Intracellular trafficking of RNA and protein molecules is critical for their maturation and function. The authors describe a method to measure mass movement of both RNA and protein molecules within a heterogeneous population of cells using multispectral imaging flow cytometry (MIFC). They use this approach to measure movement of the cytoplasmic poly(A)-binding protein C1 (PABPC1) into the nucleus of human B cells infected with Kaposi's sarcoma–associated herpesvirus (KSHV), and correlate this relocalization with the expression of the highly abundant, viral noncoding polyadenylated nuclear (PAN) RNA. This high-throughput approach will be useful for other systems in which changes in subcellular localization of RNA and protein molecules need to be monitored simultaneously.
Keywords: FACS, high-throughput, MIFC, RNA, noncoding
Abstract
We report a high-throughput application of multispectral imaging flow cytometry (MIFC) for analyzing the expression and localization of both RNA and protein molecules in a heterogeneous population of cells. The approach was developed using polyadenylated nuclear (PAN) RNA, an abundant, noncoding RNA expressed by Kaposi's sarcoma–associated herpesvirus (KSHV) during the lytic phase of infection. High levels of PAN RNA are, in part, dependent on its interaction with poly(A)-binding protein C1 (PABPC1), which relocalizes from the cytoplasm to the nucleus of lytically infected cells. We quantitatively tracked the cytoplasmic to nuclear translocation of PABPC1 and examined how this translocation relates to the expression and localization of viral RNA and protein molecules in KSHV-infected cells. This high-throughput approach will be useful for other systems in which changes in subcellular localization of RNA and protein molecules need to be monitored simultaneously.
INTRODUCTION
Kaposi's sarcoma–associated herpesvirus (KSHV) is the cause of several human cancers and lymphoproliferative disorders and is known to infect a number of different cell types, including endothelial, epithelial, and lymphatic cells (Boshoff et al. 1995; Renne et al. 1998; Dupin et al. 1999). As for other herpesviruses, the KSHV life cycle can be divided into two stages: latent and lytic infection. Restricted gene expression during latency results in the synthesis of only two polycistronic viral transcripts, yielding six proteins and 17 miRNAs (Cai et al. 2005; Samols et al. 2005; Ganem 2006). These gene products facilitate the maintenance of the viral genome and can regulate cell cycle, cell signaling, and apoptotic pathways. At any time, a small percentage of latently infected cells may enter the lytic stage, which is characterized by the temporally ordered expression of the remaining 88 viral genes (Paulose-Murphy et al. 2001). These lytic genes enable viral DNA replication and packaging of the genome into infectious particles (Ganem 2006).
The most abundant viral transcript expressed during the lytic phase is a ∼1-kb noncoding RNA called polyadenylated nuclear (PAN) (Sun et al. 1996; Zhong and Ganem 1997). PAN RNA is transcribed by RNA polymerase II and accumulates to very high levels in the host cell nucleus (about 105 copies/cell). Since PAN RNA accounts for ∼80% of all polyadenylated RNA during the lytic phase, its levels exceed those of host and viral mRNAs combined. PAN RNA does not appear to encode protein, as it localizes only to the nucleus and lacks long open reading frames.
One of the proteins that binds PAN RNA in the nucleus is poly(A)-binding protein C1 (PABPC1), an abundant host protein estimated at 8 × 106 copies per cell (Borah et al. 2011). PABPC1 binds to poly(A) stretches with an affinity of ∼7 nM. PAPBC1 normally resides in the cytoplasm, where multiple copies cooperatively bind the poly(A) tail found at the 3′ end of most mRNAs (Gorlach et al. 1994). These interactions appear to be important for the regulation of mRNA translation and stability (Mangus et al. 2003); however, PABPC1 relocalizes to the nucleus during lytic KSHV infection due to the expression of the virally encoded shutoff exonuclease (SOX) protein (Glaunsinger and Ganem 2004; Kumar and Glaunsinger 2010). SOX expression causes nuclear relocalization of PABPC1 by binding and cleaving cytoplasmic mRNAs (Covarrubias et al. 2011). PABPC1, released from its interaction with the poly(A) tail of these mRNAs, is subsequently able to interact with the nuclear receptor protein importin α, which directs its nuclear translocation (Kumar et al. 2011). Previously, the interaction of PAN RNA with nuclear relocalized PABPC1 was characterized in infected, lytically reactivated body cavity–based lymphoma-1 (BCBL1)-TReX-RTA cells (Nakamura et al. 2003; Borah et al. 2011). In these studies, PABPC1 cofractionated with PAN RNA upon affinity purification of BCBL1-TReX-RTA cell extracts using antisense oligonucleotides. Furthermore, confocal microscopy revealed very similar intranuclear patterns of PAN RNA and of relocalized PABPC1 in lytically infected cells, supporting the idea that they are included in the same nuclear complex. In uninfected, transiently transfected 293T cells, high levels of PAN RNA accumulation are dependent on the ability of cotransfected SOX protein to relocalize PABPC1 into the nucleus. Although PAN RNA is not required for the ability of SOX to carry out host shutoff in infected cells or to relocalize PABPC1, PAN RNA is required for production of high viral titers and for the expression of viral genes expressed in late lytic phase. These findings were subsequently confirmed through the generation of a recombinant virus deficient in PAN RNA (BAC36CRΔPAN), which failed to produce infectious virus upon lytic induction (Rossetto and Pari 2012). However, this subsequent study also suggested that PAN may have roles earlier in infection.
Previously, visualization of PAN RNA and cytoplasmic and nuclear-translocated PABPC1 entailed dual in situ hybridization and indirect protein immunofluorescence staining of virally infected BCBL1-TReX-RTA cells, using a laser confocal scanning microscope to collect images (Borah et al. 2011). To quantify the correlation between expression of PAN RNA and nuclear translocation of PABPC1, two individuals scored these images manually to produce averages. Manual scoring of images for RNA and protein expression and localization can potentially yield high-confidence data, since it permits judgments on variations in signal intensity, cell morphology, and cell damage for each cell. However, for the same reasons, such an approach suffers from subjectivity because each individual inevitably makes different decisions as to what level and pattern of signal constitutes RNA or protein expression and/or subcellular localization. Furthermore, manual scoring of RNA and protein signal intensities and localization is laborious and time-consuming. In the previous study, for example, collecting data from only a few hundred cells required many hours.
Here, we describe the use of multispectral imaging flow cytometry (MIFC) to study the expression and localization of PAN RNA and PABPC1 in BCBL1 cells, a B-cell-tumor-derived cell line infected with KSHV. The use of MIFC has allowed, in previous studies, the simultaneous qualitative and quantitative high-throughput analysis of individual cells comprising heterogeneous populations (Adang et al. 2006; Basiji et al. 2007; Hassman et al. 2011). The advantages of MIFC compared with the approach of manual scoring are manifold. First, it allows the analysis of many thousands of cells within a population for each experiment, providing a more comprehensive picture of changes in expression and localization of RNAs and proteins throughout an entire population. Second, it is rapid, greatly reducing the data gathering and analysis time. Third, the use of a standard MIFC template objectively scores not only all cells within an experiment by the same criteria but also cells from other experiments, reducing the subjectivity intrinsic to manual scoring. By use of MIFC, we observed robust expression of PAN RNA in the majority of KSHV lytically reactivated cells in which PABPC1 had translocated from the cytoplasm into the nucleus, and similar patterns of PAN and PABPC1 within these nuclei. We also demonstrate that detection of PAN RNA, which is expressed in early lytic phase, of K8.1 protein, which is expressed in late lytic phase, or of both PAN RNA and K8.1 protein allows us to define the degree of lytic progression of each BCBL1 cell in a heterogeneous population, and that the extent of PABPC1 nuclear relocalization increases as a function of lytic progression. Our method of staining and analysis of RNA and protein expression and localization in a heterogeneous population of cells provides a powerful and precise approach to study complex and dynamic RNA–protein interactions at the single-cell level. It, therefore, has broad potential applicability.
RESULTS
MIFC tracks RNA and protein expression and localization in each cell within a heterogeneous population of BCBL1 cells
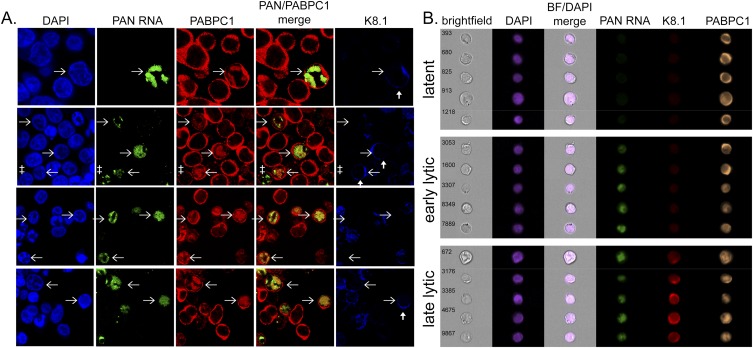
To validate MIFC as a high-throughput alternative to the manual scoring described above, we stained latent and induced BCBL1 cells for PAN RNA, PABPC1 protein, and K8.1 protein. BCBL1 cells were induced by continuous treatment with 600 μM valproic acid for 48 h. We first visualized these molecules by confocal microscopy. Figure 1A shows that this imaging approach allows clear visualization of PAN RNA, PABPC1, and the late viral protein K8.1 in cells after dual in situ hybridization and immunofluorescence staining.
FIGURE 1.
(A) Confocal images of BCBL1 cells in which DNA (DAPI), PAN RNA (FITC), PABPC1 (Alexafluor 594), and K8.1 (Alexafluor 647) are visualized. Horizontal arrows indicate the same cell in each panel, and vertical arrows indicate cells expressing K8.1 protein. A cell expressing PAN RNA but in which PABPC1 has not translocated into the nucleus is indicated (‡). (B) MIFC images of BCBL1 cells staining positive for PAN RNA and/or K8.1. The cells imaged in panel B are taken from the same culture as those images in panel A. Brightfield images as well as merged brightfield and DAPI images are shown. Numbers indicate the number of individual cells within the entire population that were analyzed in the experiment.
We then analyzed these same cells by MIFC (Fig. 1B). As expected, cells in both latent and lytic populations stained positive for PABPC1, whereas virtually no uninduced BCBL1 cells stained positive for either PAN RNA or K8.1 protein (Fig. 1B, top panel). However, after 48-h treatment with valproic acid, 38.7% ± 5.2% (SEM) of the cells expressed PAN RNA (Fig. 1B, middle and bottom panels) and 5.7% ± 0.8% (SEM) expressed K8.1 (Fig. 1B, bottom panel).
Importantly, MIFC identified the nuclear and total cell areas using the DAPI and brightfield channels, respectively. The IDEAS software then calculated the cytoplasmic area using the difference between the two, though with the high nuclear-to-cytoplasmic ratios of most B cells, including BCBL1 cells, this difference is small (Li et al. 1999; Santarelli et al. 2008). PAN RNA expression was restricted to the nucleus in these cells, whereas the expression of the viral K8.1 glycoprotein, a component of the virion particle, was restricted to the cytoplasm and plasma membrane, as previous studies reported (Sun et al. 1996; Zhong and Ganem 1997; Li et al. 1999; Zhu et al. 2005; Santarelli et al. 2008; Lefort and Flamand 2009). These results demonstrate that MIFC can simultaneously detect both viral RNAs and cellular and viral proteins of interest, while also generating images of subcellular staining patterns that agree with those from standard confocal microscopy.
MIFC allows quantitative measurements of host PABPC1 nuclear translocation in individual BCBL1 cells and its correlation with the expression of viral RNA and protein
Having demonstrated that MIFC can be used to analyze RNA and protein expression and localization simultaneously, we assessed the percentages of cells with cytoplasmic versus nuclear-translocated PABPC1 in both latent and lytically induced BCBL1 cells. To measure PABPC1 translocation, we determined the ratio of Alexafluor 594 signal in the nucleus versus the cytoplasm. For this analysis, the DAPI signal defined the nucleus and the remaining area within the overlaid brightfield image defined the cytoplasm. PABPC1 was nearly exclusively cytoplasmic in cells of the latent culture (Figs. 1B, 2A), whereas PABPC1 relocalized into the nucleus in 34.1% ± 1.8% (SEM) of the total population of cells (n = 3) (Figs. 1B, 2A) following treatment with valproic acid.
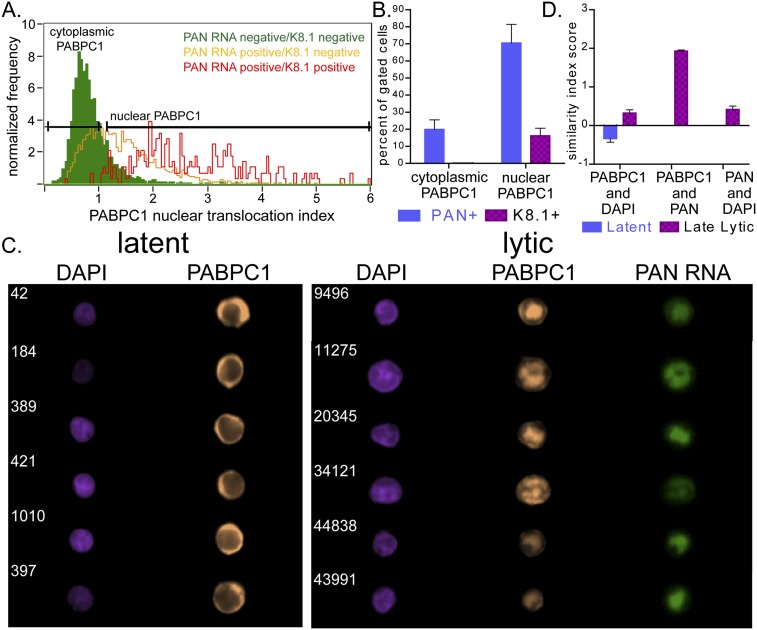
FIGURE 2.
(A) Histogram of PABPC1 nuclear translocation index in a heterogeneous population of BCBL1 cells treated with valproic acid for 48 h. Green, yellow, and red peaks represent the subpopulations of cells (1) expressing neither PAN RNA nor K8.1 protein, (2) expressing PAN RNA but not K8.1 protein, or (3) expressing both PAN RNA and K8.1 protein, respectively. (B) Percentage of cells expressing PAN RNA or K8.1 in subpopulations of cells with cytoplasmic versus nuclear-translocated PABPC1. (C) Representative images comparing similarity of DAPI and PABPC1 signals, PAN RNA and PABPC1 signals, or PAN RNA and DAPI signals in latent (left panel) or lytic (right panel) BCBL1 cells. (D) Similarity index score quantifying overlaps displayed in panel C. Columns in panels B and D represent the mean values (±SEM) of three independent experiments.
Next, using high-throughput MIFC and imaging analysis software, we measured the correlation between PABPC1 nuclear relocalization and PAN RNA expression in each cell within the entire population of cells, as well as colocalization of these molecules within each nucleus. As Figure 2B shows, 70.5% ± 6.3% of cells that displayed nuclear relocalized PABPC1 also exhibited robust expression of PAN RNA, in agreement with previous studies that calculated a similar percentage after manually scoring confocal images of cells (Borah et al. 2011). Furthermore, as evident in representative images in Figures 1B and 2C, the intranuclear patterns of PAN RNA and relocalized PABPC1 matched well, confirming the results from confocal microscopy (Fig. 1A; Borah et al. 2011). To quantify the intranuclear colocalization of PAN RNA and PABPC1 protein, we used a similarity index score (see Materials and Methods) to measure the pixel-by-pixel relative intensity for (1) PABPC1 and DAPI in latently infected cells, (2) PABPC1 and DAPI in lytically reactivated cells, and (3) PABPC1 and PAN RNA in lytically reactivated cells (Fig. 2D). A negative index score indicates poor colocalization of two molecules, while a positive index score indicates greater colocalization. The similarity index score of –0.34 ± 0.05 reflected the poor correlation of PABPC1 and DAPI signals in latently infected cells. PABPC1 and DAPI signals were more similar in lytically reactivated cells (similarity index score of +0.32 ± 0.04), reflecting the nuclear relocalization of PABPC1. However, the similarity index score of PABPC1 and PAN RNA during the lytic phase was significantly higher still (+1.93 ± 0.02). Figure 2C shows representative pairwise images of individual cells comparing DAPI and PABPC1 in latent phase (left panel), DAPI and PABPC1 in lytic phase (right panel), and PABPC1 and PAN in lytic phase (right panel). Thus, using MIFC, we were able to measure in a high-throughput quantitative manner the colocalization of RNA, DNA, and protein molecules.
Expression of PAN RNA allows for staging of lytic progression and tracking of PABPC1 nuclear translocation
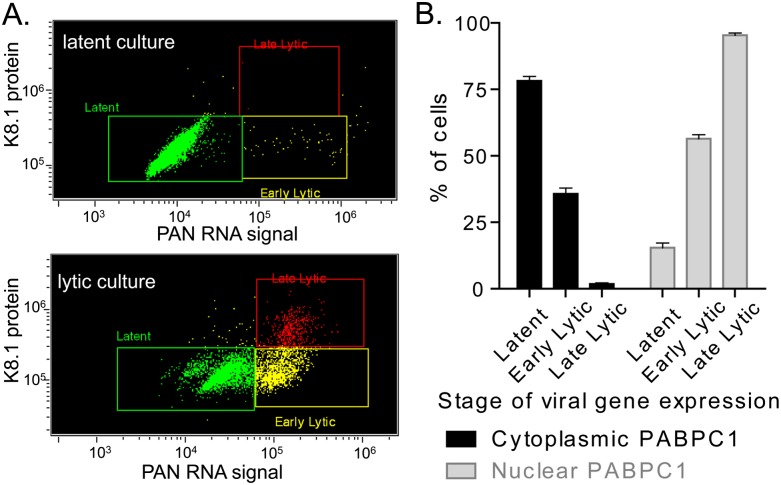
We next examined nuclear translocation of PABPC1 as a function of lytic phase progression. Although all cells in a given culture were treated with valproic acid for the same length of time, individual cells within that culture responded differently to the treatment, with some cells remaining refractory to induction and other cells progressing into various stages of the lytic phase (Daigle et al. 2010). Thus, within the same population of cells, different subpopulations could be identified as latent, early, or late stage of lytic infection. Since PAN RNA expression is evident in the early lytic phase while K8.1 protein expression occurs in late lytic phase, we designated cells with latent virus by virtue of their being both PAN-negative and K8.1-negative (60.9% ± 5.4% of the population) (Fig. 3A, top panel). Similarly, cells in early lytic state were identified as being PAN-positive and K8.1-negative (33.6% ± 6.0% of the population), and late lytic state cells were both PAN-positive and K8.1-positive (5.7% ± 0.8% of the population) (Fig. 3A, bottom panel). All cells that expressed K8.1 also expressed PAN RNA, consistent with the fact that PAN RNA is expressed earlier during lytic progression than K8.1 protein (Nakamura et al. 2003).
FIGURE 3.
(A) Scatter plots showing PAN RNA expression (x-axis) versus K8.1 protein expression (y-axis) in populations of latently infected (top panel) or lytically reactivated (bottom panel) BCBL1 cells. (B) Nuclear relocalization of PABPC1 as a function of progression into the lytic phase in a heterogeneous population of valproic acid–induced BCBL1 cells. Mean values (±SEM) of three independent experiments are shown.
Armed with the ability to distinguish cells in the early versus late state of lytic infection, we then scored for cytoplasmic versus nuclear PABPC1 localization as a function of lytic progression (for a description of the algorithm used, see Materials and Methods). Figure 3B shows that latent cells were enriched for cytoplasmic PABPC1 (78.2% ± 1.6%), whereas very few scored as having exclusively nuclear relocalized PABPC1 (15.5% ± 1.8%). Of cells in the early lytic stage of infection, about one-third (35.7% ± 2.1%) had cytoplasmic PABPC1, while just over half (56.5% ± 1.5%) of the cells in this category showed nuclear relocalization. In contrast, the overwhelming majority (95.4% ± 0.8%) of cells in the late stage of lytic infection scored positive for nuclear PABPC1. At each stage, a small percentage of cells fell between the defined cytoplasmic and nuclear gates and likely represent a transitional stage of PABPC1 localization.
DISCUSSION
MIFC has become an established technique for the high-throughput analysis of protein expression and localization in heterogeneous populations of cells (Adang et al. 2006; Basiji et al. 2007; Hassman et al. 2011). We report that use of MIFC can be expanded to include the analysis of RNA as well as protein molecules in the same cell population.
Only a few reports have used flow cytometry to study RNA expression. In one study, the investigators employed biotinylated, locked nucleic acid (LNA) probes to detect infection by Sindbis virus (Robertson et al. 2010). Although they demonstrated that labeling of the Sindbis genome by in situ hybridization was as effective a method for tracking viral infection as using a recombinant GFP virus, they were unable to study subcellular localization of the viral genome due to the intrinsic limitation of conventional flow cytometry to measure only fluorescence intensity and not localization. In another study, fluorescein-labeled peptide nucleic acid (PNA) probes were able to detect the EBV noncoding RNAs (EBERs) in EBV-infected cell lines and in T cells isolated from the blood of patients suffering from lymphoproliferative disorders (Kimura et al. 2009). In that study, immune cells were identified by staining also for cell surface markers. Similarly, a third study documented the simultaneous expression of EBERs and the viral BHLF1 mRNA in EBV-infected Raji cells by flow cytometry (Crouch et al. 1997). BHLF1 is a high-copy mRNA containing Not1 repeats; an oligonucleotide probe antisense to the repeated sequence produced an amplified in situ hybridization signal. To detect the two RNAs simultaneously in the same cells, the investigators labeled anti-EBER and anti-BHLF1 DNA oligonucleotide probes with digoxigenin and biotin, respectively, and detected with fluorescein-conjugated anti-digoxigenin or phycoerythrin-conjugated streptavidin, respectively. Finally, others have employed PNA probes to quantify telomere length in peripheral blood cells; increased telomere length yields increased binding of the anti-telomeric sequence probe, as detected by flow cytometry (Baerlocher et al. 2006).
Our study extends these previous findings in two ways. First, we used flow cytometry for high-throughput measurements of both RNA and intracellular protein expression. Previous flow cytometry studies of RNA and proteins have detected only cell surface proteins. Second, we validated the use of MIFC as a high-throughput approach to simultaneously detect and localize specific RNA and protein species within cells. This approach should be applicable to population-wide studies of RNA and protein expression and localization. For example, HIV-1 RNA serves as both the viral genome and the mRNA for producing viral proteins, with the genomic version of the RNA remaining nuclear, while the mRNA version is alternatively spliced and exported into the cytoplasm (Cullen 2003). Thus, in a population of HIV-infected cells, the progression of the viral RNA from the nuclear/genomic state to the cytoplasmic/mRNA state could be examined for each cell within the population. In these same samples, antibodies against virally encoded proteins could be used to examine the onset of viral protein expression specifically in those cells in which the HIV-1 RNA predominantly localizes in the cytoplasm. The effects of drugs or mutations deleterious to mRNA export might also be studied on a population-wide scale. Additionally, tracking the movement of lower abundance RNAs by MIFC may be facilitated by the use of tyramide-mediated signal amplification or by the tethering MBP-YFP to the 3′ UTRs of mRNAs, an elegant approach that has allowed visualization of mRNA export in live cells (Speel et al. 2006; Grunwald and Singer 2010). Such analyses will accelerate our understanding of regulatory processes in which low-abundance noncoding RNAs are increasingly implicated in critical roles.
MATERIALS AND METHODS
Growth, induction, and staining of BCBL1 cells for RNA and protein molecules
BCBL1 cells were grown in RPMI supplemented with penicillin, streptomycin, L-glutamine, and 20% fetal bovine serum. To induce KSHV lytic phase, cells were grown to a density of 0.8–1.0 million/mL, and then valproic acid was added to the culture at a final concentration of 600 μM for 48 h.
Staining of latent and lytic BCBL1 cells for confocal imaging was performed as previously described (Borah et al. 2011).
To stain latent and lytic BCBL1 cells for MIFC analysis, 100 million cells, or 30 million cells if samples were labeled only by in situ hybridization or only by protein immunofluorescence, were pelleted by centrifugation at 1800g for 10 min at room temperature. Cells were fixed with 4% formaldehyde in PBS on ice for 30 min in 15 × 85-mm borosilicate glass tubes (Fisher) that had been presiliconized using SigmaCote (Sigma). Cells were pelleted by spinning at 1800g for 5 min at 4°C in a Sorvall RC-6+ centrifuge using an SS-34 rotor fitted with rubber insulators that matched the diameter of the glass vials. Pellets were washed twice with cold PBS, resuspended in 900 μL PBST (PBS + 0.2% Triton-X) per 100 million cells, and incubated on ice for 10 min. Cells were washed twice more with cold PBS, resuspended in 900 μL PBST + 1% BSA per 100 million cells, and preblocked for 30 min on ice. Then, primary antibodies were added directly to the cell suspension at a dilution of 1:800 for the anti-PABPC1 rabbit antibody (Santa Cruz) and 1:800 or 1:1000 for the anti-K8.1 mouse antibody (Advanced Biotechnologies). Cells were incubated with primary antibody on ice for 1 h with intermittent dispersal to prevent cells from settling to the bottom of the tube. Then, cells were pelleted and washed twice with cold PBST prior to addition of secondary antibodies in PBST + 1% BSA at a dilution of 1:400 for the anti-rabbit Alexafluor 594 antibody (Invitrogen) and 1:1000 for the anti-mouse Alexafluor 647 antibody (Invitrogen). After two washes with cold PBST, cells were fixed again in 4% formaldehyde for 15 min on ice, washed twice in PBS, and then incubated in PBST for 10 min on ice.
For in situ hybridization, BCBL1 cells were washed once with room temperature PBS and once with room temperature 2× SSC (30 mM sodium citrate, 300 mM NaCl at pH 7.0). Then cells were resuspended in 600 μL prehybridization solution (50% formamide, 10% dextran sulfate, 2× SSC, 0.1% RNase-free BSA [Ambion], 500 μg/mL salmon sperm DNA [Invitrogen], 125 μg/mL Escherichia coli tRNA [Roche], and 1 mM vanadyl ribonucleoside complexes [New England Biolabs]) per 100 million cells and incubated for 1 h at 37°C. After prehybridization, 45 ng each of DIG-labeled anti-PAN RNA oligonucleotides (SB2, SB85 and SB88) in 10 μL dH2O was heated for 2 min to 95°C, snap cooled on ice, and added to 100 million cells for overnight incubation at 37°C.
The next day, 1.5 mL of RNase-free dH2O was added per tube to reduce the viscosity of the solution, and cells were pelleted as before. Pellets were washed twice at room temperature with 2× SSC, followed by two more washes with room temperature 1× SSC. Cells were fixed at room temperature with 4% formaldehyde for 15 min, followed by two washes with cold PBS. Then cells were resuspended in 900 μL PBST containing 1:1000 anti-digoxigenin antibody (Jackson Laboratories) per 100 million cells and incubated on ice for 1 h. Cells were washed twice with PBST, once with cold PBS, fixed for 10 min with 4% formaldehyde in PBS, and washed twice with cold PBS and stored at 4°C. Prior to analysis by MIFC, cells were pelleted and resuspended in 0.4 μg/mL of DAPI in PBS.
Anti-PAN RNA oligonucleotide probes for in situ hybridization were as follows: SB2, 5′-ACAAATGCCACCTCACTTTGTCGC-3′; SB85, 5′-CGCTGCTTTCCTTTCACATT-3′; SB88, 5′-GTGAAGCGGCAGCCAAGGTGACTGG-3′ (IDT). A second-generation DIG oligonucleotide labeling kit (Roche) was used as per manufacturer's instructions to label oligonucleotides used as probes for in situ hybridization.
Data collection and analysis
Prior to cytometry, images of stained BCBL1 cells were collected by confocal microscopy. For this, cells stained in solution were fixed onto glass slides pretreated with poly-L-lysine (Sigma-Aldrich), mounted with VectaShield medium (Vector Laboratories), and imaged using a 63× objective on a Leica TCS SP5 confocal microscope, as previously described (Borah et al. 2011).
Stained cells were then imaged on an Imagestream imaging cytometer (Amnis) with subsequent data analysis done using IDEAS 4.0 software. Single-plane images were analyzed, as opposed to images obtained from an extended depth of field (EDF) (as described by Ortyn et al. 2007; Hassman et al. 2011), because the use of EDF is best suited for analysis of cells with limited numbers of punctate foci each with distinct x- and y-coordinates. If, instead, signals in each cell are either too numerous such that they share x- and y-coordinates and are separated only in the z-axis (depth of cell) and/or are dimensionally large, EDF compression results in the merging of signals. Patterns for PAN RNA and nuclear-translocated PABPC1 are numerous, bright, and globular and, therefore, are best quantified by single-plane imaging over each cell population.
Single cells were identified based on aspect ratio, or roundness, and size. Focused cells were identified based on image contrast and the gradient root mean square (RMS) function of the imaging software, using brightfield images. Gradient RMS is a measure of the change in pixel intensity from the background. The greater the slope of this change, reflecting the effective contrast with the background, the more in focus that cell will be. Viable cells were gated based on their staining positively for both DAPI and PABPC1. The algorithm used to measure translocation of PABPC1 into the nucleus from the cytoplasm was designed to compare the ratio of PABPC1 intensity within the nucleus to the intensity of PABPC1 in the cytoplasm, using nuclear margins determined by the region of DAPI staining. This intensity ratio was defined as the PABPC1 “nuclear translocation index.” A PABPC1 nuclear translocation index greater than 1.2 defined cells with primarily nuclear PABPC1, while a PABPC1 nuclear translocation index less than 1.1 defined cells with primarily cytoplasmic PABPC1. These values were chosen based on a natural demarcation within the population of cells between these values, as seen in Figure 2A, as well as manual inspection of PABPC1 localization in cells with a nuclear translocation index greater than 1.2 or less than 1.1.
Colocalization of two signals (i.e., PABPC1 and PAN) was calculated by measuring the pixel-by-pixel intensity of each signal within the nucleus to obtain a similarity index score. For comparison, the colocalization of PABPC1 and PAN is compared with the colocalization of either of these signals individually with the DNA stain, DAPI.
Statistical analyses employed Prism 4.0 for Macintosh (GraphPad Software Inc.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.033126.112.
REFERENCES
- Adang LA, Parsons CH, Kedes DH 2006. Asynchronous progression through the lytic cascade and variations in intracellular viral loads revealed by high-throughput single-cell analysis of Kaposi's sarcoma-associated herpesvirus infection. J Virol 80: 10073–10082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerlocher GM, Vulto I, de Jong G, Lansdorp PM 2006. Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat Protoc 1: 2365–2376 [DOI] [PubMed] [Google Scholar]
- Basiji DA, Ortyn WE, Liang L, Venkatachalam V, Morrissey P 2007. Cellular image analysis and imaging by flow cytometry. Clin Lab Med 27: 653–670 viii [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S, Darricarrere N, Darnell A, Myoung J, Steitz JA 2011. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog 7: e1002300 doi: 10.1371/journal.ppat.1002300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff C, Schulz TF, Kennedy MM, Graham AK, Fisher C, Thomas A, McGee JO, Weiss RA, O'Leary JJ 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med 1: 1274–1278 [DOI] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci 102: 5570–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias S, Gaglia MM, Kumar GR, Wong W, Jackson AO, Glaunsinger BA 2011. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PLoS Pathog 7: e1002339 doi: 10.1371/journal.ppat.1002339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch J, Leitenberg D, Smith BR, Howe JG 1997. Epstein-Barr virus suspension cell assay using in situ hybridization and flow cytometry. Cytometry 29: 50–57 [PubMed] [Google Scholar]
- Cullen BR 2003. Nuclear RNA export. J Cell Sci 116: 587–597 [DOI] [PubMed] [Google Scholar]
- Daigle D, Megyola C, El-Guindy A, Gradoville L, Tuck D, Miller G, Bhaduri-McIntosh S 2010. Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors. J Virol 84: 993–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande JP, et al. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci 96: 4546–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu Rev Pathol 1: 273–296 [DOI] [PubMed] [Google Scholar]
- Glaunsinger B, Ganem D 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell 13: 713–723 [DOI] [PubMed] [Google Scholar]
- Gorlach M, Burd CG, Dreyfuss G 1994. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res 211: 400–407 [DOI] [PubMed] [Google Scholar]
- Grunwald D, Singer RH 2010. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467: 604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassman LM, Ellison TJ, Kedes DH 2011. KSHV infects a subset of human tonsillar B cells, driving proliferation and plasmablast differentiation. J Clin Invest 121: 752–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Miyake K, Yamauchi Y, Nishiyama K, Iwata S, Iwatsuki K, Gotoh K, Kojima S, Ito Y, Nishiyama Y 2009. Identification of Epstein-Barr virus (EBV)–infected lymphocyte subtypes by flow cytometric in situ hybridization in EBV-associated lymphoproliferative diseases. J Infect Dis 200: 1078–1087 [DOI] [PubMed] [Google Scholar]
- Kumar GR, Glaunsinger BA 2010. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol Cell Biol 30: 4996–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GR, Shum L, Glaunsinger BA 2011. Importin α-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol Cell Biol 31: 3113–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort S, Flamand L 2009. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is necessary for lytic viral gene expression, DNA replication, and virion production in primary effusion lymphoma cell lines. J Virol 83: 5869–5880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, MacKey J, Czajak SC, Desrosiers RC, Lackner AA, Jung JU 1999. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J Virol 73: 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223 doi: 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol 77: 4205–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortyn WE, Perry DJ, Venkatachalam V, Liang L, Hall BE, Frost K, Basiji DA 2007. Extended depth of field imaging for high speed cell analysis. Cytometry A 71: 215–231 [DOI] [PubMed] [Google Scholar]
- Paulose-Murphy M, Ha NK, Xiang C, Chen Y, Gillim L, Yarchoan R, Meltzer P, Bittner M, Trent J, Zeichner S 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J Virol 75: 4843–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne R, Blackbourn D, Whitby D, Levy J, Ganem D 1998. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol 72: 5182–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KL, Verhoeven AB, Thach DC, Chang EL 2010. Monitoring viral RNA in infected cells with LNA flow-FISH. RNA 16: 1679–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto CC, Pari G 2012. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog 8: e1002680 doi: 10.1371/journal.ppat.1002680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J Virol 79: 9301–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarelli R, Farina A, Granato M, Gonnella R, Raffa S, Leone L, Bei R, Modesti A, Frati L, Torrisi MR, et al. 2008. Identification and characterization of the product encoded by ORF69 of Kaposi's sarcoma-associated herpesvirus. J Virol 82: 4562–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speel EJ, Hopman AH, Komminoth P 2006. Tyramide signal amplification for DNA and mRNA in situ hybridization. Methods Mol Biol 326: 33–60 [DOI] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Miller G 1996. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci 93: 11883–11888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Ganem D 1997. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J Virol 71: 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, Chong JM, Wu L, Yuan Y 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J Virol 79: 800–811 [DOI] [PMC free article] [PubMed] [Google Scholar]