box C/D sRNP, single-particle EM, rRNA modification, RNA–protein complex, noncoding RNA
Abstract
Box C/D small (nucleolar) ribonucleoproteins [s(no)RNPs] catalyze RNA-guided 2′-O-ribose methylation in two of the three domains of life. Recent structural studies have led to a controversy over whether box C/D sRNPs functionally assemble as monomeric or dimeric macromolecules. The archaeal box C/D sRNP from Methanococcus jannaschii (Mj) has been shown by glycerol gradient sedimentation, gel filtration chromatography, native gel analysis, and single-particle electron microscopy (EM) to adopt a di-sRNP architecture, containing four copies of each box C/D core protein and two copies of the Mj sR8 sRNA. Subsequently, investigators used a two-stranded artificial guide sRNA, CD45, to assemble a box C/D sRNP from Sulfolobus solfataricus with a short RNA methylation substrate, yielding a crystal structure of a mono-sRNP. To more closely examine box C/D sRNP architecture, we investigate the role of the omnipresent sRNA loop as a structural determinant of sRNP assembly. We show through sRNA mutagenesis, native gel electrophoresis, and single-particle EM that a di-sRNP is the near exclusive architecture obtained when reconstituting box C/D sRNPs with natural or artificial sRNAs containing an internal loop. Our results span three distantly related archaeal species—Sulfolobus solfataricus, Pyrococcus abyssi, and Archaeoglobus fulgidus—indicating that the di-sRNP architecture is broadly conserved across the entire archaeal domain.
INTRODUCTION
Ribosome biogenesis occurs in all three domains of life with increasing degrees of complexity from Bacteria to Archaea to Eukarya (Henras et al. 2008; Blombach et al. 2011; Shajani et al. 2011). Fully functional ribosomes, which are essential for protein synthesis, consist of a highly organized tertiary structure of ribosomal RNA (rRNA) that is folded and assembled with ribosomal proteins. Two-thirds of the mass of the ribosome is made up of rRNA, which must be cleaved and post-transcriptionally modified before assembly with ribosomal proteins. The extent of post-transcriptional rRNA modification increases with higher organismal complexity (Decatur and Fournier 2002). rRNA modifications cluster in functionally important and phylogenetically conserved regions of the ribosome and are necessary for efficient and accurate translation (Decatur and Fournier 2002; Liang et al. 2007).
2′-O-Methylation of the ribose moiety comprises one subset of post-transcriptional modifications to the rRNA. In Eukarya and Archaea, this reaction is catalyzed by highly conserved ribonucleoprotein (RNP) enzymes in which the RNA component directs site-specific modification (Balakin et al. 1996; Kiss-Laszlo et al. 1996). In eukaryotes these enzymes are called snoRNPs (small nucleolar RNPs) and in Archaea sRNPs, because Archaea have no nucleoli. The particular class of s(no)RNPs that catalyzes 2′-O-ribose methylation, the box C/D s(no)RNPs, is named for conserved s(no)RNA sequence motifs, boxes C and D (Maxwell and Fournier 1995).
The box C/D s(no)RNA contains canonically conserved box sequences as well as unique guide sequences that base-pair with specific substrate RNAs. The conserved boxes C (RUGAUGA, where R is a purine) and D (CUGA) together form a kink-turn (k-turn) structure near the terminal stem of the s(no)RNA (sR8) (Fig. 1A; Vidovic et al. 2000; Watkins et al. 2000; Klein et al. 2001; Moore et al. 2004). In Archaea, related boxes C′ and D′ form a second k-turn, also known as a k-loop (Nolivos et al. 2005), giving the s(no)RNA a bipartite architecture (Hardin and Batey 2006). Two guide sequences provide substrate specificity through base-pairing interactions with target RNAs (Kiss-Laszlo et al. 1996; Kiss-Laszlo et al. 1998; Gaspin et al. 2000; Omer et al. 2000) and act as a “molecular ruler” directing 2′-O-methylation 5 nt upstream of the D and/or D′ box (Kiss-Laszlo et al. 1996, 1998; Tran et al. 2005). A short internal loop links boxes D′ and C′. Although the length of the loop can be highly variable in eukaryotic snoRNAs, the loop is present in both eukaryotic and archaeal domains yielding box C/D s(no)RNAs that exist as a single, unbroken ribonucleotide strand (Gaspin et al. 2000).
FIGURE 1.
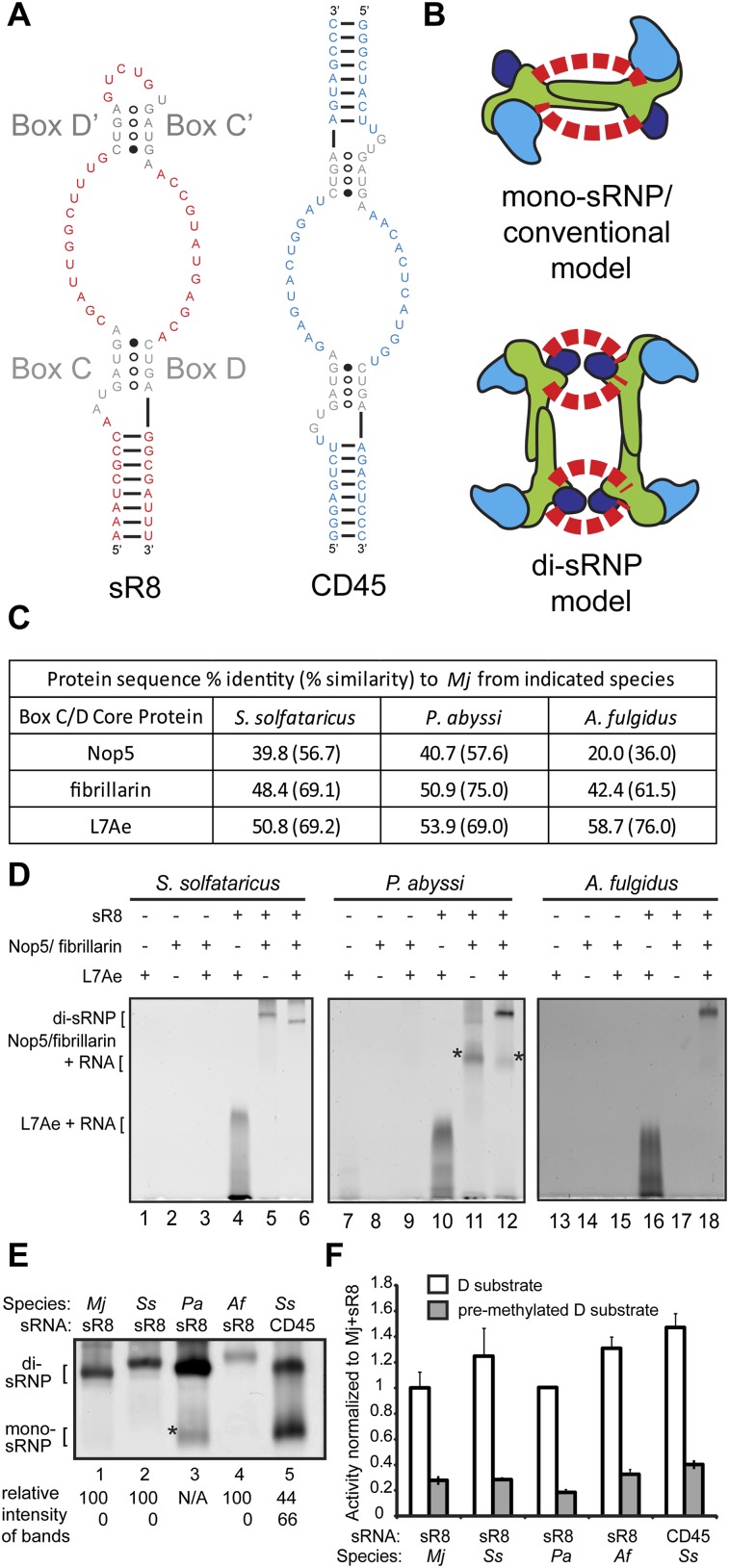
Box C/D sRNPs from four archaeal species migrate as di-sRNPs on native gels. (A) Structural characteristics of a naturally occurring box C/D sRNA, sR8, and a two-stranded box C/D sRNA mimic, CD45. (Gray) Conserved C, C′, and D, D′ boxes. Naturally occurring sRNAs such as sR8 exist as a single, continuous nucleotide strand. The CD45 RNA resembles a box C/D sRNA, but exists as two separate oligonucleotide strands instead of one continuous RNA molecule and therefore lacks an internal loop. (B) Two conflicting models for box C/D sRNP assembly. The core proteins are as follows: fibrillarin (cyan), Nop5 (green), L7Ae (dark blue). The box C/D sRNA is indicated by red dashed lines representing the two guide sequences of each sRNA; sRNA orientation is left intentionally ambiguous. (C) Sequence percent identity (and percent similarity) of box C/D core proteins from three archaeal species as compared to the homologous proteins from the Mj species. (D) Native gel electrophoresis of in vitro–assembled RNP complexes from Ss, Pa, and Af. In the absence of L7Ae, both Ss and Pa proteins Nop5 and fibrillarin bind to sR8, creating several RNP species, one of which (*, lane 11) is present in the assembly of the Pa sRNP (*, lane 12) and has the same mobility as the mono-sRNP from Ss assembled with CD45 sRNA (E, lane 5). (E) All tested archaeal box C/D sRNPs assembled with a naturally occurring box C/D sRNA migrate as di-sRNPs as assayed by native gel electrophoresis. (*) Pa Nop5-fibrillarin heterotetramer associating with the sRNA in the absence of L7Ae (see D). Quantitations of the relative intensity of di- and mono-sRNP sized bands are indicated. (F) Methylation activity assays of assembled box C/D sRNPs show that all tested archaeal sRNPs are catalytically active for site-specific methylation.
The core protein components of box C/D s(no)RNPs are also highly conserved from Eukaryotes to Archaea. In Archaea, box C/D sRNPs are composed of three core proteins: L7Ae, Nop5 (also called Nop56/58), and fibrillarin (Omer et al. 2000; Reichow et al. 2007). L7Ae is the primary sRNA binding protein and recognizes the k-turn and k-loop motifs of the sRNA (Kuhn et al. 2002; Moore et al. 2004). Nop5 forms a homodimer through its conserved coiled-coil domains and structurally links the sRNA:L7Ae complex to the methyltransferase fibrillarin (Aittaleb et al. 2003). Nop5 and fibrillarin can form a stable heterotetramer complex independent of L7Ae and sRNA (Aittaleb et al. 2003; Zhang et al. 2006; Oruganti et al. 2007). Conventional models of the box C/D sRNP consist of one sRNA bound by two copies of each core protein (a mono-sRNP), with the Nop5 coiled-coil domain running parallel to the sRNA guide sequences (Fig. 1B).
An electron microscopy (EM) structure of a box C/D sRNP from the archaeon Methanococcus jannaschii (Mj) was the first structure determined by in vitro reconstitution with a complete set of core constituents, including a naturally occurring full-length sRNA, sR8 (Fig. 1B; Bleichert et al. 2009). In contrast to the conventional model, this three-dimensional (3D) reconstruction showed that the archaeal box C/D sRNP is a di-sRNP, with four copies of each core protein and therefore two copies of the sRNA (Fig. 1B). Extensive biochemical analyses of the sRNP particle by multiple laboratories (sedimentation on glycerol gradients, native gel electrophoresis, and size-exclusion chromatography) also supported formation of a box C/D sRNP dimer (Bleichert et al. 2009; Bleichert and Baserga 2010a; Ghalei et al. 2010). By fitting atomic models of protein components into the EM density map, the di-sRNP model predicted that the Nop5 coiled-coil domains run perpendicular to the sRNAs. Corroborating this conclusion, RNase digestion dramatically reduced sRNP dimensions along one side (Bleichert et al. 2009). The presence of the mono-sRNP was not detected, and no catalytic activity was associated with fractions where a mono-sRNP would sediment in glycerol gradients. A crystal structure of a box C/D sRNP from Pyrococcus furiosus with a half-mer sRNA and short substrate was also found to be consistent with the di-sRNP model (Xue et al. 2010).
The di-sRNP model was challenged by a subsequent crystal structure of a monomeric box C/D sRNP from the archaeon Sulfolobus solfataricus (Ss) in complex with a short RNA substrate, creating a controversy over which model best represents the native sRNP architecture (Lin et al. 2011). The mono-sRNP was assembled with an artificial, two-stranded, completely symmetrical RNA, CD45, resembling a bipartite box C/D sRNA (Fig. 1A; Lin et al. 2011). This RNA was unlike naturally occurring sRNAs because it consisted of two separate oligoribonucleotides instead of a continuous, asymmetric sRNA.
To investigate the underlying causes for these two conflicting models of box C/D sRNP architecture, we have tested box C/D sRNPs from distantly related archaeal species for assembly with a natural sRNA from Mj, sR8. We have chosen to compare the Mj di-sRNP (Bleichert et al. 2009) to sRNPs assembled from S. solfataricus (Ss), P. abyssi (Pa), and A. fulgidus (Af). These species represent archaeal kingdoms from Crenarchaeota (Ss) to Euryarchaeota (Mj, Pa, Af); the evolutionary distance between the two kingdoms is approximately twice that separating Saccharomyces cerevisiae and humans (Cox et al. 2008). Using native gel electrophoresis analysis, previously shown to report sRNP assembly accurately (Bleichert and Baserga 2010a), we have determined that all archaeal sRNPs tested assemble into di-sRNPs. Mutagenesis experiments link the conserved presence of the sRNA internal loop, which is observed in all computationally predicted and experimentally verified archaeal sRNAs, to di-sRNP assembly. Negative stain EM single-particle reconstructions of catalytically active box C/D sRNPs from Ss, Pa, and Af reveal that they are all di-sRNPs. Our results demonstrate that the dimeric 3D architecture of box C/D sRNPs is broadly conserved across the archaeal domain.
RESULTS
One of the major differences between the reconstitutions that yielded the divergent di- and mono-sRNP models (Fig. 1B) was in the architecture of the sRNA component used to assemble the sRNPs (Fig. 1A). In the EM reconstruction that yielded the di-sRNP model, the sRNP was assembled with a full-length, naturally occurring box C/D sRNA, sR8, from the species M. jannaschii (Mj) and using the core protein components from Mj (Bleichert et al. 2009). In the X-ray crystallographic structure that yielded the mono-sRNP model, the sRNP was assembled with CD45, a two-stranded, artificially symmetric pair of oligonucleotides resembling a box C/D sRNA that lacks the loop between boxes C′ and D′ (Lin et al. 2011), and with the core protein components from S. solfataricus (Ss). Because the three core proteins—Nop5, fibrillarin, and L7Ae from four archaeal species (Mj, Ss, Pa, and Af)—commonly used in box C/D sRNP structural and functional studies are highly conserved (Fig. 1C), we hypothesized that architectural differences in the sRNAs used for sRNP assembly led to conflicting sRNP models derived from previous structural studies.
sRNPs from diverse archaeal species exhibit di-sRNP mobility on native gels
We first tested whether the purified protein components could be reconstituted with the sR8 sRNA, assaying sRNP assembly using native gel electrophoresis followed by silver staining as previously described (Fig. 1D; Bleichert et al. 2009). When the three box C/D core proteins from each species tested—Ss, Pa, and Af—were assembled with the sR8 RNA, a predominant RNP complex was formed (Fig. 1D, lanes 6,12,18). The Nop5/fibrillarin heterotetramer and L7Ae did not associate with each other in the absence of the sRNA, but L7Ae bound to sR8 in the absence of the Nop5/fibrillarin heterotetramer (Fig. 1D, lanes 4,10,16). These results are consistent with previous studies of box C/D core proteins (Omer et al. 2000; Kuhn et al. 2002; Aittaleb et al. 2003; Reichow et al. 2007). However, for the Pa species, Nop5/fibrillarin did associate with the sRNA in the absence of L7Ae (Fig. 1D, lane 11, *). This band runs at the same mobility as a faint band in the Pa sRNP complete assembly (Fig. 1D, lane 12, *). Because of this anomaly in assembly, we interpret this band in the complete assembly of the Pa sRNP as an Nop5/fibrillarin-sR8 complex, and not a mono-sRNP. Therefore, a single discrete architecture exists for box C/D sRNPs assembled with a native sRNA, sR8.
Because our initial reconstitution results indicated that discrete sRNPs could be assembled, we compared the relative mobilities of sRNPs from Ss, Pa, and Af via native gel electrophoresis. The Mj box C/D core proteins assembled with sR8 were used as a marker for di-sRNP assembly, because they had been shown previously to assemble solely into a di-sRNP with sR8 (Bleichert and Baserga 2010a). As a control, we also examined the Ss species assembled with the CD45 sRNA, which had been previously shown to form two differently migrating species on native gels, the faster-migrating band representing a mono-sRNP (Lin et al. 2011). We found that core proteins from each species assembled with sR8 migrated similarly to the Mj di-sRNP in native gels (Fig. 1E, lanes 1–4), with no band migrating at the size of the mono-sRNP. Although the asterisked band in the Pa assembly might be mistaken for a mono-sRNP (Fig. 1E, lane 3, *), control experiments show that this band instead represents the Pa Nop5-fibrillarin heterotetramer interacting with sR8 in the absence of L7Ae (Fig. 1D, lane 11, *). Therefore, sRNPs from all archaeal species tested predominantly formed di-sRNPs when assembled with a naturally occurring sRNA, sR8. Conversely, when the Ss box C/D core proteins were assembled with the CD45 sRNA, two distinct bands were observed (Fig. 1E, lane 5), identical to the two species previously described in native gels, where the faster-migrating band corresponds to the mono-sRNP (Lin et al. 2011). The architecture of the slower-migrating band in the CD45 assembly was not previously elucidated (Lin et al. 2011), although it runs with the mobility of a di-sRNP (Fig. 1E), suggesting that complexes containing the CD45 sRNA can assemble into both mono- and di-sRNPs. Importantly, we found that the mono-sRNP species crystallized in the Lin et al. (2011) study was not solely due to the presence of substrate RNA, because no substrate was included in this reconstitution reaction, yet the CD45 sRNA assembly yields a band that migrates as a mono-sRNP (Fig. 1E, lane 5). Quantitation and calculation of the ratio of the intensities of the di- and mono-sRNP bands yielded exclusive di-sRNP formation in the presence of the natural sR8 RNA.
The reconstituted box C/D sRNPs from each species were enzymatically active for site-specific methylation in vitro when assembled with the naturally occurring Mj sR8 RNA or the CD45 sRNA (Fig. 1F). A substrate RNA oligonucleotide premethylated at the target nucleotide was used as a negative control. Box C/D sRNPs from all four archaeal species have been shown previously to have site-specific in vitro methylation activity (Omer et al. 2002; Bortolin et al. 2003; Tran et al. 2003; Aittaleb et al. 2004).
The sRNA loop directs sRNP architecture
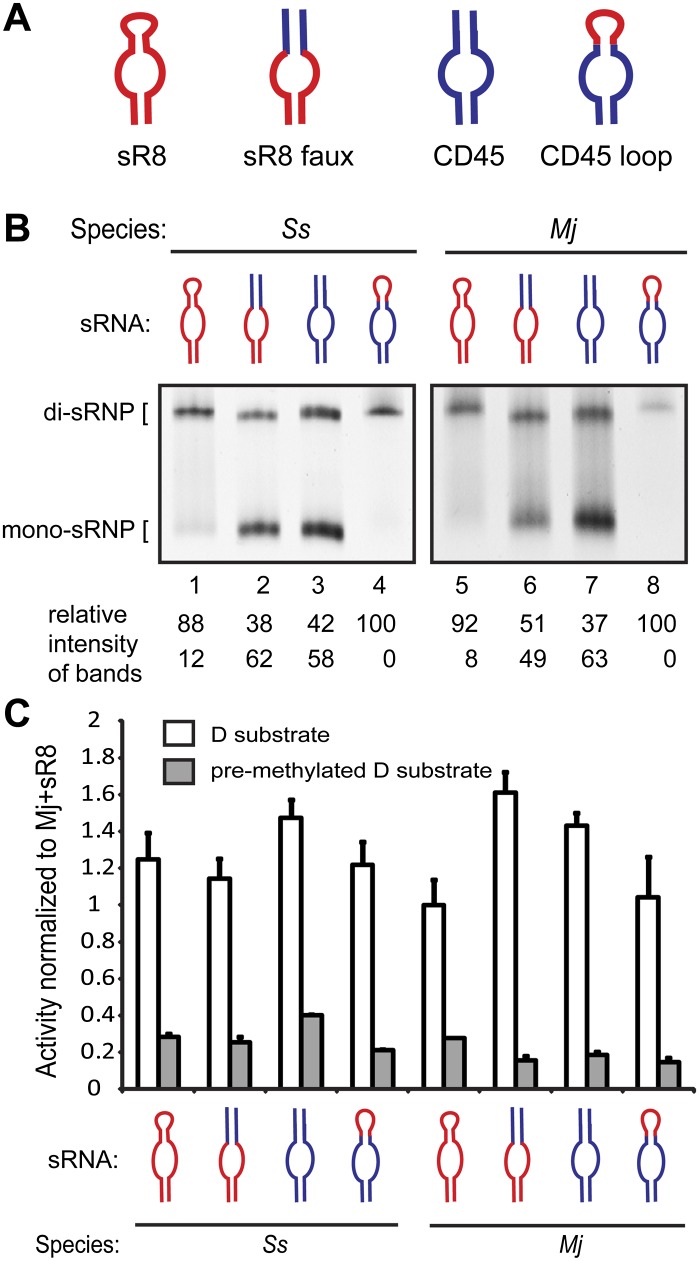
We tested whether the structural difference between the sR8 and CD45 sRNAs directly affected box C/D sRNP architecture. Using Mj sR8 as a control for di-sRNP formation and CD45 as a control for the di- and mono- sRNP mixture, we designed sRNA mutants to examine the importance of the loop between boxes C′ and D′ (Fig. 2A). The “sR8 faux” sRNA mutant is identical to the sR8 sRNA, except the internal loop between boxes C′ and D′ was replaced with a second terminal stem identical to the second terminal stem of CD45. The “CD45 loop” mutant is identical in sequence to the CD45 RNA, but instead of a second terminal stem, it contains the 5-nt loop sequence found in sR8 and links the sRNA into a single RNA strand.
FIGURE 2.
The box C/D sRNA internal loop affects sRNP architecture. (A) The naturally occurring Mj sR8 was mutagenized to contain a stem instead of an internal loop (sR8 faux) but retains all other sequences from the natural sR8 sRNA. The CD45 sRNA (Lin et al. 2011) was mutagenized to contain the internal loop from the Mj sR8 sRNA (CD45 loop). Sequences originating from sR8 (red); sequences originating from CD45 (blue). (B) Native gel electrophoresis showing sRNPs assembled as indicated using core proteins from Ss and Mj with sRNAs from A. Quantitations of the relative intensity of di- and mono-sRNP sized bands are indicated. (C) Activity assays show that all assemblies from B are catalytically active for site-specific 2′-O-ribose methylation.
Assaying sRNP reconstitution via native gel electrophoresis followed by silver staining, we tested assembly of the Ss and Mj proteins with all four sRNAs (Fig. 2B). Again, assembly of core proteins from all archaeal species with sR8 yielded a predominant band migrating at the expected size of a di-sRNP (Fig. 2B, lanes 1,5). In contrast, removing the loop from sR8 resulted in two differently migrating species corresponding to the mono- and di-sRNPs assembled with the control CD45 RNA (Fig. 2B, cf. lanes 2,3 and 6,7). However, mutating the second terminal stem of CD45 to the 5-nt loop from sR8 resulted in a single migrating species corresponding in size to the di-sRNP (Fig. 2B, lanes 4,8). Similar results were obtained with the proteins from the Pa and Af archaeal species (data not shown). Although a smear present in lanes 1 and 5 of Figure 2B affected the quantitated ratios of di-sRNP to mono-sRNP, the level of mono-sRNP present in any experiment using an sRNA containing a loop was negligible compared with the clear mono- and di-sRNP mixed population seen with sR8 faux and CD45 in lanes 2,3 and 6,7 of Figure 2B. This difference in assembly efficiency of naturally occurring sRNAs has been observed previously (Bleichert and Baserga 2010a). The clear difference in the predominant archaeal sRNP architecture when assembled with sRNAs with and without an internal loop underscores the importance of the sRNA loop in directing sRNP assembly architecture. Thus, the loop found in naturally occurring sRNAs directs predominant, if not exclusive, di-sRNP formation, because there is a one-to-one correlation between the presence of an sRNA loop and concomitant assembly into a di-sRNP.
The reconstituted sRNPs were assayed for enzymatic activity without prior purification and were found to be active in site-specific methyltransferase activity using the substrate complementary to the D guide of each sRNA (Fig. 2C).
The effect of sRNA loop size and sequence on sRNP architecture
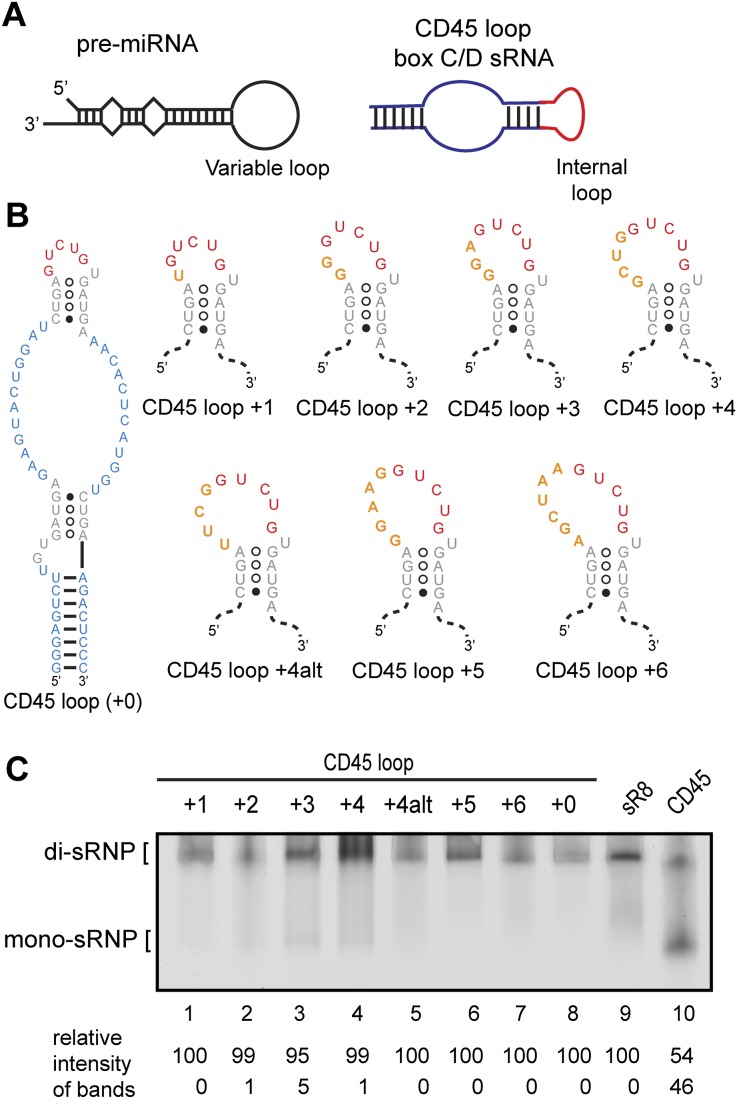
To further test the role of the sRNA loop in dictating di-sRNP assembly, we investigated the consequences of varying loop length. The secondary structure of a pre-microRNA (pre-miRNA) is reminiscent of the structure of an sRNA; the pre-miRNA exists as a hairpin containing a loop of variable length (Fig. 3A). Because of these RNA structural similarities, a recent study on Dcr-1, a Drosophila homolog of the pre-miRNA processing protein Dicer, presented a putative mode of RNA binding discrimination by box C/D proteins. Dcr-1 has been shown to recognize its substrate pre-miRNA based on its RNA secondary structure features (Tsutsumi et al. 2011). Tsutsumi et al. found that Dcr-1 recognizes the single-stranded region created by the variable-length loop of the pre-miRNA.
FIGURE 3.
sRNAs with a wide variety of internal loops confer di-sRNP assembly. (A) Hairpin stem–loops are a prevalent secondary structure in at least two classes of small noncoding RNAs: pre-microRNAs (pre-miRNAs) and box C/D s(no)RNAs. The exact number and location of base pairs vary within each class of RNAs—these diagrams are not to scale. (B) Secondary structures of CD45 loop mutants. The internal loop from the CD45 loop sRNA was mutated to include additional nucleotides of the randomized sequence. Box C/D and C′/D′ sequences (gray); sequences originating from CD45 (blue); sequences originating from sR8 (red); randomized sequences inserted in the internal loop (orange). The size of each loop in relation to the 5-nt loop of CD45 is indicated (+1, +2, etc.). (C) Assembly of Ss sRNPs with sRNAs bearing a wide variety of internal loops. Each sRNA mutant was in vitro–transcribed and assembled with Ss box C/D core proteins and assayed for di-sRNP formation by native gel electrophoresis. sRNPs assembled with sR8 and CD45 sRNAs are included as markers for di- and mono-sRNP formation. Quantitations of the relative intensity of di- and mono-sRNP sized bands are indicated.
To test whether a similar loop recognition mechanism affects box C/D sRNA assembly architecture, we mutated the internal loop of the CD45 loop sRNA. In using the CD45 loop sRNA as a template for mutagenesis, we sought to avoid any sRNP architectural bias due to the robust di-sRNP formation directed by sR8. Because archaeal sRNAs from the four species in this study contain internal loops ranging in length up to 12 nt, we designed seven CD45 loop mutants, inserting an additional 1–6 nt of randomized sequence into the internal loop of the CD45 loop sRNA (Fig. 3B). These sRNAs were transcribed and assembled in vitro with Ss proteins to assay for sRNP formation using native gel electrophoresis followed by silver staining. We found that sRNAs containing internal loops of sizes ranging from 5 nt (CD45 loop + 0) to 11 nt (CD45 loop + 6) yielded a predominant RNP species that migrated similarly to the sR8 di-sRNP control (Fig. 3C, cf. lanes 1–8 and 9), in clear contrast to the CD45 sRNA-induced mono-sRNP complex (Fig. 3C, lane 10). These results suggest that the internal loop of the box C/D sRNA plays a critical role in the obligatory formation of the di-sRNP structure.
Structural analysis of the Ss di-sRNP
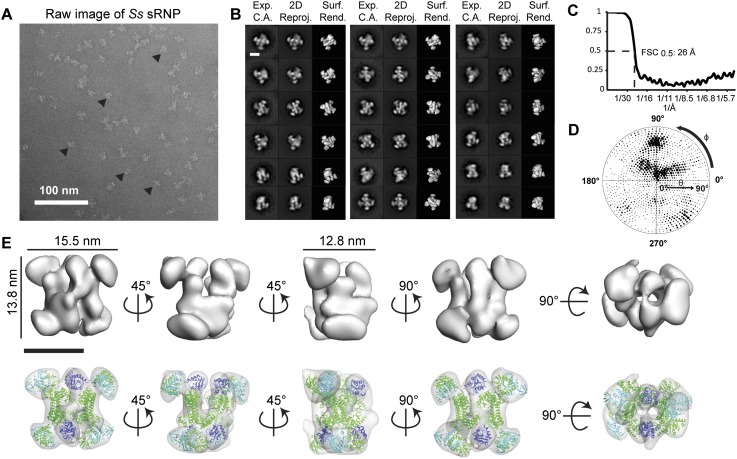
The native gel analysis in Figure 1E strongly indicated that the di-sRNP architecture is conserved to archaeal species other than Mj (Bleichert et al. 2009). To directly verify whether the box C/D di-sRNP 3D structure is conserved across Archaea, we subjected the Ss sRNP to negative stain single-particle EM analysis. To obtain a homogeneous sample suitable for single-particle analysis, the Ss core proteins were assembled with Mj sR8 sRNA and the reaction mixture was purified over a glycerol gradient. Peak fractions with fully assembled Ss sRNPs containing all three core proteins as well as the sRNA comigrated in the same peak fractions as previously described Mj di-sRNPs (Bleichert et al. 2009; Bleichert and Baserga 2010a; data not shown), confirming the initial analysis of particle size obtained by native gel electrophoresis in Figure 1E.
Glycerol gradient purified peak fractions 9–11 of the Ss sRNP were negatively stained and analyzed by single-particle EM (Fig. 4A). Particles were of similar size and shape as the Mj raw particles (Bleichert et al. 2009), consistent with our biochemical classification of the Ss box C/D sRNP as a di-sRNP. Ss sRNP particles were subjected to reference-free two-dimensional (2D) classification (Fig. 4B, experimental class averages) using the IMAGIC-5 image processing package (van Heel et al. 1996). Additionally, the particles were subjected to projection matching refinement using a low-pass filtered Mj sRNP volume as the initial model in the SPIDER image processing package (Penczek et al. 1994; Frank et al. 1996). The Ss sRNP was refined to a resolution of 26Å (0.5 Fourier Shell Correlation [FSC] criterion) (Fig. 4C), and the reconstruction represented a wide distribution of Euler angles, despite a slight preferential orientation of the Ss sRNP on the EM support grid (Fig. 4D). Surface rendered views and 2D reprojections of the 3D reconstruction at different views were generated and matched to the reference-free 2D class averages (Fig. 4B). The remarkable similarity between the reference-free 2D class averages and calculated 2D reprojections indicates that the reconstruction is in good agreement with the experimental data.
FIGURE 4.
3D single-particle EM reconstruction of the Ss box C/D di-sRNP. (A) Electron micrograph of negatively stained Ss box C/D sRNPs from glycerol gradient peak fractions. (▸) Examples of particles chosen for analysis. (B) Comparison of experimentally determined class averages (Exp. C.A.) to 2D reprojections of the reconstructed 3D volume (2D Reproj.) and to surface renderings (Surf. Rend.) of the 3D volume. Scale bar, 10 nm. (C) Fourier shell correlation (FSC) curve of the reconstructed Ss box C/D sRNP EM volume. The resolution at 0.5 FSC is indicated. (D) Distribution of the Euler angles assigned to particle images included in the 3D EM reconstruction of the Ss box C/D sRNP. Circle radii are proportional to the number of images assigned to the particular Euler angles. (E) Isodensity of the 3D volume and docking of the crystal structures of the Ss core proteins, L7Ae (dark blue), Nop5 (green), and fibrillarin (cyan), (PDB 3ID5) (Ye et al. 2009) into the isodensity map at the 3σ threshold. Scale bar, 10 nm.
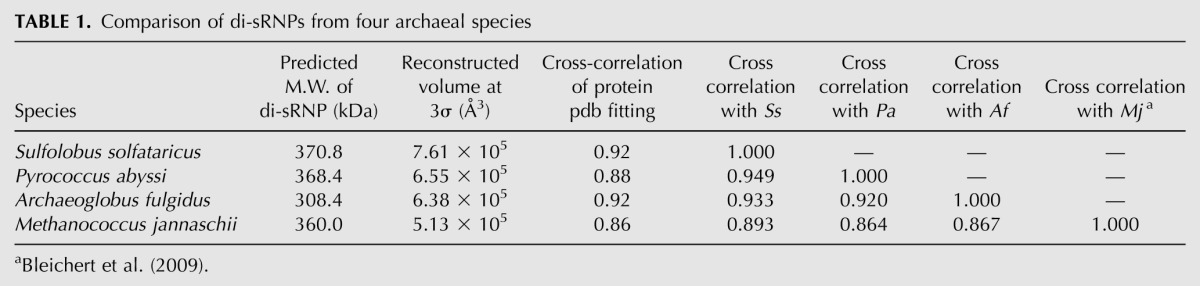
The refined Ss box C/D sRNP has dimensions of 13.8 × 15.5 × 12.8 nm (Fig. 4E). Similar to the Mj sRNP reconstruction (Bleichert et al. 2009), the Ss sRNP volume could accommodate four copies of each core protein, consistent with our biochemical classification of the sRNP as a dimeric enzyme (Fig. 4E). Atomic models of the Ss core proteins (Ye et al. 2009) were fit manually into the EM density map based on the Mj di-sRNP and computationally adjusted as rigid bodies using the program Situs (Wriggers and Birmanns 2001). The resulting cross-correlation value of 0.92 indicates a high confidence of the crystal structure docking into our 3D model (Table 1).
TABLE 1.
Comparison of di-sRNPs from four archaeal species
Although the resolution of the Ss negative stain EM structure was not sufficient for the sRNA molecules to be localized, our results are consistent with the di-sRNP model proposed in Bleichert et al. (2009). Previously, we hypothesized that the sRNAs are oriented perpendicular to the Nop5 coiled-coil domain interactions in the Mj di-sRNP structure (Bleichert et al. 2009). This hypothesis was supported by RNase digestion experiments performed in the same study. The architecture of the Ss box C/D sRNP, based on docking of atomic structures, is similar to that obtained with the Mj box C/D sRNP. We therefore hypothesize that the sRNAs adopt an orientation perpendicular to the Nop5 coiled-coil domains in the Ss di-sRNP as well. Altogether, our results are consistent with a dimeric architecture for the Ss box C/D sRNP, containing four copies of each core protein and two copies of the sRNA.
To investigate whether the di-sRNP volume could accommodate the mono-sRNP crystal structure, we docked unaltered coordinates from PDB 3PLA (Lin et al. 2011) and PDB 3ID5 (Ye et al. 2009) into the Ss di-sRNP reconstruction, first manually aligning L7Ae to its corresponding position from Figure 4E, and refining the docking using the “Fit in Map” Chimera command (Pettersen et al. 2004). The 3ID5 docking displays a poor fit of the fibrillarin protein as well as the N terminus of Nop5. To obtain the high correlation reported above for the docking of 3ID5, the N terminus of Nop5 with fibrillarin was treated as a rigid body separate from the remainder of the Nop5 protein, similarly to what was performed for the Mj di-sRNP structure (Bleichert et al. 2009). Despite aligning the Ss mono-sRNP, 3PLA (Lin et al. 2011), into the di-sRNP volume using L7Ae as an anchor, the crystal coordinates showed a poor fit into the EM density. Although it is possible the poor fit of the mono-sRNP into the di-sRNP volume is due to the conformational changes that occur upon substrate binding, the location of the sRNAs through the center of the EM volume is highly unlikely due to electrostatic repulsion between the sRNA molecules. Movie files that display the docking based on Figure 4E, 3ID5 unaltered, and 3PLA unaltered can be found in Supplemental Movies 1–3.
Distant evolutionary conservation of the di-sRNP
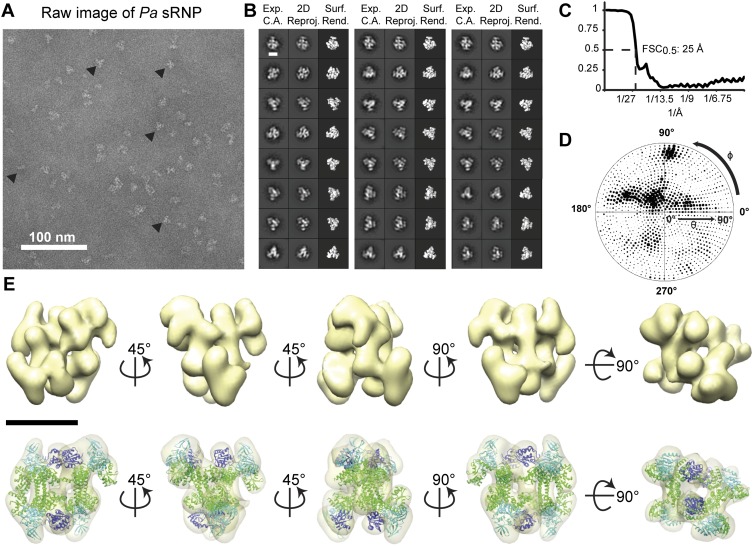
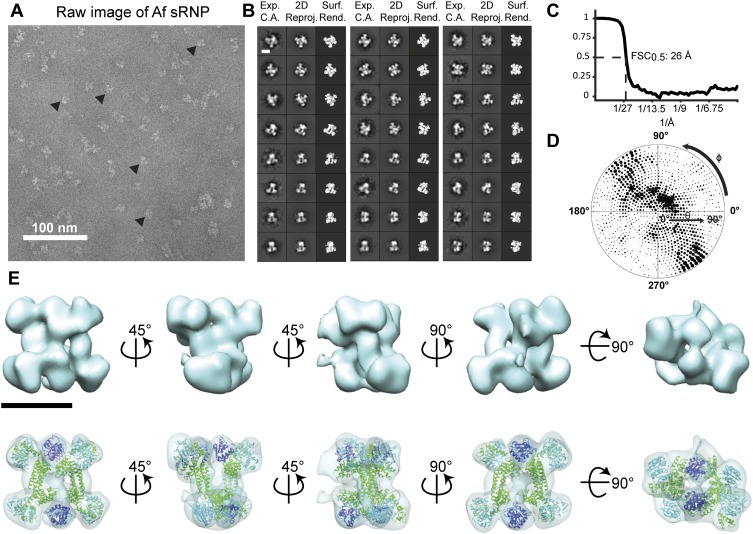
Biochemical evidence that the dimeric box C/D sRNP architecture was conserved across Archaea (Ghalei et al. 2010; Xue et al. 2010) led us to test whether the 3D structural characteristics of the di-sRNP were also retained in evolutionarily distant archaeal species (Ss is a crenarchaeote, and Mj, Pa, and Af are euryarchaeotes). We subjected box C/D sRNPs assembled from Pa and Af core proteins with sR8 to glycerol gradient purification, followed by single-particle negative stain EM analysis (Figs. 5, 6). Refined reconstructions of the Pa and Af sRNPs reached final resolutions of 25 and 26 Å, respectively (0.5 FSC criterion) and displayed a wide distribution of Euler angles (Figs. 5C,D, 6C,D). Previously solved atomic models of core proteins from each species were docked into the sRNP 3D volumes, reaching cross-correlation values of 0.88 and 0.92 for Pa and Af, respectively (Table 1). Pa and Af sRNP reconstructions were both able to accommodate four copies of each core protein, confirming the biochemical characterization of these archaeal box C/D sRNPs as di-sRNPs (Figs. 5E, 6E). Similar to the Ss sRNP reconstruction, docking the core proteins into the Pa and Af sRNP volumes is consistent with the di-sRNP model where the sRNAs run perpendicular to the Nop5 coiled-coil axis. Conservation of the overall di-sRNP 3D structure across distantly related archaeal species argues that the dimeric structure of this enzyme has functional and mechanistic implications.
FIGURE 5.
3D single-particle EM reconstruction of the Pa box C/D di-sRNP. (A) Electron micrograph of negatively stained Pa box C/D sRNPs from glycerol gradient peak fractions. (▸) Examples of particles chosen for analysis. (B) Comparison of experimentally determined class averages (Exp. C.A.) to 2D reprojections of the reconstructed 3D volume (2D Reproj.) and to surface renderings (Surf. Rend.) of the 3D volume. Scale bar, 10 nm. (C) FSC curve of the reconstructed Pa box C/D sRNP EM volume. The resolution at 0.5 FSC is indicated. (D) Distribution of the Euler angles assigned to particle images included in the 3D EM reconstruction of the Pa box C/D sRNP. Circle radii are proportional to the number of images assigned to the particular Euler angles. (E) Isodensity of the 3D volume and docking of the crystal structures of the Pa core proteins (PDB 3NMU) (Xue et al. 2010) into the isodensity map at the 3σ threshold. Colors are as in Figure 4. Scale bar, 10 nm.
FIGURE 6.
3D single particle EM reconstruction of the Af box C/D di-sRNP. (A) Electron micrograph of negatively stained Af box C/D sRNPs from glycerol gradient peak fractions. (▸) Examples of particles chosen for analysis. (B) Comparison of experimentally determined class averages (Exp. C.A.) to 2D reprojections of the reconstructed 3D volume (2D Reproj.) and to surface renderings (Surf. Rend.) of the 3D volume. Scale bar, 10 nm. (C) FSC curve of the reconstructed Af box C/D sRNP EM volume. The resolution at 0.5 FSC is indicated. (D) Distribution of the Euler angles assigned to particle images included in the 3D EM reconstruction of the Af box C/D sRNP. Circle radii are proportional to the number of images assigned to the particular Euler angles. (E) Isodensity of the 3D volume and docking of the crystal structures of the Af core proteins (PDB 1NT2) (Aittaleb et al. 2003), (1RLG) (Moore et al. 2004) into the isodensity map at the 3σ threshold. Colors are as in Figure 4. Scale bar, 10 nm.
Structural conservation of the di-sRNP suggests functional importance
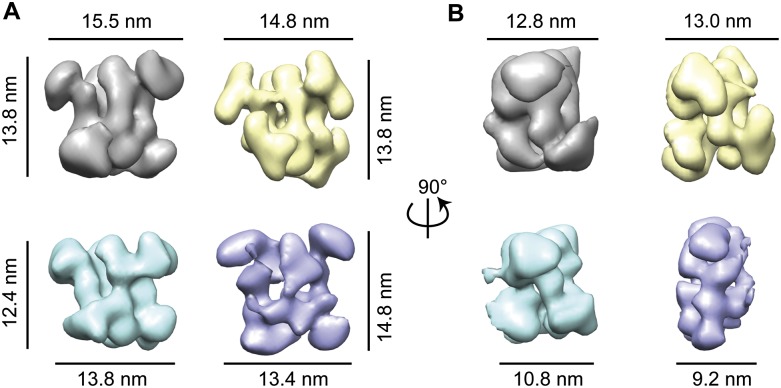
Because the overall architecture of the dimeric box C/D sRNP was confirmed through our single-particle negative stain EM analysis, we compared all four archaeal sRNP reconstructions to determine how structurally related they are to one another (Fig. 7). Rendering the threshold of each reconstruction at 3σ, we found that the Ss sRNP had the largest volume and the Mj sRNP had the smallest volume (Table 1). Taking into account the relative masses of the core proteins from each species, we were surprised that the sRNP with the smallest volume was not Af, because the Af proteins have the smallest molecular weights of all species tested. We attribute the differences in the 3σ volume to differences in staining techniques used in this study compared with the Mj study (Bleichert et al. 2009). This study used a deep-staining technique (Brenner and Horne 1959; Ohi et al. 2004), and the former Mj structural study used the carbon-sandwiching method (Valentine et al. 1968; Leschziner et al. 2007), which likely caused minor flattening of the Mj di-sRNP. This slight flattening is a known consequence of the carbon-sandwiching method (Kellenberger et al. 1982; Boisset et al. 1993; Frank 2006). The flattening can be clearly seen when comparing the side views of the reconstructed volumes from all four archaeal species (Fig. 7B), where the Mj species has the smallest thickness in the third dimension (9.2 nm compared with 10.8 nm for Af, 12.8 nm for Ss, and 13.0 nm for Pa).
FIGURE 7.
Distantly related archaeal box C/D sRNPs retain di-sRNP 3D architecture. Front (A) and side (B) views of aligned 3D EM negative stain reconstructions from four species: (Ss) gray; (Pa) yellow; (Af) blue; (Mj) purple. Ss, Pa, and Af are the three reconstructions from this study; Mj is from Bleichert et al. (2009). All reconstructions were set to 3σ thresholds. A c2 pseudosymmetry axis lies directly through the center of the front view (A) of each volume.
To assess the correlation among structures from each species, we aligned each archaeal di-sRNP in the same orientation using the Fit-Map-In-Map feature in Chimera (Pettersen et al. 2004) with the Ss sRNP as an initial model. The cross-correlation between structures was highest among the three archaeal structures reconstructed in this study, but high correlation among all four distantly related archaeal box C/D sRNPs indicates that the 3D dimeric architecture is broadly conserved across the archaeal domain (Table 1). This high degree of similarity can be easily visualized when comparing all species in aligned orientations (Fig. 7). The overall conservation of the architecture of the di-sRNP implies that the dimeric nature of the box C/D sRNP may be important for enzymatic function in vivo, because physiological sRNAs form predominantly the di-sRNP.
A structural feature that is perhaps more clear when all species are aligned to one another is the apparent c2 (twofold, 180° rotational) pseudosymmetry within the box C/D di-sRNP. In the original Mj di-sRNP random conical tilt reconstruction, pseudosymmetry was observed although no symmetry was imposed during refinement (Bleichert et al. 2009). Likewise, no symmetry was imposed in the reconstructions of the 3D volumes in this work. However, c2 pseudosymmetry was evident in each of the archaeal species box C/D sRNP 3D reconstructions when examined by eye. This axis lies directly through the center of each sRNP when viewed from the front as in Figure 7A.
To determine the robustness of this symmetry, we reconstructed all three volumes de novo assuming c2 symmetry in EMAN2 (Ludtke et al. 1999). Briefly, reference-free class averages were used as the basis for initial model generation in EMAN2. The initial model with the best match between the reprojections and class averages was used as the starting model for several rounds of projection-matching refinement while enforcing c2 symmetry. The volumes resulting from this reconstruction and refinement procedure were nearly identical to the volumes obtained from the asymmetric projection-matching reconstruction (data not shown). It is important to note that these reconstructions with symmetry imposed were performed using only the raw particle images as input into the EMAN2 pipeline; therefore, the resulting densities were reconstructed completely independently from the Mj di-sRNP initial model and the three asymmetric reconstructions reported above.
DISCUSSION
In this study, we determined that the archaeal box C/D sRNP relies on the conserved structural features of the sRNA for assembly into a dimeric architecture. All box C/D sRNPs tested from a broad range of distantly related archaeal species assemble into catalytically active di-sRNPs when reconstituted with a naturally occurring sRNA, sR8 from Mj. The internal loop that is omnipresent in archaeal box C/D sRNAs directs formation of the di-sRNP as the predominant species. Single-particle negative stain EM reconstructions of box C/D sRNPs from three evolutionarily distant archaeal species demonstrate that the di-sRNP model based on the Mj box C/D sRNP (Bleichert et al. 2009) represents a 3D sRNP architecture that is conserved across the archaeal domain in both Crenarchaeota (Ss) and Euryarchaeota (Mj, Pa, and Af) kingdoms.
Di-sRNPs are the primary architecture obtained when a naturally occurring, loop-containing sRNA is used for reconstitution. Although the quantitation in Figures 1–3 indicates variability in the ratio of di- versus mono-sRNPs from experiment to experiment and species to species, we cannot be sure that the band intensity detected in the mono-sRNP-migrating area is due to assembly of an actual mono-sRNP. Indeed, no ribose methylation activity was found associated with mono-sRNP sedimenting fractions of glycerol gradients, implying that little mono-sRNP, if any, is formed (Bleichert et al. 2009).
Limitations of single-particle negative stain electron microscopy
While single-particle negative stain electron microscopy has made possible the analysis of macromolecular structures that may not be amenable to crystallography, the technique has limitations as well. One of these limitations is resolution. At 23–27 Å, the resolution of the box C/D sRNP reconstructions from four archaeal species is not sufficient to localize the sR8 RNA. While we cannot visualize the sRNAs, the sensitivity of the di-sRNP structure to RNase implies that the sRNA is a primary structural determinant (Bleichert et al. 2009). This result, together with the information obtained through docking of the atomic models of the core sRNP protein components, is consistent with the previously proposed hypothesis that the sRNAs run perpendicular to the Nop5–fibrillarin heterotetramer (Bleichert et al. 2009). Innovations in automation (Lander et al. 2009; Voss et al. 2009; Wiedenheft et al. 2011) and in camera resolution (Bammes et al. 2012) in cryo-electron microscopy are both designed to permit resolution sufficient to visualize the sR8 RNA. Localizing the sR8 RNA in future studies will reveal how the core proteins discriminate between di- and mono-sRNP conformations through the conserved presence of the internal sRNA loop.
Internal sRNA loops are conserved and critical for di-sRNP formation
A significant finding in this study is that the internal loop of the box C/D sRNA is a primary determinant of box C/D sRNP architecture. The conserved presence of the box C/D sRNA internal loop throughout predicted archaeal sRNAs suggests that the loop is important for assembly of functional box C/D sRNPs in vivo. Indeed, the presence of these loops is used as a computational parameter to detect the presence of sRNAs in newly sequenced archaeal genomes (Lowe and Eddy 1999; Omer et al. 2000).
Internal loops of varying lengths are a prevalent RNA motif found in nature (Batey et al. 1999; Moore 1999). Tetraloops are highly represented in noncoding cellular RNAs, accounting for ∼55% of all hairpin loops in bacterial and archaeal 16S rRNAs (Woese et al. 1990). The next most prevalent loop size in 16S rRNAs is 5 nt, comprising 13% of all 16S hairpin loops (Woese et al. 1990). A similar pattern exists for the large subunit rRNA, where 4-nt and 5-nt loops occupy 38% and 24%, respectively, of the total hairpin secondary structures (Gutell and Fox 1988). Interestingly, predicted sRNAs from all four species examined in this study (Mj, Ss, Pa, and Af) contain loops of up to 12 nt (Omer et al. 2000). While the sequence of the box C/D sRNA internal loop is quite variable, the absence of loops with lengths >12 nt suggests that internal loop length is constrained in archaeal box C/D sRNAs.
Loops are a general RNA secondary structure feature important for tertiary interactions and RNP function. One example, which provided the rationale for the experiments in Figure 3, is how the two paralogs of the pre-miRNA processing protein Dicer distinguish between their closely related RNA substrates through the presence or absence of a loop (Lee et al. 2004; Tsutsumi et al. 2011). Similarly, the length and sequence of the GNRA tetraloop of the yeast mitochondrial ai5γ group II intron are critical for forming tertiary sequence-specific interactions (Abramovitz and Pyle 1997; Costa and Michel 1997). Likewise, the conserved internal loop of archaeal box C/D sRNAs imposes, through a mechanism not yet understood, a dimeric assembly of the in vitro–assembled box C/D sRNP.
Controversies in box C/D sRNP architecture
Two models have been proposed for the architecture of catalytically active archaeal box C/D sRNPs: a mono-sRNP (Lin et al. 2011) and a di-sRNP (Bleichert et al. 2009; Bleichert and Baserga 2010a; Xue et al. 2010; this study). The mono-sRNP consists of two copies of each of the three core proteins and one sRNA, while the di-sRNP consists of four copies of each of the three proteins and two sRNAs. Importantly, the two models differ in the directionality of the sRNA with respect to the Nop5/fibrillarin heterotetramer (Fig. 1B). The sRNA in the mono-sRNP runs roughly parallel to the Nop5/fibrillarin heterotetramer, while the sRNA in the di-sRNP is perpendicular. In the presence of an sRNA with an internal loop, the di-sRNP is the predominant species assembled. If, instead, a two-stranded sRNA is used (CD45), a mixture of mono- and di-sRNPs is formed. Interestingly, a predominant species with the mobility of the mono-sRNP is also observed in glycerol gradients when an sR8 RNA that is mutated in any of the conserved boxes (C, D, C′, D′) or when half-mer sRNAs are used for assembly (Bleichert and Baserga 2010a). However, archaeal sRNPs containing sRNAs with mutated or absent conserved boxes have reduced in vitro methylation activity (Tran et al. 2003). Although box C/D s(no)RNAs tolerate weak or atypical C′ or D′ boxes, especially in eukaryotes (Knox et al. 2011; van Nues et al. 2011), it is unknown how variant sRNAs function in vivo in Archaea. Taken together, reconstitution of box C/D sRNPs with any noncanonical sRNA (two-stranded, mutated in or lacking conserved sequences) causes substantial mono-sRNP assembly, revealing that the mono-sRNP is most often obtained following reconstitution with an atypical box C/D sRNA.
The mono-sRNP reconstitution that was crystallized included the substrate RNA and demonstrated positioning of the methylation enzyme, fibrillarin, to the targeted nucleotide (Lin et al. 2011). Surprisingly, no methyl group was detectable on either the substrate RNA or on the cofactor, suggesting that fibrillarin can be positioned in the absence of chemistry. In other crystal structures, the methyl-group-containing cofactor, S-adenosyl methionine (SAM), was observed complexed with fibrillarin, even though it was not added to the purification or crystallization buffers (Aittaleb et al. 2003; Oruganti et al. 2007; Ye et al. 2009; Xue et al. 2010). Because tracer experiments with 3H SAM indicated methyl group incorporation into the mono-sRNP, interpreted as fibrillarin enzymatic activity, the absence of the methyl group on the substrate RNA in the crystal structure was unexpected.
Large hinge motions of fibrillarin to position its active site for catalysis have been anticipated since the first cocrystal structures of Nop5–fibrillarin (Aittaleb et al. 2003; Oruganti et al. 2007; Bleichert et al. 2009; Ye et al. 2009; Xue et al. 2010). As expected, such conformational changes were also detected in the context of the mono-sRNP (Lin et al. 2011). However, since the conformational change is dependent on the position of the guide and substrate RNAs, it is likely that the movement of fibrillarin will be different in the context of the di-sRNP. Thus, the structural basis of site-specific ribose methylation by box C/D dimeric sRNPs is liable to be distinct from that of mono-sRNPs.
Why a di-sRNP?
Our results demonstrate that the di-sRNP model represents the predominant architecture when box C/D sRNPs are assembled with naturally occurring box C/D sRNAs. However, a remaining question is whether the di-sRNP architecture is biologically important. The four archaeal species tested so far are all hyperthermophilic organisms. At high temperatures, the di-sRNP architecture may confer greater structural stability. In addition, the presence of two sRNAs in a di-sRNP creates the possibility of up to four guide sequences assembled in one sRNP. This scenario gives a potential kinetic advantage in the process of ribosome biogenesis. A di-sRNP with two different sRNAs could bring together distal regions of the rRNA, facilitating long-range interactions in these long substrate RNAs. Although two different sRNAs assembled into a single sRNP is a possibility, this arrangement may also require a higher degree of cellular regulation in order to direct correct incorporation of different sRNAs into a single sRNP (Bleichert and Baserga 2010b).
Future structural studies at higher resolution are needed to explore the nature of di-sRNP function in the context of a natural, full-length sRNA. In addition, little is known about box C/D sRNP composition in vivo in Archaea. These avenues of investigation will give further insight into the vital role of the box C/D sRNP in making fully functional ribosomes.
MATERIALS AND METHODS
Protein expression and purification
Full-length L7Ae from S. solfataricus, P. abyssi, and A. fulgidus were cloned into pET28a (Novagen), resulting in 6xHis-tagged fusion proteins with a thrombin cleavage site at the N terminus. L7Ae proteins were expressed and purified as in Bleichert et al. (2009).
Nop5 (Ss amino acid residues 1–380 [full-length 412 amino acids], Pa amino acid residues 1–373 [full-length 404 amino acids], and full-length Af) and full-length fibrillarin from Ss, Pa, and Af were cloned into pET21a and pET28a (Novagen), respectively, after adding appropriate restriction sites during PCR amplification. Both proteins from each species were coexpressed and purified as in Bleichert et al. (2009), with the addition of S200 size-exclusion chromatography as a final purification step.
In vitro transcription and assembly of sRNPs
The CD45 and sR8 faux sRNA oligoribonucleotides were chemically synthesized (Dharmacon). All of the other sRNA genes were cloned into the pCR4-TOPO vector (Invitrogen), adding a 5′ T7 polymerase promoter sequence, and were in vitro–transcribed as in Bleichert et al. (2009). Oligonucleotide sequences used for cloning and amplification can be found in Supplemental Table 1. All sRNPs were assembled as in Bleichert et al. (2009).
Native gel analysis of sRNPs
Ten microliters of assembled box C/D sRNPs was loaded onto native polyacrylamide gels (6% polyacrylamide, 375 mM Trizma-base at pH 8.8, 1.5 mM MgCl2) and resolved by electrophoresis in a buffer containing 25 mM Tris-glycine (pH 8.3) and 1.5 mM MgCl2 for 3–4 h at 4°C. The gels were stained with ethidium bromide to visualize the sRNA followed by silver staining to visualize the proteins.
Glycerol gradient centrifugation
Assembled sRNPs were purified on 10%–25% glycerol gradients and analyzed as in Bleichert et al. (2009).
In vitro methylation activity assays
Methylation activity assays were performed similarly to Tran et al. (2003), Gagnon et al. (2006), and Bleichert et al. (2009). Sixty microliters of unpurified sRNP assembly reaction was mixed with 15 μL of reaction mix to a final concentration of 20 mM HEPES (pH 7.5), 500 mM NaCl, 10% glycerol, 1.5 mM MgCl2, 9 μM substrate RNA, 27 μM S-adenosyl-L-methionine (SAM), and 0.795 μCi/μL S-adenosyl-L-[methyl-3H]-methionine (3H-SAM) (PerkinElmer). Reactions were incubated for 30 min at 70°C, then 25-μL aliquots were spotted in triplicate onto a double layer of Whatman 3MM and DE83 filters. Filters were washed once with 5 mL of 10% TCA, three times with 5 mL of 5% TCA, and three times with 5 mL of 100% ethanol. Filters were dried, and the incorporation of tritiated methyl groups into substrate RNAs was quantified by liquid scintillation counting. Sequences for substrate RNAs as well as negative controls can be found in Supplemental Table 1.
Electron microscopy and single-particle analysis
Preparation of EM grids and data collection
Archaeal sRNPs purified from peak fractions of glycerol gradients were buffer-exchanged into buffer E (20 mM HEPES at pH 7.5, 500 mM NaCl, 1.5 mM MgCl2, 6% trehalose) to remove glycerol. Four microliters of sample was applied to a glow-discharged C-flat holey carbon grid (Electron Microscopy Sciences) with a layer of thin film carbon for support. After adsorption for 1 min, samples were negatively stained with 2% uranyl formate, gently blotted to remove excess stain, and allowed to air-dry.
Specimens were examined using an FEI Tecnai T12 electron microscope equipped with a LaB6 filament and operated at 120 kV. Images were recorded at a nominal magnification of 42,000× using a GATAN Ultra4000 4k × 4k CCD camera using low-dose conditions with an exposure dose of 20–30 e− Å−2. The defocus range for all species examined was −0.6 to −1.1 μm. The CCD-recorded micrographs had a pixel size of 2.7Å and were used directly for image processing.
Image processing and volume reconstruction
Particles of 128 × 128 square pixels were picked manually using the boxer program in the EMAN suite (Ludtke et al. 1999), normalized, band-pass filtered, and centered in preparation for classification. The total number of particles used in each reconstruction varied: Ss had 6149, Pa had 9248, and Af had 6336. The IMAGIC-5 (van Heel et al. 1996) image processing package was used for reference-free two-dimensional alignment and classification through several iterations of multivariate statistical analysis and multireference alignment for each species to generate 100 classes (60–90 particles per class). The Mj box C/D sRNP reconstruction (Bleichert et al. 2009) was low-pass filtered to 120 Å. This volume served as the initial model for projection matching refinement (Penczek et al. 1994) using the program SPIDER (Frank et al. 1996) to obtain the refined structure of the Ss box C/D sRNP. The Ss volume was low-pass filtered to 50 Å and used as an initial model for projection matching refinement of the Pa and Af single-particle data. No symmetry was imposed at any stage of the reconstruction.
Visual inspection of reference-free class averages and asymmetric reconstructions suggested that all three species displayed c2, or possibly even d2, symmetry. The EMAN2 (Ludtke et al. 1999) pipeline was used to create independent reference-free class averages and generate initial models assuming either c2 or d2 symmetry from these class averages. Reprojections of the initial models with c2 symmetry imposed matched more closely to the experimental class averages than reprojections of those with d2 symmetry imposed (data not shown); consequently, c2 symmetry was enforced during the subsequent projection matching refinement procedure for all species using their respective c2 symmetrized initial models. The final symmetrical reconstructions of Ss, Pa, and Af sRNPs had estimated resolutions of 26, 23, and 27 Å, respectively (0.5 FSC criterion) (data not shown).
Docking of atomic models into the 3D EM density map
The docking of corresponding species' atomic models was first done by visual inspection and manual docking of either individual proteins or cocomplexes as rigid bodies using Chimera (Pettersen et al. 2004). Crystal structures (3ID5 [Ye et al. 2009] into Ss, 3NMU [Xue et al. 2010] into Pa, 1RLG [Moore et al. 2004] and 1NT2 [Aittaleb et al. 2003] into Af) were placed in analogous locations to the protein fitting of Bleichert et al. (2009) and guided by the Fit-Model-In-Map function in Chimera. To minimize steric interference and to maximize correlation, simultaneous multi-fragment refinement of manual docking was performed with the Collage program within Situs 2.6 (Wriggers and Birmanns 2001). The 3D volumes were displayed using the UCSF-Chimera software package (Pettersen et al. 2004).
DATA DEPOSITION
The 3D EM volumes from this study have been deposited into the EMDataBank (http://www.emdatabank.org) and are available under accession numbers EMD-5419, EMD-5420, and EMD-5421.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank J. Archibald (Dalhousie University) for helpful guidance on determining evolutionary distances; P. Blum (University of Nebraska) for the kind gift of Ss genomic DNA; and current and former members of the Baserga, Wang, and J. Steitz laboratories for technical assistance, helpful discussions, and for critical reading of this manuscript. We thank staff in the Yale CryoEM facility and High-Performance Computing facility for their maintenance of these facilities. This work was supported in part by NIH GM052581 (to S.J.B.), NRSA NIH/NIGMS T32 GM007223 Training Grant (to K.R.B.-P.), and NIH 5 T32 GM008283 Training Grant (to D.W.T.). K.R.B.-P. and D.W.T. are NSF Graduate Research Fellows. H.-W.W. is the recipient of a Smith Family Award for Excellence in Biomedical Research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.033134.112.
REFERENCES
- Abramovitz DL, Pyle AM 1997. Remarkable morphological variability of a common RNA folding motif: The GNRA tetraloop–receptor interaction. J Mol Biol 266: 493–506 [DOI] [PubMed] [Google Scholar]
- Aittaleb M, Rashid R, Chen Q, Palmer JR, Daniels CJ, Li H 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat Struct Biol 10: 256–263 [DOI] [PubMed] [Google Scholar]
- Aittaleb M, Visone T, Fenley MO, Li H 2004. Structural and thermodynamic evidence for a stabilizing role of Nop5p in S-adenosyl-L-methionine binding to fibrillarin. J Biol Chem 279: 41822–41829 [DOI] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834 [DOI] [PubMed] [Google Scholar]
- Bammes BE, Rochat RH, Jakana J, Chen DH, Chiu W 2012. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. J Struct Biol 177: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Rambo RP, Doudna JA 1999. Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl 38: 2326–2343 [DOI] [PubMed] [Google Scholar]
- Bleichert F, Baserga SJ 2010a. Dissecting the role of conserved box C/D sRNA sequences in di-sRNP assembly and function. Nucleic Acids Res 38: 8295–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Baserga SJ 2010b. Ribonucleoprotein multimers and their functions. Crit Rev Biochem Mol Biol 45: 331–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleichert F, Gagnon KT, Brown BA II, Maxwell ES, Leschziner AE, Unger VM, Baserga SJ 2009. A dimeric structure for archaeal box C/D small ribonucleoproteins. Science 325: 1384–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach F, Brouns SJ, van der Oost J 2011. Assembling the archaeal ribosome: Roles for translation-factor-related GTPases. Biochem Soc Trans 39: 45–50 [DOI] [PubMed] [Google Scholar]
- Boisset N, Radermacher M, Grassucci R, Taveau JC, Liu W, Lamy J, Frank J, Lamy JN 1993. Three-dimensional immunoelectron microscopy of scorpion hemocyanin labeled with a monoclonal Fab fragment. J Struct Biol 111: 234–244 [DOI] [PubMed] [Google Scholar]
- Bortolin ML, Bachellerie JP, Clouet-d'Orval B 2003. In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNATrp. Nucleic Acids Res 31: 6524–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Horne RW 1959. A negative staining method for high resolution electron microscopy of viruses. Biochim Biophys Acta 34: 103–110 [DOI] [PubMed] [Google Scholar]
- Costa M, Michel F 1997. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J 16: 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM 2008. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci 105: 20356–20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ 2002. rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351 [DOI] [PubMed] [Google Scholar]
- Frank J. 2006. Three-dimensional electron microscopy of macromolecular assemblies: Visualization of biological molecules in their native state. Oxford University Press, New York. [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A 1996. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Gagnon KT, Zhang X, Agris PF, Maxwell ES 2006. Assembly of the archaeal box C/D sRNP can occur via alternative pathways and requires temperature-facilitated sRNA remodeling. J Mol Biol 362: 1025–1042 [DOI] [PubMed] [Google Scholar]
- Gaspin C, Cavaille J, Erauso G, Bachellerie JP 2000. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: Lessons from the Pyrococcus genomes. J Mol Biol 297: 895–906 [DOI] [PubMed] [Google Scholar]
- Ghalei H, Hsiao HH, Urlaub H, Wahl MC, Watkins NJ 2010. A novel Nop5–sRNA interaction that is required for efficient archaeal box C/D sRNP formation. RNA 16: 2341–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Fox GE 1988. A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Res (Suppl) 16: r175–r269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin JW, Batey RT 2006. The bipartite architecture of the sRNA in an archaeal box C/D complex is a primary determinant of specificity. Nucleic Acids Res 34: 5039–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger E, Haner M, Wurtz M 1982. The wrapping phenomenon in air-dried and negatively stained preparations. Ultramicroscopy 9: 139–150 [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T 1996. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z, Henry Y, Kiss T 1998. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J 17: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA 2001. The kink-turn: A new RNA secondary structure motif. EMBO J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox AA, McKeegan KS, Debieux CM, Traynor A, Richardson H, Watkins NJ 2011. A weak C′ box renders U3 snoRNA levels dependent on hU3-55K binding. Mol Cell Biol 31: 2404–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JF, Tran EJ, Maxwell ES 2002. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res 30: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. 2009. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR, Nogales E 2007. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc Natl Acad Sci 104: 4913–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ 2007. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell 28: 965–977 [DOI] [PubMed] [Google Scholar]
- Lin J, Lai S, Jia R, Xu A, Zhang L, Lu J, Ye K 2011. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature 469: 559–563 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171 [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W 1999. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ 1995. The small nucleolar RNAs. Annu Rev Biochem 64: 897–934 [DOI] [PubMed] [Google Scholar]
- Moore PB 1999. Structural motifs in RNA. Annu Rev Biochem 68: 287–300 [DOI] [PubMed] [Google Scholar]
- Moore T, Zhang Y, Fenley MO, Li H 2004. Molecular basis of box C/D RNA–protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure 12: 807–818 [DOI] [PubMed] [Google Scholar]
- Nolivos S, Carpousis AJ, Clouet-d'Orval B 2005. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res 33: 6507–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T 2004. Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online 6: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer AD, Lowe TM, Russell AG, Ebhardt H, Eddy SR, Dennis PP 2000. Homologs of small nucleolar RNAs in Archaea. Science 288: 517–522 [DOI] [PubMed] [Google Scholar]
- Omer AD, Ziesche S, Ebhardt H, Dennis PP 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci 99: 5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oruganti S, Zhang Y, Li H, Robinson H, Terns MP, Terns RM, Yang W 2007. Alternative conformations of the archaeal Nop56/58-fibrillarin complex imply flexibility in box C/D RNPs. J Mol Biol 371: 1141–1150 [DOI] [PubMed] [Google Scholar]
- Penczek PA, Grassucci RA, Frank J 1994. The ribosome at improved resolution: New techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy 53: 251–270 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G 2007. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res 35: 1452–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajani Z, Sykes MT, Williamson JR 2011. Assembly of bacterial ribosomes. Annu Rev Biochem 80: 501–526 [DOI] [PubMed] [Google Scholar]
- Tran EJ, Zhang X, Maxwell ES 2003. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J 22: 3930–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Zhang X, Lackey L, Maxwell ES 2005. Conserved spacing between the box C/D and C′/D′ RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA 11: 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y 2011. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol 18: 1153–1158 [DOI] [PubMed] [Google Scholar]
- Valentine RC, Shapiro BM, Stadtman ER 1968. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry 7: 2143–2152 [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M 1996. A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- van Nues RW, Granneman S, Kudla G, Sloan KE, Chicken M, Tollervey D, Watkins NJ 2011. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J 30: 2420–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell 6: 1331–1342 [DOI] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B 2009. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 166: 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466 [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E 2011. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477: 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Winker S, Gutell RR 1990. Architecture of ribosomal RNA: Constraints on the sequence of “tetra-loops.” Proc Natl Acad Sci 87: 8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Birmanns S 2001. Using Situs for flexible and rigid-body fitting of multiresolution single-molecule data. J Struct Biol 133: 193–202 [DOI] [PubMed] [Google Scholar]
- Xue S, Wang R, Yang F, Terns RM, Terns MP, Zhang X, Maxwell ES, Li H 2010. Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle. Mol Cell 39: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Jia R, Lin J, Ju M, Peng J, Xu A, Zhang L 2009. Structural organization of box C/D RNA-guided RNA methyltransferase. Proc Natl Acad Sci 106: 13808–13813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Champion EA, Tran EJ, Brown BA II, Baserga SJ, Maxwell ES 2006. The coiled-coil domain of the Nop56/58 core protein is dispensable for sRNP assembly but is critical for archaeal box C/D sRNP-guided nucleotide methylation. RNA 12: 1092–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]