Abstract
Recent advances in high-throughput biotechnologies have led to the rapid growing research interest in reverse engineering of biomolecular systems (REBMS). ‘Data-driven’ approaches, i.e. data mining, can be used to extract patterns from large volumes of biochemical data at molecular-level resolution while ‘design-driven’ approaches, i.e. systems modeling, can be used to simulate emergent system properties. Consequently, both data- and design-driven approaches applied to –omic data may lead to novel insights in reverse engineering biological systems that could not be expected before using low-throughput platforms. However, there exist several challenges in this fast growing field of reverse engineering biomolecular systems: (i) to integrate heterogeneous biochemical data for data mining, (ii) to combine top–down and bottom–up approaches for systems modeling and (iii) to validate system models experimentally. In addition to reviewing progress made by the community and opportunities encountered in addressing these challenges, we explore the emerging field of synthetic biology, which is an exciting approach to validate and analyze theoretical system models directly through experimental synthesis, i.e. analysis-by-synthesis. The ultimate goal is to address the present and future challenges in reverse engineering biomolecular systems (REBMS) using integrated workflow of data mining, systems modeling and synthetic biology.
Keywords: reverse engineering biological systems, high-throughput technology, –omic data, synthetic biology, analysis-by-synthesis
INTRODUCTION
Recent advances in high-throughput biotechnologies have impacted the progress in reverse engineering biological systems, especially for biochemical pathways and cells. The large volume of biochemical data at molecular-level resolution has made ‘data-driven’ approaches possible, where patterns may be extracted directly from the data to reveal new insight [1–6]. Such data-driven approaches are also known as ‘data mining’. At the same time, ‘design-driven’ approaches are also increasingly used to replicate features of biological systems such as scale-free organization [7], motif distribution [8] and feedback [9] so as to simulate and abstract emergent properties of biological systems [10, 11]. Such design-driven approaches may also be called ‘systems modeling’. Thus, high-throughput data mining coupled with design-driven systems modeling for biochemical pathways and cells represents a new era of reverse engineering biological systems, which we refer to as ‘reverse engineering biomolecular systems’ (REBMS) in this context (Figure 1).
Figure 1.
Integrated pipeline for reverse engineering biomolecular systems (REBMS) using high-throughput –omic data that combines data mining, modeling and simulation and testing and validation
On the whole, the key challenges in REBMS are to (i) integrate heterogeneous biochemical data for data mining [12–16], (ii) combine top–down and bottom–up approaches for systems modeling [17–22] and (iii) validate system models experimentally [23–28]. In this article, we first present an overview of common high-throughput platforms to recap existing data acquisition issues in Section 2. In Section 3, we review challenges and strategies for data mining and systems modeling, and in Section 4, we illustrate some recent progress made in the field of REBMS using two case studies. We close in Section 5 by exploring the emerging field of synthetic biology, which is an exciting approach to analyze and validate theoretical system models directly through experimental synthesis.
DATA ACQUISITION AND HANDLING
In this section, we review high-throughput data acquisition technologies for genomic, proteomic, metabolomic and cytometry data such as microarrays [29], next-generation sequencing (NGS) [30], mass spectrometry [31] and microfluidics [32]. It is important to recap existing issues encountered during data acquisition in the hope of recognizing the potential ‘garbage in—garbage out’ problem during downstream mining, modeling and validation. Then, while recognizing that ever-increasing data volumes present associated challenges in data handling such as storage security, retrieval efficiency, manageability of databases [33], we also review minimum information standards in particular because of its development as a community-driven effort to manage and integrate different data types and formats through standardized database schema and visualization [15].
High-throughput omic data acquisition
DNA microarrays [29] are a common high-throughput technology to acquire genomic data, namely gene expression, at relatively low cost. Data artifacts in microarrays regularly arise due to hybridization and spatial effects [34], slide variation [35] and dye-related signal correlation bias in two-color assays [36]. Subsequently, various engineering and statistical solutions have been developed to handle these artifacts [37, 38]. Recently, NGS technologies, such as Illumina/Solexa, Roche/454 and SOLiD, have also emerged as high-throughput genomic data acquisition platforms but not without some technical and analytical issues. NGS data volume is much larger than that of DNA microarray data, leading to issues in data storage, transfer and analysis [39]. However, it is still difficult to determine expressions from NGS data because of the low specificity of sequence alignment and the presence of splice variants [40]. Accordingly, several algorithms and tools have also been developed to address such NGS-specific issues, namely to improve the accuracy of sequence alignment, quantifying gene expression and to present sequence data visually [41–44]. Two common types of transcriptome assembly strategies are: (i) reference-based algorithms, such as SpliceMap [45], Blat [46] and TopHat [47], that work with a reference genome for assembly, (ii) the so called ‘de novo’ strategies, such as Trans-ABySS [48] and Trinity [49], that work by traversing collapsed De Bruijn graphs constructed from all substrings of reads with a specific length. In recent years, the advent of these methods in conjunction with high performance computing frameworks provided us with a complete landscape of full set of transcripts (Figure 1).
For proteomic and metabolomic data, mass spectrometry is a popular technology with applications ranging from proteomic characterization to perturbation analysis of model organisms [31, 50]. Furthermore, recent research in tissue imaging mass spectrometry (TIMS) effectively adds a second dimension to traditional mass spectrometry data by acquiring multiple mass spectra at different spatial locations of the same tissue sample. Consequently, TIMS may be used to study spatial progression by identifying and quantifying biomarkers of interest that are associated with particular tissue regions. For instance, the human cancer proteome may be analyzed in situ using MALDI TIMS [51] and TIMS image pixels may be correlated spatially with mass spectra clustering to highlight functionally similar regions in tumors [52], among other applications [53–55]. Detection sensitivity and data reproducibility [56–58] remain problematic for mass spectrometry.
On the level of whole cells, microfluidics is another high-throughput platform that enables the study of live cells, including unicellular organisms, in both population and single-cell settings by manipulating various chemical and physical conditions of the micro-environment. The functionality of microfluidics that enables a variety of experiments to be performed on a single device is also sometimes referred to as ‘lab-on-a-chip’. While microfluidics facilitates real-time observation of live cells under real and artificial conditions [32, 59], it remains challenging to design and manufacture appropriate chip features so as to capture the phenotype or behavior of interest from a given group of cells.
Minimum information standards
The development of minimum information standards is a community-driven effort to provide guidelines for sharing high-throughput –omic data in the challenge of large datasets. The goal of these standards is to guide researchers to systematically annotate experimental protocols and results. Different standards have been proposed for different types of data and different studies. For example, the MIAME (Minimum Information About a Microarray Experiment) and MIFlowCyt (Minimum Information about a Flow Cytometry Experiment) standards are based on specific data acquisition modalities [60, 61], while MIAMET (i.e. Minimum Information About a Metabolomics Experiment) and MINI (i.e. Minimum Information about a Neuroscience Investigation) are based on specific research areas [62, 63]. For REBMS, MIRIAM (Minimum Information Requested In the Annotation of biochemical Models) provides standards for describing and sharing computational models [64], while MIASE (i.e. Minimum Information About a Simulation Experiment) provides a framework to document in silico, or simulation, experiments performed using a model [65].
Some minimum information standards are now well-accepted by the community-at-large. For example, adherence to MIAME is required by a number of high-profile journals, including Nature and Science [66]. Additionally, some curated biological databases encourage data submissions adhering to these community standards; Table 1 presents a selected list of these databases. Accepted minimum information standards may also guide the development of other community standards; for example, accommodation of MIRIAM annotations in Systems Biology Markup Language (SBML) [67, 68]. However, because minimum information standards are developed by various interest groups, there is some uncertainty with regard to the appropriate standards for interdisciplinary research. The MIBBI portal (http://mibbi.org) makes an attempt to address this problem by providing a resource hub for minimum information standards. The MIBBI project aims to promote the accessibility, transparency and collaborative nature of interdisciplinary research by providing a central resource, from which standards can be tracked, compared, coordinated and adopted [61]. At the time of writing, 32 minimum information projects are registered in the MIBBI portal.
Table 1:
Resources for REBMS using –omic data
| Type | Name | URL | Description |
|---|---|---|---|
| Genomic data | GenBank | www.ncbi.nlm.nih.gov/genbank | Annotated collection of all publicly available DNA sequences; international collaboration among USA, Europe and Japan |
| EMBL Nucleotide Sequence Database | www.ebi.ac.uk/embl | ||
| DNA Database of Japan (DDBJ) | www.ddbj.nig.ac.jp | ||
| Proteomic data | UniProt | www.uniprot.org | Protein sequence and function |
| BRENDA | www.brenda-enzymes.org | Enzyme functional data | |
| SABIO-RK | sabio.villa-bosch.de | Biochemical reactions, kinetic equations with parameters and experimental conditions under which these parameters were measured | |
| Metabolomic data | LIPIDMAPS | www.lipidmaps.org | Comprehensive resource for lipid biology, e.g. structure and lipid-associated proteins |
| Pathways | KEGG | www.genome.jp/kegg | Integrated resource for building blocks and functions of biological systems |
| Reactome | www.reactome.org | Open-source, manually curated and peer-reviewed pathway database | |
| System models | BioModels | www.ebi.ac.uk/biomodels-main | Peer-reviewed, published, computational models from systems biology |
| Portal sites | Pathguide | www.pathguide.org | Pathway- / molecular interaction-related |
| ExPASy | expasy.org | Bioinformatics |
DATA MINING AND SYSTEMS MODELING
Data mining and systems modeling are complementary efforts for REBMS. Data mining processes large datasets by extracting correlated data features, while systems modeling creates a unified theoretical framework, through time and space, by aggregating and associating relevant data features to predict other system behaviors.
Challenges and strategies for data mining
Data mining typically involves (i) exploratory analysis, i.e. discovering biologically interesting relationships in the data, (ii) feature selection, i.e. mining for biomarkers or interesting features and (iii) prediction, i.e. building statistical models for either diagnostic/prognostic applications or predicting system behavior. Exploratory analysis methods can reveal the natural organization of high-throughput data and identify patterns [69]. The most common unsupervised clustering method applied to high-throughput data is hierarchical clustering [70], others include k-means clustering [71], principal component analysis (PCA) [72] and bi-clustering [73]. For example, unsupervised clustering identifies groups of genes with similar expression patterns, or groups of samples with similar molecular profiles [74]. On the other hand, many high-throughput data experiments generate samples with known underlying grouping or clustering information. Thus, we can use this information to supervise the identification of features that are important for modeling. Many supervised feature selection methods exist for mining gene expression data, but these methods may be applied to high-throughput data in general [75]. Exploratory analysis and feature selection ultimately lead to prediction analysis, an important aspect of systems modeling. Regardless of the biological application (e.g. clinical diagnosis/prognosis or systems modeling), statistical modeling must be systematically evaluated to overcome challenges and avoid common pitfalls [76].
A well-known issue in –omic data mining is ‘curse-of-dimensionality’, where the number of biological samples is significantly smaller than the number of feature dimensions [77]. Especially for REBMS, the power of data mining is limited by this issue. Pooling several datasets across different studies for training to increase sample size and filling in missing data may improve data quality and quantity [78]. Extensive cross-validation during training is also proposed to overcome the limits of poor data such as small sample size and sample noise [79, 80]. In a recent issue of Nature Biotechnology, using DNA microarrays as a common high-throughput platform to measure gene expression, multiple challenges for data mining such as ‘batch effect’, ‘cross-platform difference’, ‘classifier selection’, ‘performance metric’ and ‘prevalence’ etc., and strategies to counter these challenges were studied by an international consortium consisting of 100 s of data mining researchers across 36 institutions. The outcome of this study is the publication of sensible ‘good-practice’ guidelines for each of these issues [78, 81–84].
Models must be predictive in nature in order to be scientifically useful [85]. Recently, omic-based predictive models have received increased attention in the scientific literature because of their potential for clinical applications [84, 86, 87]. That is, for high-throughput measurement such as genomic (or proteomic) expression, data mining is expected to lead to diagnostic and prognostic models that can predict disease state. Thus, another challenge in data mining is to ensure consistent performance of prediction models across both training and testing datasets (i.e. the cross-validation performance within training datasets is similar as the external-validation performance with testing datasets). Simon et al. and Quackenbush [76, 86] examine key steps and common pitfalls involved in building and evaluating predictive models. Simon stresses the importance of correctly estimating the accuracy of prediction models on future samples. This involves proper division of samples into training and testing sets before any analysis, so that none of the test samples are used in training predictors. Michiels et al. [88] reinforce this recommendation after re-analyzing several large cancer prediction studies. Their results show that many of these studies predict no better than random chance, and the selection of features greatly depends on the samples. They recommend a method of repeated random sampling to better estimate the mean and variance of prediction error. Besides, the process for building and evaluating predictive models, and identifying factors that affect predictive performance has been studied extensively [76, 84, 89, 90]. The observation is that, for genomic-based diagnostic models, prediction performance is difficult to evaluate. It involves either (i) retrospective evaluation after collecting an adequate number of future samples, or (ii) estimation of predictive performance using cross validation or other statistical sampling methods [91–93]. Despite the difficulties and pitfalls involved in building predictive models, -omic predictor models are feasible for diagnostic or prognostic in clinical use [94]. Once the features are extracted from the high-throughput data, they can be analyzed to build networks for REBMS.
Systems modeling—static network inference
Static network inference is the process of reconstructing the topology of biological systems to represent data features (e.g. genes, proteins or metabolites) as nodes and interactions as edges to predict new interactions. Static networks can be deterministic or probabilistic. However, they are only able to model a snapshot of complex dynamics of biological systems. The availability of extensive databases and sharing of high-throughput datasets has promoted genome-scale static network modeling. The modeling methods include correlation networks and Bayesian networks etc. The correlation network is one of the simplest models [95] and has low computational cost: two genes are predicted to interact if their expression profiles are highly correlated. Several other network inference algorithms such as ARCANE [96] and RELNET [97], which are based on mutual information have also been proposed. The limitation of correlation networks is that only instances of pairwise interaction are captured, and epistasis, where one gene interacts with several other genes, is not. Bayesian networks are also increasingly used for static network inference. The power of Bayesian networks comes from the provision to capture uncertainty in any contributing factor, including model parameters, hidden variables and observations, using probabilistic methods. This makes Bayesian inference a useful framework to model inherent uncertainty in biological data, which may be a result of biological or technical variability [98]. Bayesian networks may also be used to model epistasis where standard statistical tools such as multivariate logistic regression models have failed [99, 100].
In recent disease-centered applications of static network inference, Sebastiani et al. [100] used Bayesian networks to identify SNPs predictive of stroke in sickle cell patients; Tran et al. [101] used Bayesian networks in combination with copy number variation analysis to identify potential driver genes in breast cancer; and Carro et al. [102] used the mutual information-based algorithm ARACNE to identify key transcriptional regulators of the mesenchymal cellular phenotype in gliomas, which is associated with more aggressive disease and poorer prognosis. In another study, Faith et al. [103] used both their own microarray data and data from nine other publications to perform genome-scale network inference on Escherichia coli, a model organism for which substantial information about transcriptional regulatory interactions is known. Existing data was thereby used as a test-bed for evaluating the performance of network inference algorithms. The authors showed that at 60% precision, the predictions of the best-performing algorithm, based on mutual information, included 338 known interactions documented in the database RegulonDB and 741 novel interactions. Of the predictions at all precision levels, 268 interactions were experimentally tested via ChIP and 21 were confirmed. In another recent study, Zhu et al. [104] performed large-scale gene network inference by integrating genotypic, gene expression, transcription factor-binding site and protein–protein interaction datasets in yeast, which is one of the few model organisms for which comprehensive high-throughput datasets of multiple types are available. The authors showed that in terms of predicting known causal regulators in gene sets, the Bayesian networks constructed using multiple data types were superior to the Bayesian network based on gene expression data only; in addition, five previously unknown interactions made by the Bayesian network built using all four data types were experimentally confirmed.
While static network inference has produced promising results, it still faces several challenges: (i) when performing genome-scale analysis, the number of possible parameters to be found can greatly exceed the number of data points, which leads to high computational complexity; (ii) limited data availability from the available measurement may require simplifying assumptions about the complex, non-linear biological mechanisms [105], and (iii) algorithm performance can vary greatly, even for the same algorithm applied to different datasets. Marbach et al. [106] recently published the results of the DREAM3 in silico community-wide challenge, in which simulated network data was used to perform double-blind tests of gene network inference methods. Notably, a large proportion of the tested methods (11 of 29) performed poorly, with results not significantly improved over random guessing. Additionally, systemic errors in prediction were associated with ‘fan-in’, ‘fan-out’, ‘cascade’ and ‘feed-forward loop’ motifs, which describe situations including coregulation and combinatorial regulation. Similar variation in performance was linked to the properties of different biological networks by Ooi and Phan [107], who recently used known gene– and protein–scale interactions from enriched pathways of different classes in keloid fibroblasts to compare the performance of ARACNE [96] and BANJO (Bayesian Network Inference with Java Objects) [108]. Their findings indicated that the transcriptional networks were better suited to network inference analysis [107]. These results indicate that there are substantial opportunities in researching static network inference of REBMS such as to develop increasingly practicable methods that take into account fundamental biochemical mechanisms and motifs.
Systems modeling—network dynamics
REBMS is not complete without modeling system dynamics in addition to static network topology. A fully deterministic model that can be used to capture the complex dynamics of biological systems is the Boolean network. It is noteworthy to mention that models can be categorized in different types. They can be considered as static versus dynamic or deterministic versus probabilistic. For example, the Bayesian network is a probabilistic network that is static, but the Boolean network is a deterministic rule-based model that can capture dynamics. The Boolean network model [109] is a major effort in gene regulatory network inference, where gene expression is quantized to only two states: ON and OFF. The Boolean state of each gene is functionally related to other genes using Boolean functions. This assumes a fully deterministic environment that does not account for innate uncertainty in biological systems, data and model selection [110]. Beside this, the use of Boolean networks for network topology inference is limited by two major challenges: (i) gene expression cannot be described adequately by only two states and (ii) inference of Boolean functions for a large number of genes is computationally expensive. Introducing probabilistic Boolean networks was a major breakthrough to capturing not only dynamics of biological systems but also uncertainty in data, model selection and biological systems [110]. As the probabilistic Boolean network is the ‘unrolled’ version of Boolean networks in time, the dynamic Bayesian network [111] is the counterpart for the standard Bayesian network. Besides probabilistic Boolean networks and dynamic Bayesian networks, Ordinary Differential Equations (ODEs) have been used as an alternative to model gene regulatory networks. Tyson et al. [112] and Goodwin [113] have used non-linear differential equations to reverse engineer a kinetic model of gene regulation process. There are some issues in modeling biological systems dynamics using linear and non-linear ODEs. First, these models need a detailed a priori knowledge about the form of reaction rate functions usually not available in practice. Second, the limited number of samples commonly available for these studies makes inference of parameters used in reaction mechanisms very difficult. Piecewise-linear differential equations have been proposed to alleviate the aforementioned problems for modeling reaction mechanisms [18, 114]. Savageau [20, 115] has proposed power-law simplification of non-linear differential equations as an alternative approach. Furthermore, the stochastic master equation has been used to capture the delays induced by various reaction rates of biochemical processes [21].
Because reaction rate parameters are critical to these traditional models, the methods for modeling systems dynamics are sometimes reduced to parameter estimation problems. Examples of reaction rate parameters include rate constants for mass action kinetics [116], kinetic orders in biochemical systems theory [20, 115], stoichiometric coefficients and flux rates for flux balance analysis [117] and control coefficients and elasticities for metabolic control analysis [118]. Parameter estimation cannot be avoided and can easily lead to the problem of data over-fitting. In cases of over-fitting, parameters are overly sensitive to data noise, and fitted values differ clearly from measured values [119, 120]. In biological systems measured by high-throughput ‘-omics’ technologies, significantly more parameters are required than measurements available in terms of temporal points and in terms of number of samples. Thus, the parameter estimation problem is worse. In addition, dynamic biochemical systems need to be able to predict the behavior of real systems and to extrapolate components of the system at future time points.
Based on the observation that certain emergent properties in biological systems, namely robustness, resemble properties of engineering systems, this has led to a recent (or renewed) wave of efforts in REBMS in terms of cybernetics and control theory [10, 11, 121]. Cybernetics is the study of structural complexity in animal and machine that enables communication and control [122] and is closely related to control theory. In the context of biological systems, this approach was first applied to study organ systems in physiology, for instance, in circulation [123], immunology [124] and the central and peripheral nervous systems [125, 126]. More recently, cybernetics has been applied to study metabolic pathways [17, 127] at molecular levels. While cybernetics emerged as the science for effective organization within systems, control theory was developed to guide the behavior of dynamic systems. On the other hand, control, i.e. regulation of motion, is sufficient to achieve system stability and robustness. For instance, bacterial chemotaxis in E. coli is an example of relatively simple but robust cell behavior that may be modeled in terms of classical control theory. Specifically, exact adaptation in bacterial chemotaxis is observed to be robust and not affected by changes in protein levels [128, 129]. Consequently, such behavior may be modeled as the result of single-variable integral control that ensures convergence to steady state without error [22], which is a global systems property. Furthermore, details of possible mechanisms that enable the robust behavior can be probed using time-varying stimuli [130]. Modern control theory may also be useful to model regulatory behaviors in larger metabolic pathways [19]. Although recent studies do show promise in the use of cybernetics and control theory to model dynamics in REBMS, at the same time, the key challenge remains: conditions for robustness established using cybernetics and control theory are sufficient but not necessary. Thus, substantial experimental validation is required to prove the uniqueness of proposed system models for REBMS.
PROGRESS IN REBMS: TWO CASE STUDIES
In this section, we present two case studies to illustrate recent progress in the field of REBMS. In the first case study, we discuss the data mining and analysis of genomic interactions in breast cancer by integrating gene expression, SNP, CNV and DNA methylation data. In the second case study, we discuss the systems modeling of robust adaptation in bacterial chemotaxis by integrating bottom–up and top–down approaches. Ultimately, for REBMS using –omic data, integrating various data and methods is necessary and most likely fruitful.
Integrative analysis of genomic interactions in REBMS of breast cancer
To date, there are several studies that attempt to use integrative omic analysis to understand human diseases, most of which were focused on cancer due to the availability of disease tissue besides blood samples [131–137].
A recent published paper by the Cancer Genome Atlas Research Network [137] conducted integrative analysis of the single nucleotide polymorphism (SNP), copy number variation (CNV), expression profiling of coding and non-coding RNAs and global DNA methylation profiling datasets with 309 clinically annotated high grade serous ovarian cancer patients and an additional 180 patients with all but SNP data. The authors mapped the original DNA reads to the human genome, excluding duplicate reads, used CHASM [138] and MutationAssessor to identify functional mutations, used GISTIC analysis to extract quality copy number data, used consensus clustering [139] to analyze mRNA, miRNA and methylation data, used HotNet [140] to analyze protein–protein interactions and used PARADIGM [141] to estimate differential integrated pathway activity. They found that late stage ovarian cancer tumors have specific mutations of interest at both high and low prevalence; TP53 mutations were found in 96% of tumors and somatic mutations in nine genes including BRCA1, BRCA2, CDK12, NF1, RB1 and were statistically recurrent, despite low prevalence. They went on to identify 113 significant copy number aberrations in DNA and 168 genes with promoter methylation events. Furthermore, they characterized four subtypes of ovarian cancer transcription, three subtypes of miRNA and four subtypes of promoter methylation. Survival duration was found to be associated with a transcriptional signature, explaining the impact of BRCA1/2 and CCNE1 aberrations on tumor survival. Lastly, in about half of the tumors considered, they identified defects in the homologous recombination pathways, suggesting a role for Notch and FOXM1 in the pathophysiology of serous ovarian cancer. The outcomes of this case study show that in the era of high-throughput data, it is possible to integrate different data to derive a holistic view of the biomolecules for REBMS.
Modeling robust adaptation in bacterial chemotaxis
The E. coli chemotaxis network is a model system that has been successfully reverse engineered in terms of (i) establishing a framework to describe robustness of a biological system and (ii) elucidating the mechanisms that may be responsible to achieve the robustness. That is, in bacterial chemotaxis, it is observed that the steady-state tumbling frequency in a homogeneous ligand environment is insensitive to the value of ligand concentration [129]. Previously, researchers thought that the rates of specific network reactions need to be fine-tuned in order to maintain the same steady-state behavior under different conditions [142]. However, more recent studies suggest that adaptation is a robust network property that is independent of particular rates of reaction [128, 129] and the robust adaptation in chemotaxis may be the result of integral feedback [22], which is a common setup in engineering modeling that ensures the convergence to the desired steady-state.
The robust adaptation in bacterial chemotaxis by feedback has led to the integration of bottom–up and top–down approaches in systems modeling. From a bottom–up perspective, individual network components (i.e. biochemical reactions) are quantified and characterized using rates of reaction. Because these individual reactions constitute the network, the observed system properties arise directly from adding all pieces of biochemical reactions together with parameter estimation. On the other hand, from a top–down perspective, the system demonstrates robust behavior even when the system input is perturbed. Thus, the bottom–up approach may be viewed as an ‘open-loop’ system based on mass action modeling and parameter estimation, while the top–down approach may be viewed as a ‘closed-loop’ system. In this case study, both bottom–up and top–down approaches are integrated to describe robust adaptation in bacterial chemotaxis.
MODEL VALIDATION AND ANALYSIS-BY-SYNTHESIS: DUAL ROLES OF SYNTHETIC BIOLOGY
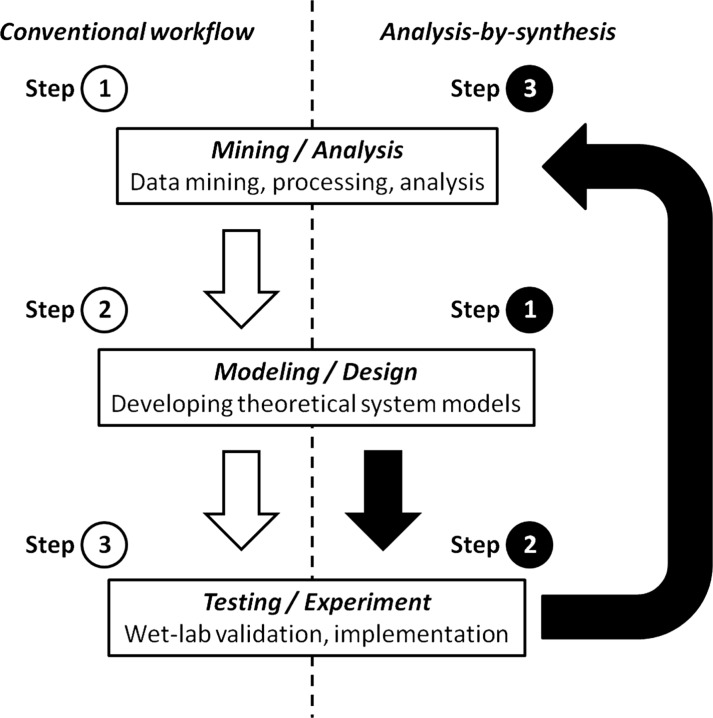
Recently, research in synthetic biology has emerged as an exciting opportunity for model validation and analysis-by-synthesis, serving dual roles in REBMS (Figure 2). Synthetic biology, i.e. the creation of novel biological components (macromolecules), devices (pathways) and systems (organisms) by modifying existing ones or using artificial materials, is promising but also challenging because of the unpredictability and poor reproducibility of experimental results.
Figure 2.
Dual role of synthetic biology in reverse engineering biomolecular systems (REBMS): (left) to test and validate models as a result of upstream mining and modeling, i.e. as the third step in a conventional workflow and (right) to create biomolecular components, devices and systems for analysis-by-synthesis, i.e. synthetic biology is the driver for REBMS
Analysis-by-synthesis is gaining more interest in REBMS because of its potential to design, manufacture and observe viable biomolecular components and devices within predictive, scalable and reproducible systems. It is an active process, common in the field of speech and communications, to analyze signals where only and all the signals to be analyzed can be generated [143]. Biomolecular components and devices refer not only to individual biochemical metabolites but also to stable self-contained modules. Then, biomolecular systems may mean any process, computational or otherwise, that is the result of clever combinations of biomolecular components and devices. Furthermore, such systems may also give rise to other components and devices. Thus, in the context of synthetic biology, the premise of analysis-by-synthesis is to be able to generate a representative variety of artificial biomolecular components, devices and systems.
Successful methods for generating biomolecular components and devices need to be scalable with fast growing high-throughput technologies. For instance, ontology mining tools such as GoMiner [144] are scalable to handle large ontology datasets so as to mine for biomolecular components and devices from linked ontology terms. On the other hand, the synthesis of artificial biomolecular components and devices using equivalent circuits in systems modeling and synthetic biology is not yet scalable. The efficacy and efficiency of assembling metabolites is directly correlated to the dexterity with which researchers may manipulate the biochemistry of the particular metabolites. Additionally, because of the diversity and heterogeneity of biomolecular systems and signals, individual (groups of) efforts in analysis-by-synthesis must be communicated effectively to make any collective progress as a community and to create impact through clinical translation. Examples include the IUPS Physiome Project (www.physiome.org.nz), where biosensors, bioreactors and multi-scale models for diagnosis and treatment. Thus, through an iterative process of data acquisition, mining, modeling and validation, emergent properties of reverse engineered biomolecular systems predicted by mathematical models may be verified and validated in synthetic biology for REBMS.
Synthetic biomolecular components (macromolecules), devices (pathways) and systems (organisms)
The primary challenge of synthetic biology for REBMS is to create and reproduce synthetic biomolecular components (macromolecules), devices (pathways) and systems (organisms) that can replicate the functions of its natural counterparts in vivo. For example, synthetic enzymes that mimic metabolism have been created [145] even though most results to date are relatively simple proof-of-concept creations; toehold reactions have been created to implement DNA circuit control mechanisms without enzymes [146], which has potential uses in automatic drug delivery; and synthetic pathways such as gene networks have also been created to mimic pathways for pathogenic virulence [147]. However, the inability to manufacture and splice DNA reliably, together with the need for extensive post-synthesis modification, has hindered the synthesis of larger, more intricate gene networks. On the other hand, a number of advancements have been made in terms of regulating gene networks, for instance, by modulating the expression of specific proteins [148] with synthetic ribosome-binding sites [149] and selectively introducing mutations in the TATA boxes of promoters to reduce unwanted network interactions (or noise) [150]. At the level of whole cells, including unicellular organisms, a number of experiments have recently demonstrated the potential of synthetic biology to modify the existing genome of bacterial cells to perform novel functions, e.g. to detect light–dark edges in images [151], implement a genetic clock [152] and inhibit biofilm formation [153]. More recently, Gibson et al. [154] reported the creation of a bacterial cell controlled by a synthetic genome, which opens the possibilities of creating minimal cells with desired properties [25].
The reliability and reproducibility of synthetic biology may be improved by in silico modeling and simulation, such as in computational structural biology for synthetic macromolecules to simulate folding and tertiary interactions. Blake et al. [23] described a method for scalable, cost-effective, sequence-independent assembly to construct large DNA components. For synthetic pathways, there are a variety of simulation software using BioBricks to construct standardized models of biochemical networks to predict kinetic outcomes [24], such as SynBioSS [155], Eugene [156] and TinkerCell [157]. While computer-aided design (CAD) tools may be useful to design and simulate synthetic macromolecules and pathways in silico, the challenge remains to: (i) validate such designs experimentally and (ii) close the simulation-experimentation information loop by incorporating CAD knowledge to improve synthetic biology in practice. Crosstalk, i.e. noise, can also occur between host and synthetic circuitry that violates the modularity of components and devices in expressing synthetic biomolecular systems through host organisms. This problem is difficult to fully address because of the complexity of intracellular signaling, but an optimization protocol, which was tested in E. coli, has been developed to allow targeted modification of existing circuits that simplifies design and testing [27]. Wang et al. [28] also developed multiplex automated genome engineering (MAGE), which enables large-scale cell programming and evolution.
The challenges and opportunities make synthetic biology an exciting topic for REBMS, where theoretical outcomes from data mining and systems modeling may be verified and validated directly through analysis-by-synthesis. While there is some progress toward addressing the overall key challenge of model validation directly through synthesis in terms of macromolecules, devices and systems, issues remain to ensure the fidelity, reproducibility and scalability of such synthetic biomolecular components, devices and systems [24–26, 28, 158].
Synthetic biology: a case study
Recent breakthroughs in synthetic biology made it possible to synthesize and implant an entire genome to create living cells. Specifically, a bacterial genome can be synthesized and transplanted to another bacterium [154]. This bottom–up approach involves constructing a very small genome and re-engineering a reduced version of the entire genome for implantation and viability testing. In this study, Gibson et al. resequenced the Mycoplasma mycoides genome and disassembled it into 1078 overlapping cassettes of 1080-bp long. Then they implanted the chemically synthesized cassettes into yeast, whose DNA-repair enzymes linked the short sequences together. These generated medium-sized stings of DNA were then transferred into E. coli and back into yeast. By repeating this procedure three times, a complete genome of M. mycoides, comprising 1.08 Mb was constructed.
The complete M. mycoides genome was then transplanted into another bacterium, namely Mycoplasma capricolum. Although few genes were deleted or disrupted in this genome insertion, they observed that properties of the transplanted M. capricolum are the same as M. mycoides. While this first construct of a living cell is just a proof of concept, the methods and technologies developed hold great promise for many other applications.
CONCLUDING REMARKS
Data-driven mining and design-driven modeling have led to significant progress in REBMS using high-throughput –omic data. In addition to theoretical analyses, the next step is to combine analysis with synthesis through synthetic biology. Research in synthetic biology is shown to serve dual roles for REBMS in model validation and analysis-by-synthesis. Thus, the overall key challenges in REBMS are to (i) integrate heterogeneous biochemical data at molecular-level resolution for data mining, (ii) combine top–down and bottom–up approaches for systems modeling and (iii) validate theoretical system models directly by experiment, in particular synthesis (Table 2). While significant progress has been made toward addressing specific problems, key issues remain to (i) increase the accuracy of feature extraction in data mining, (ii) improve the simulation and abstraction of emergent system properties in systems modeling and (iii) ensure the fidelity of assembled macromolecules and pathways in synthetic biology. By incorporating data mining, systems modeling and synthetic biology to create an integrated and iterative research pipeline, we are hopeful that these remaining issues in REBMS will be overcome.
Table 2:
Summary of challenges, progress and remaining issues and opportunities for REBMS
| Key challenge | Progress | Issues/opportunities | |
|---|---|---|---|
| Data mining | Integrate and analyze high-throughput data | Increase accuracy of feature extraction | |
| Systems modeling | Use top–down with bottom–up approaches |
|
Improve integration of top–down and bottom–up models |
| Synthetic biology | Validate systems models | Ensure fidelity of assembled synthetic components and devices |
Key Points.
To deal with high-throughput data in REBMS, data-driven mining approaches can extract patterns and features for downstream modeling.
Motivated by similarities between biological stability and engineering robustness, design-driven approaches can simulate and abstract emergent properties of complex biomolecular systems.
Challenges in data analysis and modeling for REBMS may be overcome by integrating heterogeneous datasets for mining and combining both top–down and bottom–up approaches for modeling.
Synthetic biology is an emerging field that proves to be an exciting opportunity for REBMS, serving dual roles in validation and analysis-by-synthesis.
FUNDING
This work was supported in part by grants from the National Institutes of Health Bioengineering Research Partnership R01CA108468, Center for Cancer Nanotechnology Excellence U54CA119338, and 1RC2CA148265; Georgia Cancer Coalition Distinguished Cancer Scholar Award to MD Wang, and grants 5R21DA025168-02, 1R01HG004836-01 and 4R00LM009826-03 to G. Alterovitz.
Biographies
Chang F. Quo received his BSc in Biomedical Engineering from the Georgia Institute of Technology in 2005 and is currently a doctoral student in Bioengineering at the Georgia Institute of Technology. His research is focused on the use of modern control theory to model regulation in metabolic systems.
Chanchala Kaddi received BSc in Biomedical Engineering from the Georgia Institute of Technology in 2008 and is currently a doctoral student in Bioengineering at the Georgia Institute of Technology. She is an NSF Graduate Research Fellow. Her research is focused on high-throughput data analysis for tissue imaging mass spectrometry.
John H. Phan, PhD, received BSc in Computer Engineering from the University of Oklahoma in 2003 and PhD in Biomedical Engineering from the Georgia Institute of Technology and Emory University in 2009. He is currently a Postdoctoral Fellow at Emory University. His research is focused on knowledge-driven mining of high-throughput biological data, high-performance and grid computing and translational bioinformatics.
Amin Zollanvari, PhD, received PhD from Texas A&M University in 2010. He is currently a Postdoctoral Fellow at the Harvard-MIT Division of Health Sciences and Technology. His research is focused on modeling of complex diseases and behaviors using genome-wide association data, Bayesian theory, statistical pattern recognition, asymptotic analysis of statistics in large dimensionality space and ontological mapping and inference.
Mingqing Xu, PhD, is an Full Professor of Genetics at Bio-X Institutes jointly established by the Chinese Academy of Sciences and Shanghai Jiao Tong University. He is joint professor at Shanghai Jiao Tong University School of Medicine. He earned his PhD degree in Human Genetics at Shanghai Jiao Tong University in 2005 and then did his Postdoctoral training in Statistical Genetics and Epidemiology at Harvard University. He has been serving as Director of the Translational Research Center for Integrative Omics at Bio-X Institutes of the Chinese Academy of Sciences and Shanghai Jiao Tong University since 2011. His research interests are focused on translational omics and epidemiology, including high-throughput omic data analysis and its application to personalized medicine; statistical genomics; and environmental and molecular epidemiology in human complex diseases.
May D. Wang, PhD, is a tenured Associate Professor in the Wallace H. Coulter Department of Biomedical Engineering jointly established by Georgia Institute of Technology and Emory University in Atlanta, Georgia. She is a Georgia Cancer Coalition Distinguished Cancer Scholar, and has served as Director of Biocomputing and Bioinformatics Core in Emory-Georgia Tech Cancer Nanotechnology Center focusing on Bio-Nan-Info Integration for Personalized Oncology. Dr. Wang has also served as a co-Director in the Georgia Tech Tissue Imaging Mass Spectrometry Center since 2005. Her primary research interests are translational bioinformatics, imaging informatics, and health informatics for personalized systems medicine: including high-throughput −omic data quality control and analysis for clinical biomarker identification; tissue imaging informatics; pathway modeling; and delivering personalized health informatics solution by mobile health.
Gil Alterovitz, PhD, is an Assistant Professor at the Harvard/MIT Health Sciences and Technology Division Children's Hospital Informatics Program (CHIP). He is affiliated with the Department of Electrical Engineering and Computer Science at MIT and Harvard Medical School Partners’ Center for Genetics and Genomics (HPCGG). His research interests involve development of novel, interdisciplinary approaches that bridge engineering and medicine. Specifically, he is involved in developing methods for studying biological networks and signal processing within proteomics.
References
- 1.Cherry JM, Ball C, Weng S, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387(6632):67–73. [PMC free article] [PubMed] [Google Scholar]
- 2.Harris TW, Antoshechkin I, Bieri T, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–7. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter PJ, Borg TK. Integration from proteins to organs: the Physiome Project. Nat Rev Mol Cell Biol. 2003;4(3):237–43. doi: 10.1038/nrm1054. [DOI] [PubMed] [Google Scholar]
- 4.Kidd JM, Cooper GM, Donahue WF, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabilloud T, Hochstrasser D, Simpson RJ. Is a gene-centric human proteome project the best way for proteomics to serve biology? Proteomics. 2010;10(17):3067–72. doi: 10.1002/pmic.201000220. [DOI] [PubMed] [Google Scholar]
- 6.Tweedie S, Ashburner M, Falls K, et al. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–9. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeong H, Tombor B, Albert R, et al. The large-scale organization of metabolic networks. Nature. 2000;407(6804):651–4. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 8.Kaluza P, Vingron M, Mikhailov AS. Self-correcting networks: function, robustness, and motif distributions in biological signal processing. Chaos. 2008;18(2):026113. doi: 10.1063/1.2945228. [DOI] [PubMed] [Google Scholar]
- 9.Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322(5900):390–5. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitano H. Systems biology: a brief overview. Science. 2002;295(5560):1662–4. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 11.Lauffenburger DA. Cell signaling pathways as control modules: complexity for simplicity? Proc Natl Acad Sci USA. 2000;97(10):5031–3. doi: 10.1073/pnas.97.10.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campain A, Yang Y. Comparison study of microarray meta-analysis methods. BMC Bioinformatics. 2010;11(1):408. doi: 10.1186/1471-2105-11-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conlon EM, Song JJ, Liu JS. Bayesian models for pooling microarray studies with multiple sources of replications. BMC Bioinformatics. 2006;7:247. doi: 10.1186/1471-2105-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamid JS, Hu P, Roslin NM, et al. Data integration in genetics and genomics: methods and challenges. Hum Genomics Proteomics. 2009;2009:869093. doi: 10.4061/2009/869093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li A. Facing the challenges of data integration in biosciences. Engineer Lett. 2006;13(3):EL_13_3_13. [Google Scholar]
- 16.Rhodes DR, Yu JJ, Shanker K, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101(25):9309–14. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behre J, Wilhelm T, von Kamp A, et al. Structural robustness of metabolic networks with respect to multiple knockouts. J Theor Biol. 2008;252(3):433–41. doi: 10.1016/j.jtbi.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 18.Glass L, Hill C. Ordered and disordered dynamics in random networks. Europhys Lett. 1998;41(6):599–604. [Google Scholar]
- 19.Quo CF, Moffitt RA, Merrill AH, Jr, et al. Adaptive control model reveals systematic feedback and key molecules in metabolic pathway regulation. J Comput Biol. 2011;18(2):169–82. doi: 10.1089/cmb.2010.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savageau MA. Biochemical systems analysis. 1. Some mathematical properties of rate law for component enzymatic reactions. J Theor Biol. 1969;25(3):365–9. doi: 10.1016/s0022-5193(69)80026-3. [DOI] [PubMed] [Google Scholar]
- 21.van Kampen NG. Stochastic Processes in Physics and Chemistry. North Holland: Elsevier; 1997. [Google Scholar]
- 22.Yi TM, Huang Y, Simon MI, et al. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci USA. 2000;97(9):4649–53. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blake WJ, Chapman BA, Zindal A, et al. Pairwise selection assembly for sequence-independent construction of long-length DNA. Nucleic Acids Res. 2010;38(8):2594–602. doi: 10.1093/nar/gkq123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotech. 2009;27(5):465–71. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foley PL, Shuler ML. Considerations for the design and construction of a synthetic platform cell for biotechnological applications. Biotechnol Bioengineer. 2010;105(1):26–36. doi: 10.1002/bit.22575. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Gallego F, Schmidt-Dannert C. Multi-enzymatic synthesis. Curr Opin Chem Biol. 2010;14(2):174–83. doi: 10.1016/j.cbpa.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 27.Norville JE, Derda R, Gupta S, et al. Introduction of customized inserts for streamlined assembly and optimization of biobrick synthetic genetic circuits. J Biol Engineer. 2010;4(1):1–11. doi: 10.1186/1754-1611-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang HH, Isaacs FJ, Carr PA, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460(7257):894–8. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schena M, Shalon D, Davis RW, et al. Quantitative monitoring of gene-expression patterns with a complementary-DNA microarray. Science. 1995;270(5235):467–70. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Ruotti V, Stewart RM, et al. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26(4):493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11(6):427–39. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 32.Bennett MR, Pang WL, Ostroff NA, et al. Metabolic gene regulation in a dynamically changing environment. Nature. 2008;454(7208):1119–22. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandras C, Weaver T, Zouberakis M, et al. Models for financial sustainability of biological databases and resources. Database. 2009;2009 doi: 10.1093/database/bap017. bap017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian JA, Kluger Y, Yu HY, et al. Identification and correction of spurious spatial correlations in microarray data. BioTechniques. 2003;35(1):42–44. doi: 10.2144/03351bm03. 46, 48. [DOI] [PubMed] [Google Scholar]
- 35.Yang YH, Dudoit S, Luu P, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30(4):e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox WG, Beaudet MP, Agnew JY, et al. Possible sources of dye-related signal correlation bias in two-color DNA microarray assays. Anal Biochem. 2004;331(2):243–54. doi: 10.1016/j.ab.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Nadon R, Shoemaker J. Statistical issues with microarrays: processing and analysis. Trends in Genetics. 2002;18(5):265–71. doi: 10.1016/s0168-9525(02)02665-3. [DOI] [PubMed] [Google Scholar]
- 38.Stokes TH, Moffitt RA, Phan JH, et al. chip artifact CORRECTion (caCORRECT): A bioinformatics system for quality assurance of genomics and proteomics array data. Ann Biomed Engineer. 2007;35(6):1068–80. doi: 10.1007/s10439-007-9313-y. [DOI] [PubMed] [Google Scholar]
- 39.Richter BG, Sexton DP. Managing and analyzing next-generation sequence data. PLoS Comput Biol. 2009;5(6):e1000369. doi: 10.1371/journal.pcbi.1000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pop M, Salzberg SL. Bioinformatics challenges of new sequencing technology. Trends Genet. 2008;24(3):142–9. doi: 10.1016/j.tig.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kriseman J, Busick C, Szelinger S, et al. BING: biomedical informatics pipeline for next generation sequencing. J Biom Inform. 2010;43(3):428–34. doi: 10.1016/j.jbi.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11(5):473–83. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milne I, Bayer M, Cardle L, et al. Tablet-next generation sequence assembly visualization. Bioinformatics. 2010;26(3):401–2. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics. 2009;25(8):1026–32. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Au KF, Jiang H, Lin L, et al. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38(14):4570–8. doi: 10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12(4):656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson G, Schein J, Chiu R, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7(11):U909–62. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 49.Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat Rev Genet. 2009;10(9):617–27. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 51.Deininger SO, Ebert MP, Futterer A, et al. MALDI Imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J Proteome Res. 2008;7(12):5230–6. doi: 10.1021/pr8005777. [DOI] [PubMed] [Google Scholar]
- 52.Alexandrov T, Kobarg JH. Efficient spatial segmentation of large imaging mass spectrometry datasets with spatially aware clustering. Bioinformatics. 2011;27(13):I230–8. doi: 10.1093/bioinformatics/btr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Djidja MC, Claude E, Snel MF, et al. MALDI-ion mobility separation-mass spectrometry imaging of glucose-regulated protein 78 kDa (Grp78) in human formalin-fixed, paraffin-embedded pancreatic adenocarcinoma tissue sections. J Proteome Res. 2009;8(10):4876–84. doi: 10.1021/pr900522m. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, Chen Y, Momin A, et al. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol Cancer. 2010;9:186. doi: 10.1186/1476-4598-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rauser S, Marquardt C, Balluff B, et al. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J Proteome Res. 2010;9(4):1854–63. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- 56.Greving MP, Patti GJ, Siuzdak G. Nanostructure-initiator mass spectrometry metabolite analysis and imaging. Anal Chem. 2011;83(1):2–7. doi: 10.1021/ac101565f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDonnell LA, van Remoortere A, van Zeijl RJM, et al. Mass spectrometry image correlation: quantifying colocalization. J Proteome Res. 2008;7(8):3619–27. doi: 10.1021/pr800214d. [DOI] [PubMed] [Google Scholar]
- 58.Peterson DS. Matrix-free methods for laser desorption/ionization mass spectrometry. Mass Spectrometry Rev. 2007;26(1):19–34. doi: 10.1002/mas.20104. [DOI] [PubMed] [Google Scholar]
- 59.Luo J, Wang J, Ma TM, et al. Reverse engineering of bacterial chemotaxis pathway via frequency domain analysis. PLoS One. 2010;5(3):e9182. doi: 10.1371/journal.pone.0009182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME) - toward standards for microarray data. Nat Genet. 2001;29(4):365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 61.Taylor CF, Field D, Sansone S-A, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat Biotech. 2008;26(8):889–96. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bino RJ, Hall RD, Fiehn O, et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004;9(9):418–25. doi: 10.1016/j.tplants.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 63.Gibson F, Overton PG, Smulders TV, et al. Minimum information about a neuroscience investigation (MINI): electrophysiology. Nat Precedings. 2008 hdl:10101/npre.12008.11720.10101. [Google Scholar]
- 64.Le Novere N, Finney A, Hucka M, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat Biotech. 2005;23(12):1509–15. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- 65.Waltemath D, Adams R, Beard DA, et al. Minimum information about a simulation experiment (MIASE) PLoS Comput Biol. 2011;7(4):e1001122. doi: 10.1371/journal.pcbi.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoeckert CJ, Quackenbush J, Brazma A, et al. Minimum information about a functional genomics experiment: the state of microarray standards and their extension to other technologies. DDT: Targets. 2004;3(4):159–64. [Google Scholar]
- 67.Hucka M, Finney AM, Hoops S, et al. Systems Biology Markup Language (SBML) Level 2: structures and facilities for model definitions. Nat Precedings. 2007 doi: 10.2390/biecoll-jib-2015-271. http://hdl.nature.com/10101/npre.2007.58.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laibe C, Le Novere N. MIRIAM resources: tools to generate and resolve robust cross-references in Systems Biology. BMC Sys Biol. 2007;1:58. doi: 10.1186/1752-0509-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jain AK. Data clustering: 50 years beyond K-means. Pattern Recognit Lett. 2010;31(8):651–66. [Google Scholar]
- 70.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu FX. Genetic weighted k-means algorithm for clustering large-scale gene expression data. BMC Bioinformatics. 2008;9(Suppl 6):S12. doi: 10.1186/1471-2105-9-S6-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ringnér M. What is principal component analysis? Nat Biotechnol. 2008;26(3):303–4. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 73.Preli A, Bleuler S, Zimmermann P, et al. A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics. 2006;22(9):1122–9. doi: 10.1093/bioinformatics/btl060. [DOI] [PubMed] [Google Scholar]
- 74.Freyhult E, Landfors M, Önskog J, et al. Challenges in microarray class discovery: a comprehensive examination of normalization, gene selection and clustering. BMC Bioinformatics. 2010;11(1):503. doi: 10.1186/1471-2105-11-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–17. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 76.Simon R, Radmacher M, Dobbin K, et al. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95(1):14–8. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 77.Phan JH, Moffitt RA, Stokes TH, et al. Convergence of biomarkers, bioinformatics and nanotechnology for individualized cancer treatment. Trends Biotechnol. 2009;27(6):350–8. doi: 10.1016/j.tibtech.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Luo J, Schumacher M, Scherer A, et al. A comparison of batch effect removal methods for enhancement of prediction performance using MAQC-II microarray gene expression data. Pharmacogenomics J. 2010;10(4):278–91. doi: 10.1038/tpj.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boulesteix AL, Strobl C. Optimal classifier selection and negative bias in error rate estimation: an empirical study on high-dimensional prediction. BMC Med Res Methodology. 2009;9(1):85. doi: 10.1186/1471-2288-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lusa L. Class prediction for high-dimensional class-imbalanced data. BMC Bioinformatics. 2010;11(1):523. doi: 10.1186/1471-2105-11-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boulesteix AL. Over-optimism in bioinformatics research. Bioinformatics. 2010;26(3):437. doi: 10.1093/bioinformatics/btp648. [DOI] [PubMed] [Google Scholar]
- 82.Fan X, Lobenhofer E, Chen M, et al. Consistency of predictive signature genes and classifiers generated using different microarray platforms. Pharmacogenomics J. 2010;10(4):247–57. doi: 10.1038/tpj.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parry R, Jones W, Stokes T, et al. k-Nearest neighbor models for microarray gene expression analysis and clinical outcome prediction. Pharmacogenomics J. 2010;10(4):292–309. doi: 10.1038/tpj.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi L, Campbell G, Jones WD, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotech. 2010;28(8):827. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dougherty ER. On the epistemological crisis in genomics. Curr Genomics. 2008;9(2):69–79. doi: 10.2174/138920208784139546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quackenbush J. Microarray analysis and tumor classification. N Eng J Med. 2006;355(9):960. doi: 10.1056/NEJMc061813. [DOI] [PubMed] [Google Scholar]
- 87.Weigelt B, Baehner FL, Reis JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220(2):263–80. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 88.Michiels S, Koscielny S, Hill C. Prediction of cancer outcome with microarrays: a multiple random validation strategy. Lancet. 2005;365:488–92. doi: 10.1016/S0140-6736(05)17866-0. [DOI] [PubMed] [Google Scholar]
- 89.Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci USA. 2002;99(10):6562–6. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics. 2006;7:91. doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Braga-Neto UM, Dougherty ER. Is cross-validation valid for small-sample microarray classification? Bioinformatics. 2004;20(3):374–80. doi: 10.1093/bioinformatics/btg419. [DOI] [PubMed] [Google Scholar]
- 92.Fu WJJ, Carroll RJ, Wang SJ. Estimating misclassification error with small samples via bootstrap cross-validation. Bioinformatics. 2005;21(9):1979–86. doi: 10.1093/bioinformatics/bti294. [DOI] [PubMed] [Google Scholar]
- 93.Isaksson A, Wallman M, Goransson H, et al. Cross-validation and bootstrapping are unreliable in small sample classification. Pattern Recognition Lett. 2008;29(14):1960–5. [Google Scholar]
- 94.Fan X, Shi L, Fang H, et al. DNA microarrays are predictive of cancer prognosis: a re-evaluation. Clin Cancer Res. 2010;16(2):629. doi: 10.1158/1078-0432.CCR-09-1815. [DOI] [PubMed] [Google Scholar]
- 95.Stuart JM, Segal E, Koller D, et al. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–55. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 96.Margolin AA, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Butte AJ, Kohane IS. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pac Sym Biocomput. 2000:418–29. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 98.Wilkinson DJ. Bayesian methods in bioinformatics and computational systems biology. Brief Bioinform. 2007;8(2):109–16. doi: 10.1093/bib/bbm007. [DOI] [PubMed] [Google Scholar]
- 99.Lee S, Abbott P, Johantgen M. Logistic regression and Bayesian networks to study outcomes using large datasets. Nurs Res. 2005;54(2):133–8. doi: 10.1097/00006199-200503000-00009. [DOI] [PubMed] [Google Scholar]
- 100.Sebastiani P, Ramoni MF, Nolan V, et al. Genetic dissection and prognostic modeling of overt stroke in sickle cell anemia. Nat Genet. 2005;37(4):435–40. doi: 10.1038/ng1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tran LM, Zhang B, Zhang Z, et al. Inferring causal genomic alterations in breast cancer using gene expression data. BMC Sys Biol. 2011;5:121. doi: 10.1186/1752-0509-5-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):U318–68. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Faith JJ, Hayete B, Thaden JT, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. Plos Biol. 2007;5(1):54–66. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu J, Zhang B, Smith EN, et al. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat Genet. 2008;40(7):854–61. doi: 10.1038/ng.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayete B, Gardner TS, Collins JJ. Size matters: network inference tackles the genome scale. Mol Sys Biol. 2007;3 doi: 10.1038/msb4100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marbach D, Prill RJ, Schaffter T, et al. Revealing strengths and weaknesses of methods for gene network inference. Proc Natl Acad Sci USA. 2010;107(14):6286–91. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ooi BNS, Phan TT. Insights gained from the reverse engineering of gene networks in keloid fibroblasts. Theor Biol Med Model. 2011;8:13. doi: 10.1186/1742-4682-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu J, Smith VA, Wang PP, et al. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004;20(18):3594–603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
- 109.Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol. 1969;22(3):437–67. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- 110.Shmulevich I, Dougherty ER, Kim S, et al. Probabilistic Boolean networks: a rule-based uncertainty model for gene regulatory networks. Bioinformatics. 2002;18(2):261–74. doi: 10.1093/bioinformatics/18.2.261. [DOI] [PubMed] [Google Scholar]
- 111.Murphy K, Mian S. University of California at Berkeley; Modelling gene expression data using dynamic Bayesian networks. 1999. unpublished technical report. [Google Scholar]
- 112.Tyson JJ, Othmer HG, editors. The Dynamics of Feedback Control Circuits in Biochemical Pathways. New York: Academic Press; 1978. [Google Scholar]
- 113.Goodwin BC. New York, NY: Academic Press; 1963. Temporal organization in cells. [Google Scholar]
- 114.Mestl T, Plahte E, Omholt SW. A mathematical framework for describing and analyzing gene regulatory networks. J Theor Biol. 1995;176(2):291–300. doi: 10.1006/jtbi.1995.0199. [DOI] [PubMed] [Google Scholar]
- 115.Savageau MA. Biochemical systems analysis. 2. Steady-state solutions for an N-pool system using a power-law approximation. J Theor Biol. 1969;25(3):370–9. doi: 10.1016/s0022-5193(69)80027-5. [DOI] [PubMed] [Google Scholar]
- 116.Waage P, Gulberg CM. Studies concerning affinity (translated) J Chem Education. 1986;63(12):1044–7. [Google Scholar]
- 117.Varma A, Palsson BO. Metabolic flux balancing - basic concepts, scientific and practical use. Nat Biotech. 1994;12(10):994–8. [Google Scholar]
- 118.Fell DA. Metabolic control analysis - a survey of its theoretical and experimental development. Biochem J. 1992;286:313–30. doi: 10.1042/bj2860313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alvarez-Vasquez F, Sims KJ, Cowart LA, et al. Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature. 2005;433(7024):425–30. doi: 10.1038/nature03232. [DOI] [PubMed] [Google Scholar]
- 120.Gupta S, Maurya MR, Merrill AH, Jr, et al. Integration of lipidomics and transcriptomics data towards a systems biology model of sphingolipid metabolism. BMC Sys Biol. 2011;5:26. doi: 10.1186/1752-0509-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stelling J, Sauer U, Szallasi Z, et al. Robustness of cellular functions. Cell. 2004;118(6):675–85. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 122.Wiener N. 2nd edn. Cambridge MA, USA: MIT Press; 1965. Cybernetics: or control and communication in the animal and the machine. [Google Scholar]
- 123.Guyton AC, Granger HJ, Coleman TG. Circulation - overall regulation. Ann Rev Physiol. 1972;34:13–46. doi: 10.1146/annurev.ph.34.030172.000305. [DOI] [PubMed] [Google Scholar]
- 124.Tauber AI. The immune system and its ecology. Philos Sci. 2008;75(2):224–45. [Google Scholar]
- 125.Bianchi AL, Gestreau C. The brainstem respiratory network: an overview of a half century of research. Resp Physiol Neurobiol. 2009;168(1–2):4–12. doi: 10.1016/j.resp.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 126.French AS. The systems analysis approach to mechanosensory coding. Biol Cyber. 2009;100(6):417–26. doi: 10.1007/s00422-008-0262-9. [DOI] [PubMed] [Google Scholar]
- 127.Guzun R, Saks V. Application of the principles of systems biology and Wiener's cybernetics for analysis of regulation of energy fluxes in muscle cells in vivo. Int J Mol Sci. 2010;11(3):982–1019. doi: 10.3390/ijms11030982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Alon U, Surette MG, Barkai N, et al. Robustness in bacterial chemotaxis. Nature. 1999;397(6715):168–71. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 129.Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387(6636):913–7. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- 130.Shimizu TS, Tu Y, Berg HC. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol Sys Biol. 2010;6:382. doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11(7):476–86. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hicks C, Asfour R, Pannuti A, et al. An integrative genomics approach to biomarker discovery in breast cancer. Cancer Inform. 2011;10:185–204. doi: 10.4137/CIN.S6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu ET. Integrative biology - a strategy for systems biomedicine. Nat Rev Genet. 2009;10(1):64–8. doi: 10.1038/nrg2488. [DOI] [PubMed] [Google Scholar]
- 134.Rose AE, Poliseno L, Wang J, et al. Integrative genomics identifies molecular alterations that challenge the linear model of melanoma progression. Cancer Res. 2011;71(7):2561–71. doi: 10.1158/0008-5472.CAN-10-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sun Z, Asmann YW, Kalari KR, et al. Integrated analysis of gene expression, CpG island methylation, and gene copy number in breast cancer cells by deep sequencing. PLoS One. 2011;6(2):e17490. doi: 10.1371/journal.pone.0017490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang K, Diskin SJ, Zhang H, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469(7329):216–20. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bell D, Berchuck A, Birrer M, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Carter H, Samayoa J, Hruban RH, et al. Prioritization of driver mutations in pancreatic cancer using cancer-specific high-throughput annotation of somatic mutations (CHASM) Cancer Biol Ther. 2010;10(6):582–7. doi: 10.4161/cbt.10.6.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chin L, Meyerson M, Aldape K, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comp Bio. 2011;18(3):507–22. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 141.Vaske CJ, Benz SC, Sanborn JZ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26(12):i237–45. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hauri DC, Ross J. A model of excitation and adaptation in bacterial chemotaxis. Biophys J. 1995;68(2):708–22. doi: 10.1016/S0006-3495(95)80232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bell CG, Stevens KN, House AS, et al. Reduction of speech spectra by analysis-by-synthesis techniques. J Acoustical Soc Am. 1961;33(12):1725–8. [Google Scholar]
- 144.Zeeberg BR, Feng WM, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4(4):R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dueber JE, Wu GC, Malmirchegini GR, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotech. 2009;27(8):753–9. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 146.Zhang DY, Winfree E. Control of DNA strand displacement kinetics using toehold exchange. J Am Chem Soc. 2009;131(47):17303–14. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- 147.Lin K, Husmeier D, Dondelinger F, et al. Reverse engineering gene regulatory networks related to quorum sensing in the plant pathogen pectobacterium atrosepticum. In: Fenyo D, editor. Computational Biology. Vol. 673. New York: Humana Press; 2010. pp. 253–81. [DOI] [PubMed] [Google Scholar]
- 148.Menolascina F, di Bernardo M, di Bernardo D. Analysis, design and implementation of a novel scheme for in-vivo control of synthetic gene regulatory networks. Automatica. 2011;47(6):1265–70. [Google Scholar]
- 149.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotech. 2009;27(10):946–50. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Murphy KF, Adams RM, Wang X, et al. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Res. 2010;38(8):2712–26. doi: 10.1093/nar/gkq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tabor JJ, Salis HM, Simpson ZB, et al. A synthetic genetic edge detection program. Cell. 2009;137(7):1272–81. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tigges M, Denervaud N, Greber D, et al. A synthetic low-frequency mammalian oscillator. Nucleic Acids Res. 2010;38(8):2702–11. doi: 10.1093/nar/gkq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2011;469(7329):212–5. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gibson DG, Glass JI, Lartigue C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329(5987):52–6. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 155.Weeding E, Houle J, Kaznessis YN. SynBioSS designer: a web-based tool for the automated generation of kinetic models for synthetic biological constructs. Brief Bioinform. 2010;11(4):394–402. doi: 10.1093/bib/bbq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bilitchenko L, Liu A, Cheung S, et al. Eugene - A domain specific language for specifying and constraining synthetic biological parts, devices, and systems. PLoS One. 2011;6(4):e18882. doi: 10.1371/journal.pone.0018882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chandran D, Bergmann FT, Sauro HM. TinkerCell: modular CAD tool for synthetic biology. J Biol Engineer. 2009;3(19) doi: 10.1186/1754-1611-3-19. doi:10.1186/1754-1611-1183-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Silva-Rocha R, de Lorenzo V. Noise and robustness in prokaryotic regulatory networks. Ann Rev Microbiol. 2010;64:257–75. doi: 10.1146/annurev.micro.091208.073229. [DOI] [PubMed] [Google Scholar]
- 159.Friedman N, Linial M, Nachman I, et al. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7(3–4):601–20. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- 160.Keller AD, Schummer M, Hood L, et al. Bayesian classification of DNA array expression data. Technical Report UW-CSE-2000-08-01. 2000 [Google Scholar]
- 161.Malovini A, Nuzzo A, Ferrazzi F, et al. Phenotype forecasting with SNPs data through gene-based Bayesian networks. BMC Bioinformatics. 2009;10:S7. doi: 10.1186/1471-2105-10-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pachter L, Alexandersson M, Cawley S. Applications of generalized pair hidden Markov models to alignment and gene finding problems. J Comput Biol. 2002;9(2):389–99. doi: 10.1089/10665270252935520. [DOI] [PubMed] [Google Scholar]
- 163.Guldberg CM, Waage P. Studies concerning affinity (translated) J Chem Education. 1986;63(12):1044–7. [Google Scholar]
- 164.Varma A, Palsson BO. Metabolic flux balancing - basic concepts, scientific and practical use. Bio-Technology. 1994;12(10):994–8. [Google Scholar]
- 165.Afonso B, Silver PA, Ajo-Franklin CM. A synthetic circuit for selectively arresting daughter cells to create aging populations. Nucleic Acids Res. 2010;38(8):2727–35. doi: 10.1093/nar/gkq075. [DOI] [PMC free article] [PubMed] [Google Scholar]