Abstract
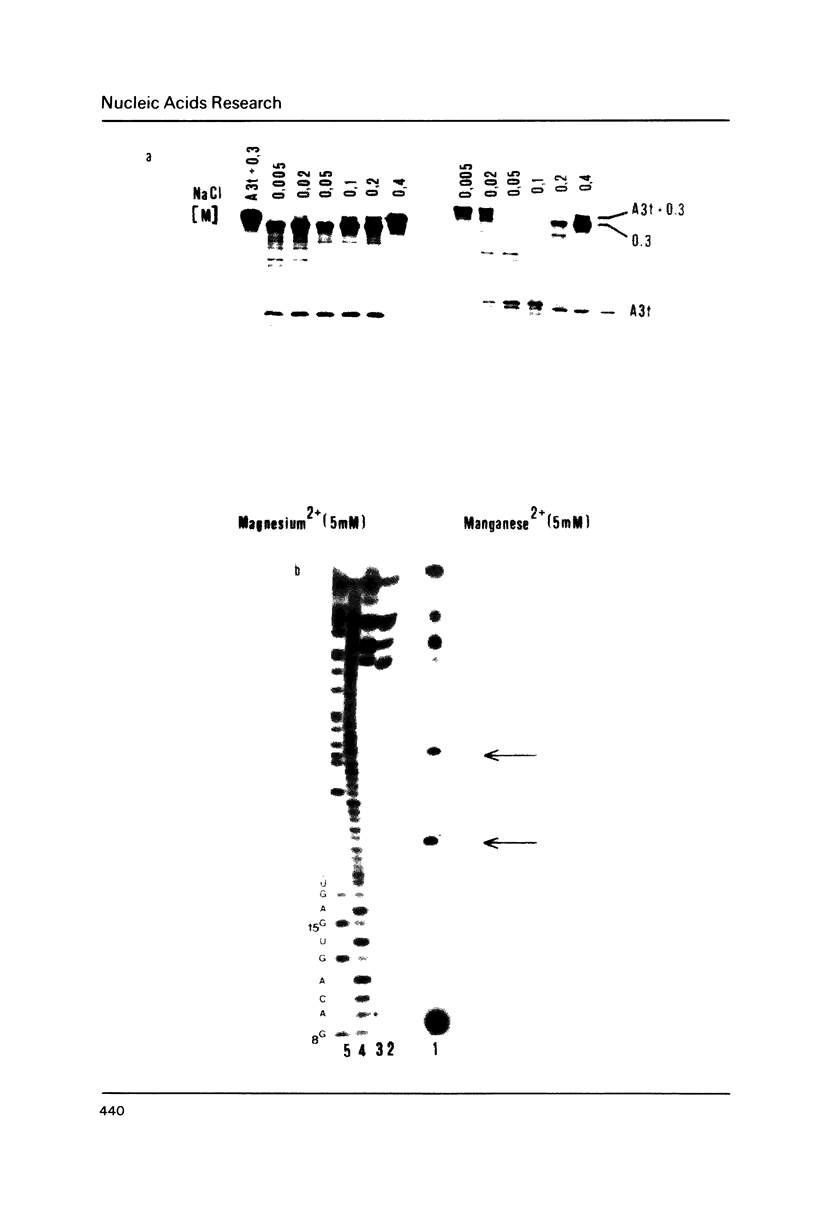
Five 'secondary' cleavage sites of E. coli RNAaseIII within the initiator RNA A3t (141 nucleotides) from bacteriophage T7 are described. Cleavage takes place apparEntly within or at the bottom of a short double-stranded stretch of RNA. Three secondary sites are efficiently cleaved in the presence of magnesium 2+ ions, two additional sites are cleaved in the presence of manganese 2+ ions at low monovalent salt concentrations (less than 0.05 M).
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellofatto V., Amemiya K., Shapiro L. Purification and characterization of an RNA processing enzyme from Caulobacter crescentus. J Biol Chem. 1983 May 10;258(9):5467–5476. [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Darlix J. L., Spahr P. F., Bromley P. A. Analysis of Rous sarcoma virus (RSV) RNA structure by means of specific nucleases. Virology. 1978 Oct 15;90(2):317–329. doi: 10.1016/0042-6822(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fideltiy of cleavage. J Biol Chem. 1976 Jun 25;251(12):3807–3814. [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Vournakis J. N., Donis-Keller H., Chaconas G., Dougall D. K., Kafatos F. C. End labeling of enzymatically decapped mRNA. Nucleic Acids Res. 1977 Dec;4(12):4165–4174. doi: 10.1093/nar/4.12.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. J., Dunn J. J., Wimmer E. Identification of specific fragments containing the 5' end of poliovirus RNA after ribonuclease III digestion. Nucleic Acids Res. 1978 Nov;5(11):4039–4054. doi: 10.1093/nar/5.11.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kram R., Tomkins G. M. Pleiotypic control by cyclic AMP: interaction with cyclic GMP and possible role of microtubules. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1659–1663. doi: 10.1073/pnas.70.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev N., Silengo L., Schlessinger D. Synthesis of a large precursor to ribosomal RNA in a mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3361–3365. doi: 10.1073/pnas.70.12.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto A., Lee Y. F., Babich A., Jacobson A., Dunn J. J., Wimmer E. Restricted fragmentation of poliovirus type 1, 2 and 3 RNAs by ribonuclease III. J Mol Biol. 1979 Feb 25;128(2):165–177. doi: 10.1016/0022-2836(79)90124-4. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Whiteley H. R. Bacillus subtilis RNAase III cleavage sites in phage SP82 early mRNA. Cell. 1983 Jul;33(3):907–913. doi: 10.1016/0092-8674(83)90033-8. [DOI] [PubMed] [Google Scholar]
- Robertson H. D., Dickson E., Dunn J. J. A nucleotide sequence from a ribonuclease III processing site in bacteriophage T7 RNA. Proc Natl Acad Sci U S A. 1977 Mar;74(3):822–826. doi: 10.1073/pnas.74.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]