Abstract
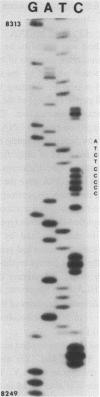
A specific segment of mitochondrial DNA from 18 people was examined by two methods of direct DNA sequencing. This segment includes a small noncoding region (V) shown before by restriction analysis to exhibit length polymorphism. All 11 of the human mtDNAs previously reported to have a deletion in this region proved to lack one of the two adjacent copies of a 9-base-pair sequence normally present in human mtDNAs. Phylogenetic analysis suggests that this deletion occurred only once during the evolution of modern types of human mtDNA and that it will be a valuable anthropological marker for peoples of East Asian origin. The one human mtDNA reported to have an addition in region V differs from the wild type by two mutations in the first copy of the 9-base-pair sequence: one transition and an addition of four cytosines, thereby producing a run of 11 cytosines. One of the direct DNA sequencing methods uses a single oligonucleotide primer to facilitate dideoxy sequencing from purified mtDNA templates. The second, more successful, method first amplifies this mtDNA segment enzymatically with two flanking primers (the "polymerase chain reaction") and then uses a third primer for DNA sequencing. This latter method, which works on the DNA extracted from small amounts of blood as well as on purified mtDNA, is shown to be a rapid means of defining sequence variants without purifying and cloning the same DNA segment from many individuals.
Full text
PDF


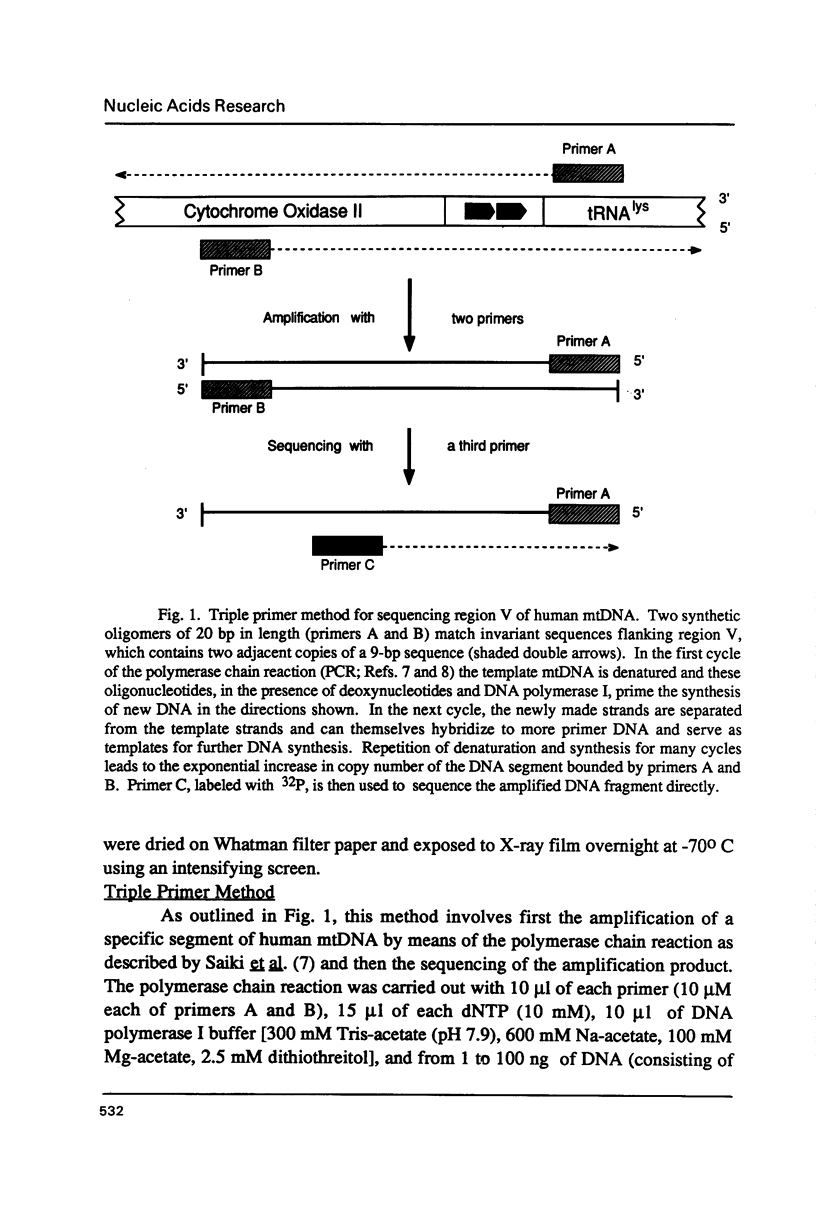
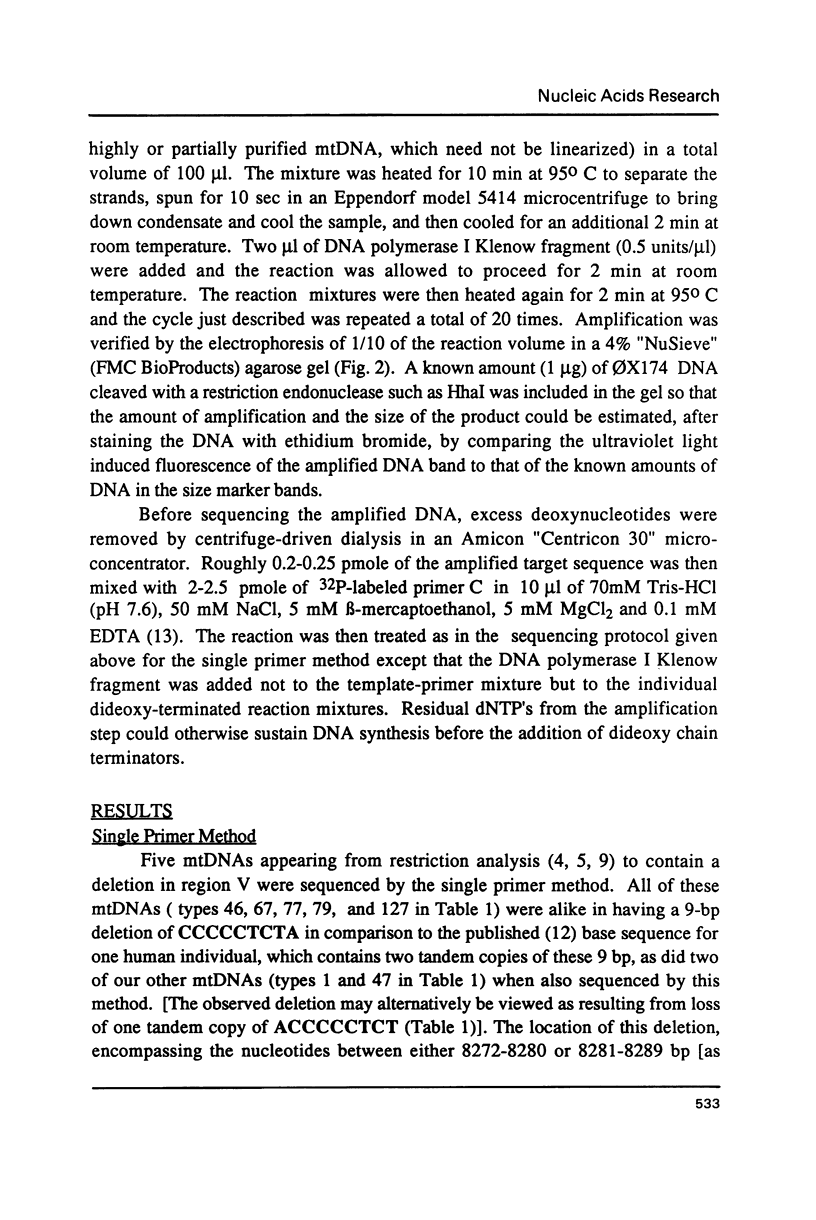




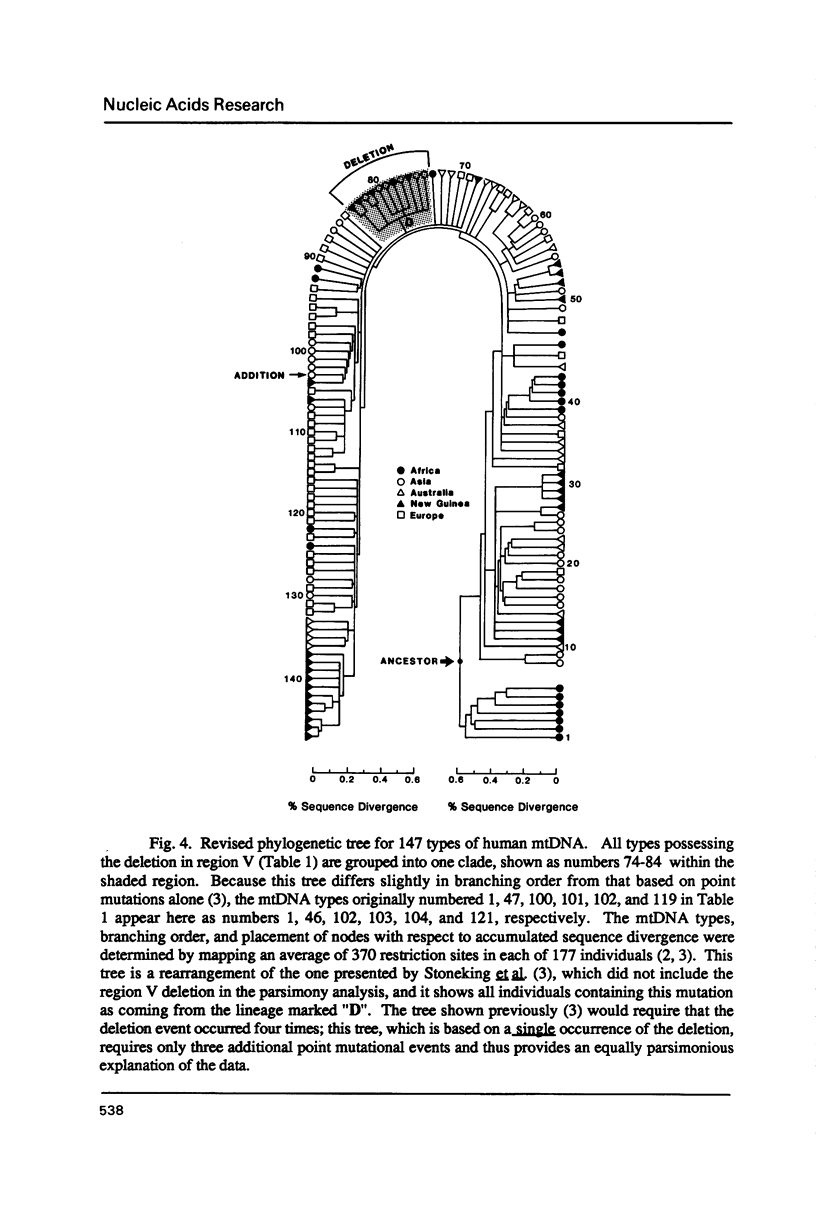




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Brown G. G., DesRosiers L. J. Rat mitochondrial DNA polymorphism: sequence analysis of a hypervariable site for insertions/deletions. Nucleic Acids Res. 1983 Oct 11;11(19):6699–6708. doi: 10.1093/nar/11.19.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Brown W. M., Wilson A. C. Polymorphic sites and the mechanism of evolution in human mitochondrial DNA. Genetics. 1984 Mar;106(3):479–499. doi: 10.1093/genetics/106.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann R. L., Wilson A. C. Length mutations in human mitochondrial DNA. Genetics. 1983 Aug;104(4):699–711. doi: 10.1093/genetics/104.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles R. E., Blanc H., Cann H. M., Wallace D. C. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg B. D., Newbold J. E., Sugino A. Intraspecific nucleotide sequence variability surrounding the origin of replication in human mitochondrial DNA. Gene. 1983 Jan-Feb;21(1-2):33–49. doi: 10.1016/0378-1119(83)90145-2. [DOI] [PubMed] [Google Scholar]
- Hauswirth W. W., Clayton D. A. Length heterogeneity of a conserved displacement-loop sequence in human mitochondrial DNA. Nucleic Acids Res. 1985 Nov 25;13(22):8093–8104. doi: 10.1093/nar/13.22.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W. W., Van de Walle M. J., Laipis P. J., Olivo P. D. Heterogeneous mitochondrial DNA D-loop sequences in bovine tissue. Cell. 1984 Jul;37(3):1001–1007. doi: 10.1016/0092-8674(84)90434-3. [DOI] [PubMed] [Google Scholar]
- Horai S., Matsunaga E. Mitochondrial DNA polymorphism in Japanese. II. Analysis with restriction enzymes of four or five base pair recognition. Hum Genet. 1986 Feb;72(2):105–117. doi: 10.1007/BF00283927. [DOI] [PubMed] [Google Scholar]
- Jones C. W., Kafatos F. C. Accepted mutations in a gene family: evolutionary diversification of duplicated DNA. J Mol Evol. 1982;19(1):87–103. doi: 10.1007/BF02100227. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Bugawan T. L., Horn G. T., Mullis K. B., Erlich H. A. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature. 1986 Nov 13;324(6093):163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf S. J., Horn G. T., Erlich H. A. Direct cloning and sequence analysis of enzymatically amplified genomic sequences. Science. 1986 Sep 5;233(4768):1076–1078. doi: 10.1126/science.3461561. [DOI] [PubMed] [Google Scholar]
- Stoneking M., Bhatia K., Wilson A. C. Rate of sequence divergence estimated from restriction maps of mitochondrial DNAs from Papua New Guinea. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):433–439. doi: 10.1101/sqb.1986.051.01.052. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Suggs S. V., Miyoshi K., Bhatt R., Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981 Dec;16(1-3):21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Shaffer J., Murphy R. F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979 Aug 10;6(11):3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]