Abstract
The dynamics of neutrophil chemotaxis under competing chemoattractant gradients was studied using a microfluidic platform. This microfluidic platform, which establishes a stable and dynamic gradient of chemoattractants across a cell culture chamber, enabled the investigation of human neutrophil migration patterns in the presences of four different chemoattractants (leukotriene B4, chemokine C-X-C motif ligands 2 and 8, and fMLP) and competing gradients of all pairwise combinations. The migration patterns for individual cells were tracked and quantitatively analyzed, and the results suggest a hierarchy among these chemoattractants of fMLP > CXCL8 > CXCL2 > leukotriene B4. In all conditions, over 60 % of neutrophils exposed to a competing gradient move toward the stronger signal though the weaker chemoattractant still influences neutrophil motility. These results yield insight about how each chemoattractant contributes to overall neutrophil chemotaxis within complex physiological environments.
INTRODUCTION
Chemotaxis is an important cellular process in immune response by which cells migrate toward a site of interest, such as the site of infection,1, 2 inflammation,3, 4 injury,5, 6 or even cancer metastasis.7–9 In chemotaxis, cells are guided by signaling molecules called chemoattractants. In unusual circumstances, such as inflammation or infection, specific chemoattractants are up/down-regulated or newly introduced. Fifty years of studies on chemotaxis has identified various chemoattractants and revealed that each cell type within the immune system responds to a unique set of chemoattractant signals.10–16 However, conventional assay techniques have limited ability to delineate the relative strength of each chemoattractant for a given cell type and the way a chemoattractant influences that cell. Conventional chamber-based assay platforms, such as the Boyden chamber17–19 or the Dunn chamber20–21, rely on diffusion of asymmetrically introduced molecules to establish a chemical gradient; as such, the platforms do not allow precise control over concentrations of the gradient, and the resulting gradient degrades with time. Also, traditional methods are typically not quantitative and provide no insight on the cellular behavior at the single cell level because they monitor ensemble average characteristics. While true that cells behave in an ensemble in vivo, individual cells of the same cell type show heterogeneity in their response to a stimulus. This heterogeneity may be important in the biological function of the cells and is clearly demonstrated by histogram analysis of neutrophil populations herein. More importantly, experiments using conventional assays are commonly performed in an over-simplified environment, such as static buffer conditions or a single chemoattractant gradient18–25, which is insufficient to reflect the real chemotactic environment of the cells in vivo as the chemical signaling events in the in vivo milieu occur under complex and dynamic environments. A few research efforts have endeavored to take such environmental aspects into account25–28, and these studies have revealed several characteristics of chemotaxis not obvious using classical methods, such as the effect of dynamic gradients on chemotaxis or cellular integration of multiple chemotactic signals.
To address the challenge of introducing complexity into chemotaxis studies, this research utilizes microfluidics to form competing gradients of two chemoattractants while monitoring the resulting behavior of individual human neutrophils. Microfluidic platforms enable creation and control of dynamic microenvironments while providing other advantages, such as small volume sample requirements, device optical transparency, and biocompatibility, based on the small device dimensions and the device media (polydimethylsiloxane or PDMS in this research)29, 30. Using a microfluidic platform, this work investigates all pairwise combinations of four chemoattractants to quantitatively analyze neutrophil chemotaxis and reveals a hierarchical preference of neutrophils among these chemoattractants.
Neutrophils are granular leukocytes that play important roles in the immune system. Several diseases, including lung cancer and asthma, have shown abnormalities in their neutrophil populations or the levels of neutrophil chemoattractants3, 4, 9, 13, 31–35, and therefore, understanding their chemotaxis will enable deeper understanding of the immune response, both fundamentally and in these diseases. A variety of chemoattractants have been shown to induce neutrophil chemotaxis, including chemokine C-X-C motif ligands 2 and 8 (CXCL2 and CXCL8), leukotriene B4 (LTB4), and formyl-met-leu-phe (fMLP), the four chemoattractants considered herein. CXCL2 and CXCL8 are part of a chemokine sub-family produced by many cell types (including neutrophils, T-cells, endothelial cells, epithelial cells, and others)13, 36 with known roles in several inflammatory diseases. A major lipid product of several cell types, LTB4, is reported as an indicator of neutrophil activation32, 35, 37, and is known to play a role in cell adhesion, oxygen metabolite production, and degranulation of immune cells.32. Lastly, fMLP is believed to originate from bacterial protein degradation and is known to activate several cell types, including neutrophils, to produce tissue-destructive oxygen-derived free radicals in phagocytic cells as well as induce neutrophil chemotaxis.38 In short, CXCL2, CXCL8, and LTB4 are host-derived chemoattractants while fMLP is a bacterially derived chemoattractant for neutrophils. The first three are used in this study to mimic a situation in which only host-derived signals exist, and fMLP is employed to mimic a situation in which host- and bacterially derived signals conflict. While a couple of recent studies have investigated the chemotaxis of neutrophils in complex environment,25, 39–41 situational and hierarchical preference of neutrophils has not yet been studied. To be noted, as the goal of this research is to reveal the influence of various types of chemoattractants (hierarchical preference of neutrophils for chemoattractants), other contributing characteristics that can also affect neutrophil chemotaxis, such as mean chemoattractant concentration and steepness of a chemoattractant gradient, are finely controlled in this research.
EXPERIMENTAL SECTION
1. Cell Preparation
Ethylenediaminetetraacetic acid (EDTA)-anticoagulated whole human blood samples were obtained from Memorial Blood Center (St. Paul, MN) according to approved IRB protocol E&I ID#07809. All blood samples were collected from healthy donors by a skilled professional at the Memorial Blood Center on the day that experiments were performed and were used immediately after the samples were obtained. Neutrophils were isolated from 5mL of blood by density gradient centrifugation (Solon, OH) following a previously reported protocol.42 Neutrophils were resuspended in Hank’s Buffered Salt Solution (HBSS, Sigma-Aldrich, St. Louis, MO) containing 2% human serum albumin (HSA, Sigma-Aldrich, St. Louis, MO) that was kept at 37 °C.
2. Device Fabrication
The microfluidic devices were fabricated using photolithography techniques. Mask designs were printed on transparent film (Cad/Art Service Inc., Bandon, OR) and transferred onto a chrome mask plate with a positive photoresist (Nanofilm, Westlake Village, CA). The transferred designs were developed using a developer solution (351 developer, Rohm and Hass Electronic Materials LLC, Marlborough, MA) to expose the chrome layer underneath the photoresist. Finally, the exposed chrome was etched down in a chrome etchant solution (Cyantek Corporation, Fremont, CA); the remaining photoresist was removed, and the plate was washed and dried. To fabricate a device master, a silicon wafer was cleaned using 1:10 hydrofluoric acid (Avantor Performance Materials, Phillipsburg, NJ):DI water solution and spin-coated with a negative photoresist, SU-8 50 (Microchem, Newton, MA). The mixing channel dimensions were 50 µm (W) × 100 µm (D) × 2330 µm (L) while the cell culture chamber was 450 µm (W) × 100 µm (D) × 2500 µm (L). After a baking step, the device design was transferred onto the photoresist layer on a silicon wafer through the previously prepared mask. Following a second baking step, the device design on the silicon wafer was developed in SU-8 developer (Microchem, Newton, MA). Upon completion of the master, a Sylgard 184 elastomer and curing agent mixture (10:1 ratio, Ellsworth Adhesives, Germantown, WI) was cast onto the master and cured overnight at 95 °C. After punching inlet and outlet holes, the PDMS layer was bonded onto a glass coverslip via oxygen plasma treatment at 100 W for 10 seconds. The complete device was then brought into a biosafety cabinet where the device was exposed to UV light for an hour and kept in the biosafety cabinet to promote sterility until use.
3. Device preparation for experiments
Before use, the channels were washed using a 70% v/v ethanol solution in sterilized Milli-Q water (Millipore, Billerica, MA). After rinsing the channels with sterilized Milli-Q water, a 250 µg/mL solution of human fibronectin (Invitrogen, Carlsbad, CA) in sterilized Milli-Q water was introduced through the channels, and the device was incubated under 5 % CO2 at 37 °C for an hour. . Following fibronectin incubation, 50 µL of neutrophil suspension (2 – 8 × 106 cells/mL) in HBSS containing 2% HSA were injected into the cell culture chamber inlet, and the device was kept in the incubator for another hour to promote neutrophil adhesion. The device was then connected to a syringe pump, and HBSS containing 2% HSA was injected through the channel for 2 minutes to remove non-adherent neutrophils. Then, the desired combination of chemoattractants (e.g. HBSS containing 2% HSA vs. CXCL2 in HBSS containing 2% HSA) was introduced through the two chemical inlets. CXCL2, CXCL8, and fMLP were purchased from Sigma Aldrich (St. Louis, MO) and LTB4 was purchased from Cayman Chemical (Ann Arbor, MI). All chemicals were used as received.
4. Gradient Confirmation
To ensure that the device was appropriate to generate the expected chemoattractant gradients, computational fluid dynamics (CFD) modeling was performed using COMSOL Multiphysics ver. 4.1. To reduce the time necessary for modeling, stationary analysis was performed on a single mixing channel geometry in 3-D. Flow rates used for modeling were 50, 75, and 100 µL/h, and diffusion coefficients for molecules introduced into the channels ranged from 1.5 × 10−5 to 1.0 × 10−6 cm2/s, values simulating both small molecule and small protein diffusion.43, 44
5. Time-Lapse Microscopy
Time-lapse images of neutrophils migrating inside the microfluidic cell culture chamber were obtained using Metamorph Ver. 7.7.5 imaging software on an inverted microscope equipped with a 10 × objective (Nikon, Melville, NY) and a CCD camera (QuantEM, Photometrics, Tucson, AZ). Images of neutrophils in the chamber were acquired every 10 seconds for 30 minutes.
6. Analysis of data
For experiments with a single chemoattractant gradient of CXCL2, CXCL8, LTB4, or fMLP, 15 or more neutrophils were randomly chosen from each device and analyzed. For experiments with competing gradients of chemoattractants, 10 or more neutrophils were randomly chosen from both the left and right sides of a channel (total of 20 or more neutrophils per device), and separately analyzed. For each condition, neutrophils from 3 different donors were used (3 independent experiments per condition). MetaMorph, the imaging software, tracked positions of each cell in each frame, and the tracked data was further analyzed using Microsoft Excel. From the obtained data, the migration patterns of neutrophils were described using 3 numeric parameters: motility index (MI), chemotactic index (CI), and effective chemotactic index (ECI). Similar interpretation has been previously done to describe neutrophil chemotaxis40. MI indicates how much of total possible neutrophil movement was in the overall dominant xy direction and is defined as the ratio of final displacement (d) and maximum displacement (dmax) of a neutrophil:
MI = d/dmax
where dmax is defined as the product of the average migration speed of the tracked neutrophil and the total migration time.
The orientation of neutrophils during migration is quantified by CI which is defined as:
CI = x/dtotal
where × is the final displacement along the x-axis (the direction of the gradient) and dtotal is the total migration distance.
Finally, ECI (the product of MI and CI) is used to quantify how effective chemotaxis was in the neutrophil migration. In this report, movement toward the right side of the channel results in positive CI and ECI values. These three parameters for individual cells are plotted as histograms, and averaged values of these parameters are plotted per condition. T-test and analysis of variance (ANOVA) without assumption of a Gaussian distribution (also called Kruskal-Wallis test) are used for statistical comparison, and the error in all figures in this manuscript are represented as the standard error of the mean.
RESULTS AND DISCUSSION
Building a gradient using microfluidic mixing channels
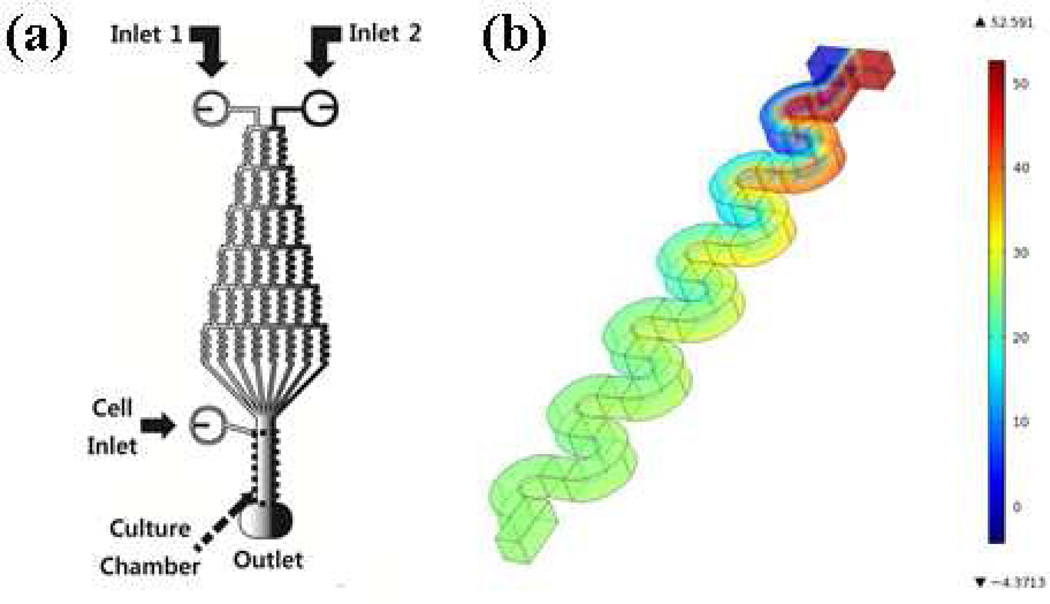
The microfluidic platform consists of many serpentine channels and an observation channel (the cell culture chamber) (Figure 1-a). Chemoattractant-containing solutions introduced into the series of serpentine channels for mixing establish a stable gradient of chemoattractant in the observation channel.45 The key to establishing such a gradient in the observation channel is a complete mixing process in each serpentine channel; therefore, a CFD module in COMSOL multiphysics 4.1 was used to verify that the device operated as desired (Figure 1-b). In the simulations, 1.5 × 10−5 cm2/s and 1.0 × 10−6 cm2 /s diffusion coefficients were used to model small molecules and small proteins, respectively, diffusing through the channels.43, 44 Figure 1-b shows an example result for 1.5 × 10−5 cm2 /s diffusion coefficient and 100 µL/h flow rate, and all other model results showed a complete mixing process like that depicted in figure 1-b, confirming that neutrophils will be exposed to the desired concentration gradient of chemoattractants as specified below. Final gradients can be further evaluated using fluorescent dyes with the molecular weight similar to the chemoattractants of interest in this study; work done by others26, 45 has demonstrated gradient instability less than 10 % as long as the flow rate is maintained.26 For the purpose of this study, the investigation of neutrophil chemotaxis in a complex environment, the effect of flow itself was also evaluated. The shear stress in the main observation channel generated by the flow conditions was around 0.01 N/m2 at the maximum (calculated using COMSOL multiphysics simulation, data not shown), which is significantly smaller than the shear stress levels in in vivo environments (~ 1 N/m2 – 6 N/m2).46 Therefore, it is likely that the effect of the flow itself on neutrophil chemotaxis is minimized in this research.
Figure 1.
(a) Schematic of the device used in this work (not to scale), and (b) an example result for the mixing process modeled by COMSOL multiphysics (flow rate of 100 µL/h and a diffusion coefficient of 1.5 × 10−6 cm2s−1, molecule introduced to the right side inlet with the concentration of 50 ng/mL).
Neutrophil migration under single gradients
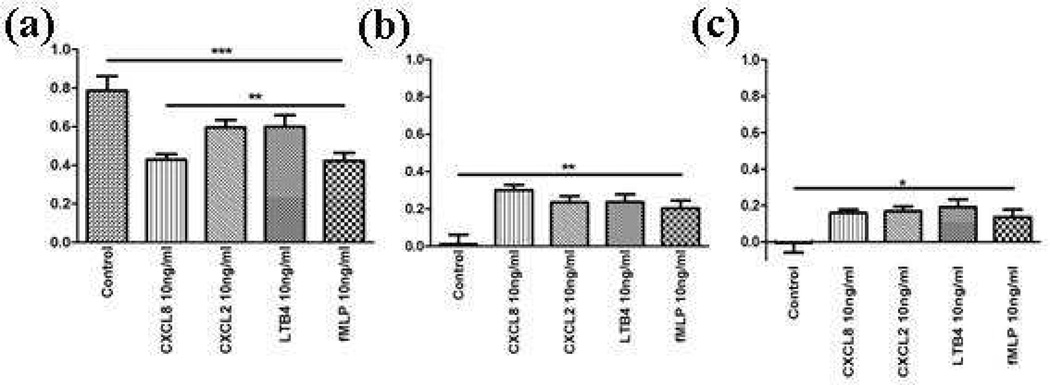
Neutrophil chemotaxis under single gradients of CXCL2, CXCL8, LTB4, and fMLP was studied using histogram analysis and three numerical parameters: MI to indicate motility of neutrophils’ migration, CI to quantify the orientation of neutrophils during migration, and ECI (the product of MI and CI) to quantify how effective chemotaxis is in the neutrophil migration. The receptors for each chemoattractant are different (CXCR1 and CXCR2 for CXCL2 and CXCL8, BLT1 and BLT2 for LTB4, and FLPR1 for fMLP)13, 32, 35–38, 47 so the expectation from this investigation was that neutrophils would respond differently to different gradients. First, control experiments were performed with media containing no chemoattractant. As shown in figure 2, cells showed the smallest effectiveness of chemotaxis (the smallest ECI) in their movement under this condition. Then, several gradients of different, individual chemoattractants were tested to find the maximum concentration in a gradient that resulted in indistinguishable ECI values for the four individual chemoattractants. The goal of this study was to reveal preference of neutrophils among the considered chemoattractants; therefore, using gradients resulting in similar ECI values eliminates the possible effect on neutrophil chemotaxis from the gradient itself.26, 48 Different gradients for different chemoattractants were tested, keeping all other conditions the same. The results from different gradients of the same chemoattractant (Figure S-1) and the same gradient of different chemoattractants showed that neutrophil responses are dependent both on the type and the concentration of chemoattractants, which is in agreement with previously reported results.26, 48–50 Results showed that the gradient with 10 ng/mL maximum concentration of each chemoattractant gave indistinguishable ECI values (Figure 2-c) (LTB4: 0.191 ± 0.0420, fMLP: 0.136 ± 0.0421, CXCL8: 0.159 ± 0.0191, CXCL2: 0.168 ± 0.0263, by Kruskal-Wallis test). To be noted, MI values for the 10 ng/mL gradient of chemoattractants are different (Figure 2-a). Previous studies have reported that the phosphatidylinositol 3-kinase (PI3K) signaling pathway is a critical pathway in neutrophil chemotaxis against host-derived chemoattractant, while mitogen-activated protein (MAP) kinase p38 is used in fMLP-induced neutrophil chemotaxis.24–25,41 Considering this and various receptors for each chemoattractant, it is interesting to note that MI for the CXCL8 condition was different from the MI values for either CXCL2 or LTB4 conditions. These data confirm that, while CXCL2 and CXCL8 share receptors, and LTB4 and CXCL8 use the same signaling cascade (PI3K), the resulting chemotaxis is not necessarily the same. In addition to MI, CI, and ECI parameter differences, different chemoattractants resulted in varied distributions of cells responding to the chemoattractant (Figure S-2). Interestingly, cells responding to the LTB4 gradient, compared to those of who were exposed to CXCL8, CXCL2, or fMLP gradients, showed a wider population distribution in the MI histogram (0.51 vs. 0.23, 0.28, or 0.31 standard deviation). Cells responding to LTB4 or fMLP showed a wider distribution in their ECI histogram than cells exposed to CXCL8 or CXCL2 (0.35, 0.32 vs. 0.16, 0.20 standard deviation). These distributions indicate that the mechanism of neutrophil response to different chemoattractants is different and results in distinctive characteristics in their migration patterns.
Figure 2.
(a) MIs, (b) CIs, and (c) ECIs under the control condition and single gradient conditions (N = 50 for control, 58 for CXCL8, 55 for CXCL2, 57 for LTB4, and 60 for fMLP). Results were statistically tested using Kruskal-Wallis test (*: p < 0.1, **: p < 0.01, ***: p < 0.0001)
Neutrophil migration under fMLP-containing competing gradients
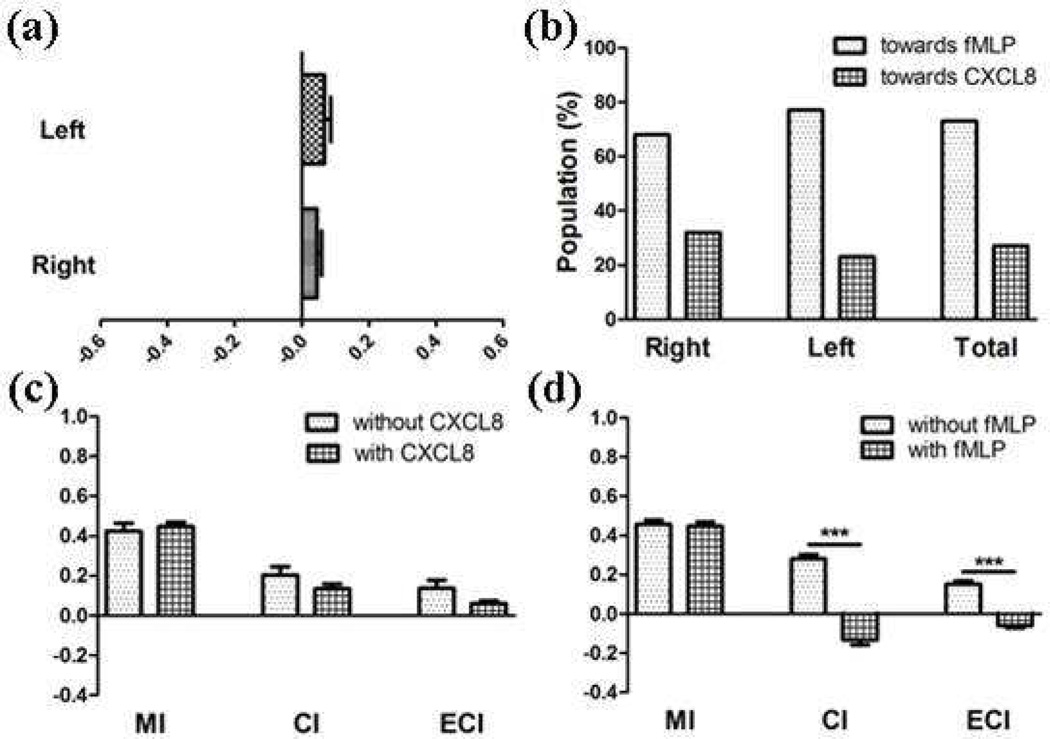
Neutrophils were exposed to CXCL8-fMLP (Figure 3), CXCL2-fMLP (Figure S-2), or LTB4-fMLP (Figure S-3) competing gradients, and their chemotaxis was monitored (Table 1). As mentioned, in each device, 10 or more cells from left side of the culture chamber were chosen and analyzed separately from 10 or more cells randomly chosen from the right side of the culture chamber. In all cases, overall migration of the cells exposed to the three pairwise competing gradients was toward fMLP, indicating that fMLP is the strongest chemoattractant among the four chemoattractants (Table 1). For example, under the CXCL8-fMLP competing gradient condition in which a 10 ng/mL CXCL8 solution was introduced from the left side inlet while a 10 ng/mL fMLP solution was introduced from the right side inlet, the overall trend for migration from both sides of the channel was toward fMLP (Figure 3-a). Precisely, 77% of cells from the left side of the channel and 68% of cells from the right side of the channel moved toward a higher concentration of fMLP (Figure 3-b). The population of cells moving away from fMLP was 27%, which was different from 16% observed during single gradient fMLP-mediated chemotaxis, indicating that the cells sensed and responded to both CXCL8 and fMLP simultaneously. Ultimately, however, fMLP-induced chemotaxis overwhelmed the CXCL8, and 73% of cells in the entire channel moved towards fMLP. Similar trends were observed in all fMLP-containing competing gradient conditions (Figure 3-b). Considering that the gradient for each chemoattractant (10 ng/mL maximum concentration in a gradient) and the effectiveness of chemotaxis under the single gradient condition for each chemoattractant (Figure 2-c) were kept the same, these data reveal that there is an hierarchy among the chemoattractants considered herein, with the strongest neutrophil response to fMLP. This is in agreement with previous reports that showed that activation of the p38 MAP kinase pathway overwhelms the PI3K pathway, and the chemotaxis of neutrophils in the system is dominated by p38 MAP kinase pathway (activated by fMLP in this research).24, 25
Figure 3.
Results from CXCL8-fMLP-mediated chemotaxis (***: p < 0.0001 using unpaired T-test). (a) ECIs from each sides of the channel with Nleft = 31 and Nright = 36 (positive sign indicates that the chemotaxis was toward fMLP while negative sign indicates that the chemotaxis was toward CXCL8), (b) Populations of the cells moved toward a chemoattractant, (c) MI, CI, and ECI comparison to those from fMLP-mediated chemotaxis (positive sign indicates that the chemotaxis was toward fMLP while negative sign indicates that the chemotaxis was toward CXCL8), and (d) MI, CI, and ECI comparison to those from CXCL8-mediated chemotaxis (positive sign indicates that the chemotaxis was toward CXCL8 while negative sign indicates that the chemotaxis was toward fMLP).
Table 1.
Result summary for all single and competing gradient conditions.
| % of cells moved toward right* (%) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Conditions | Ntotal | Left side |
Right Side |
Total | MI | CI | ECI | ECI (CI) toward |
| CXCL8-fMLP | 67 | 77 | 68 | 73 | 0.447 ± 0.0224 | 0.133 ± 0.0251 | 0.060 ± 0.0131 | fMLP |
| CXCL2-fMLP | 61 | 91 | 69 | 80 | 0.713 ± 0.0705 | 0.176 ± 0.0355 | 0.137 ± 0.0352 | fMLP |
| LTB4-fMLP | 60 | 81 | 93 | 88 | 0.606 ± 0.0444 | 0.313 ± 0.0402 | 0.209 ± 0.0327 | fMLP |
| LTB4-CXCL8 | 65 | 89 | 38 | 66 | 0.631 ± 0.0558 | 0.130 ± 0.0415 | 0.112 ± 0.0505 | CXCL8 |
| CXCL2-CXCL8 | 68 | 66 | 78 | 73 | 0.456 ± 0.0350 | 0.116 ± 0.0282 | 0.087 ± 0.0241 | CXCL8 |
| CXCL2-LTB4 | 61 | 74 | 63 | 69 | 0.278 ± 0.0314 | 0.106 ± 0.0208 | 0.053 ± 0.0181 | LTB4 |
| fMLP | 60 | N/A | N/A | 84 | 0.423 ± 0.0413 | 0.204 ± 0.0406 | 0.136 ± 0.0421 | fMLP |
| CXCL8 | 58 | N/A | N/A | 89 | 0.430 ± 0.0274 | 0.301 ± 0.0277 | 0.159 ± 0.0191 | CXCL8 |
| LTB4 | 57 | N/A | N/A | 79 | 0.599 ± 0.0603 | 0.237 ± 0.0418 | 0.191 ± 0.0420 | LTB4 |
| CXCL2 | 55 | N/A | N/A | 82 | 0.596 ± 0.0375 | 0.234 ± 0.0347 | 0.168 ± 0.0263 | CXCL2 |
Indicates the chemoattractants in bold.
The three numerical parameters, MI, CI, and ECI, were used to quantitatively analyze neutrophil migration patterns in detail (Table 1). MI, CI, and ECI values from competing gradient conditions were statistically compared to those from the single gradient experiments. In the CXCL8-fMLP competing gradient condition, for example, MI, CI, and ECI values obtained from CXCL8-fMLP competing gradient experiments (0.447 ± 0.0224, 0.133 ± 0.0251, and 0.0602 ± 0.0131, respectively) were compared to the values from both the fMLP single gradient condition (0.423 ± 0.0413, 0.204 ± 0.0406, and 0.136 ± 0.0421, respectively, Figure 3-c), and the CXCL8 single gradient condition (0.430 ± 0.0274, 0.301 ± 0.0277, and 0.159 ± 0.0191, respectively, Figure 3-d). A general trend was found from the three fMLP-containing competing gradient conditions; the numerical indices from the competing gradient experiments were comparable to those from the stronger chemoattractant (fMLP in this case) single gradient experiments (Figure 3-c and 3-d). MI, CI, and ECI values from all fMLP-containing gradient conditions, except the MIs under the CXCL2-fMLP condition and LTB4-fMLP condition, were statistically indistinguishable from those of the fMLP single gradient conditions (c and d for Figure 3, S-3, and S-4). These results reveal that chemoataxis under the fMLP-containing competing gradient conditions are dominated by the fMLP-induced chemotaxis behavior. While the existence of a dominant signal has been shown from all competing gradient conditions above based on the direction of overall chemotaxis (both population and sign of CI and ECI values, Table 1), the same trend can also be found based on overall population of cells moving away from the stronger signal source. 27%, 20%, and 12% of cells moved away from fMLP under CXCL8-fMLP, CXCL2-fMLP, and fMLP-LTB4 experiments, respectively, and this results in the same fMLP > CXCL8 > CXCL2 > LTB4 hierarchical order. Again, this order reflects that fact that the p38 MAP kinase-dependent signaling pathway (for the fMLP signal) overwhelms PI3K-dependent signaling pathway (for the host-derived chemoattractants).24 However, the results also indicate that, while PI3K pathway is overwhelmed, it still influences neutrophil chemotaxis, and a stronger signal opposite fMLP interferes more with chemotaxis toward the dominant signal. Evidence showing that neutrophils use an hierarchy to prioritize certain chemical signals are accumulating in the literature,39, 41 and the results in this study also suggest that the cells in a complex environment use a hierarchy to prioritize particular signals from the surrounding environment while every signal present in the environment contributes to their chemotaxis. The MI results from CXCL2-fMLP and LTB4-fMLP conditions that do not match the MI values from the dominant signal gradient experiments (fMLP in this case) will be further explored in the discussion section.
Neutrophil migration under a competing gradient of host-derived chemoattractants
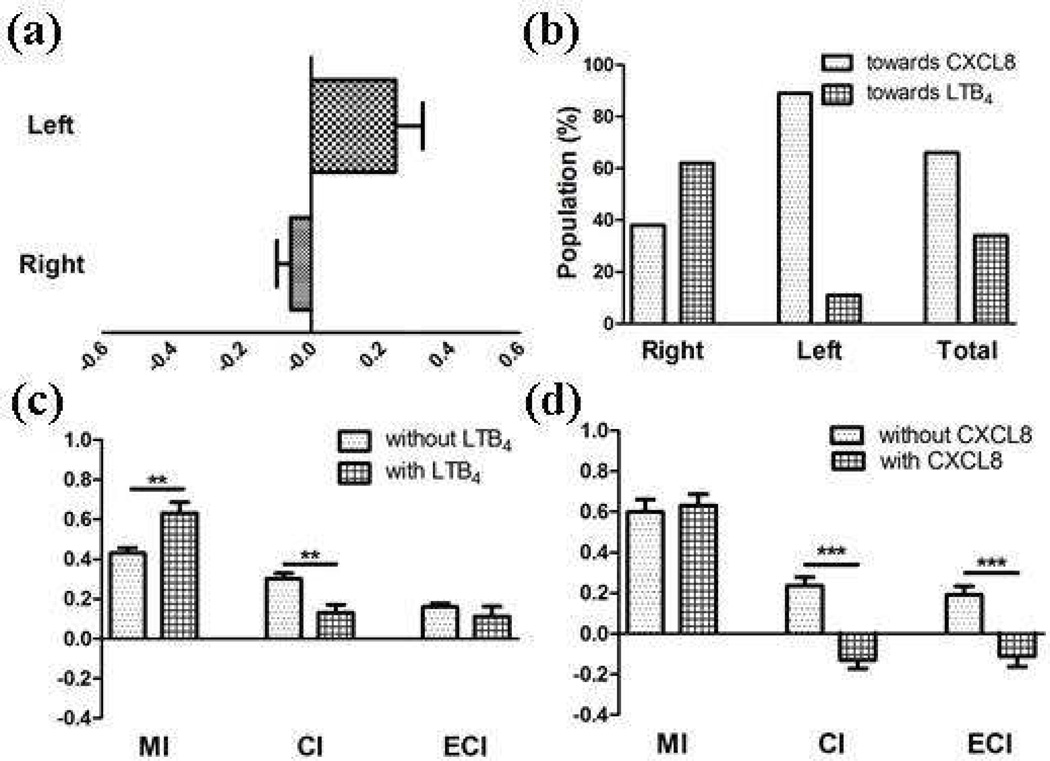
Neutrophils were also exposed to competing gradients that are combinations of only host-derived chemoattractants (LTB4-CXCL8, CXCL2-CXCL8, and CXCL2-LTB4). This investigates the hierarchy of chemoattractants that, whether they share receptors or not, trigger the same chemical signaling cascades (PI3K-dependent).13, 51 In LTB4-CXCL8 and CXCL2-CXCL8 conditions, it was common for the cells to migrate preferentially toward CXCL8 (Table 1). For example, under the LTB4-CXCL8 competing gradient condition (a 10 ng/mL LTB4 solution on the left side and a 10 ng/mL CXCL8 solution on the right side), overall chemotaxis of the cells was toward the right side of the chamber (toward CXCL8, Figure 4-a), with 38% of cells in the right side of the channel and 89% of cells in the left side of the channel moving toward CXCL8 (Figure 4-b); a total of 66% of cells in the entire channel moved toward CXCL8. These results suggest that CXCL8 is the most potent chemoattractant among the three host-derived chemoattractants (CXCL8, CXCL2, and LTB4), and, even though the chemoattractant shares either receptor or signaling pathway with another, each chemoattractant possesses its own strength for induction of neutrophil chemotaxis.
Figure 4.
Results from LTB4-CXCL8-mediated chemotaxis (**: p < 0.01 and ***: p < 0.0001 using unpaired T-test). (a) ECIs from each side of the channel with Nleft = 32 and Nright = 33 (positive sign indicates that the chemotaxis was toward CXCL8 while negative sign indicates that the chemotaxis was toward LTB4), (b) Populations of the cells moving toward a chemoattractant, (c) MI, CI, and ECI comparison to those from CXCL8-mediated chemotaxis (positive sign indicates that the chemotaxis was toward CXCL8 while negative sign indicates that the chemotaxis was toward LTB4), and (d) MI, CI, and ECI comparison to those from LTB4-mediated chemotaxis (positive sign indicates that the chemotaxis was toward LTB4 while negative sign indicates that the chemotaxis was toward CXCL8).
The three numerical parameters, MI, CI, and ECI, were used to quantitatively analyze the neutrophil migration patterns (Table 1). MI, CI, and ECI values from competing gradient conditions were statistically compared to those from single gradient experiments (Figure 4 and S-5). In the LTB4-CXCL8 competing gradient condition (Figure 4) as an example, the obtained MI, CI, and ECI values were compared to the values from both the CXCL8 single gradient condition (Figure 4-c), and the LTB4 single gradient condition (Figure 4-d). CIs and ECIs under the two competing gradient conditions were significantly lower than the CIs and ECIs obtained under single gradient conditions (c and d for Figure 4 and S-5). This indicates that neutrophil chemotaxis under host-derived chemoattractant only competing gradient conditions cannot be represented as the stronger chemoattractant (CXCL8 in this case)-mediated chemotaxis.
In the CXCL2-LTB4 condition (Figure S-6), the results suggested that LTB4 was more potent than CXCL2, and it was observed that the MI, CI, and ECI values were significantly reduced compared to those from either single CXCL2 or LTB4 single gradient conditions. This indicates that neutrophil chemotaxis under the CXCL2-LTB4 competing gradient condition cannot be represented by neutrophil chemotaxis under the more potent chemoattractant (LTB4); however, as stated, it was found that there was still a hierarchy that neutrophils use to prioritize the LTB4 signal.
The results under competing gradients of only host-derived chemoattractants indicate an hierarchy of CXCL8 > LTB4 > CXCL2 and support the conclusion that neutrophil migration patterns in complex environments cannot be explained by simple vector addition; there is a dominant signal in each pair of conditions. Like the results from fMLP-containing competing gradient conditions, neutrophils appear to integrate signals from each chemoattractant with a hierarchy to prioritize a certain signal.
Discussion
Neutrophil chemotaxis: Against both bacterially derived and host-derived chemoattractants
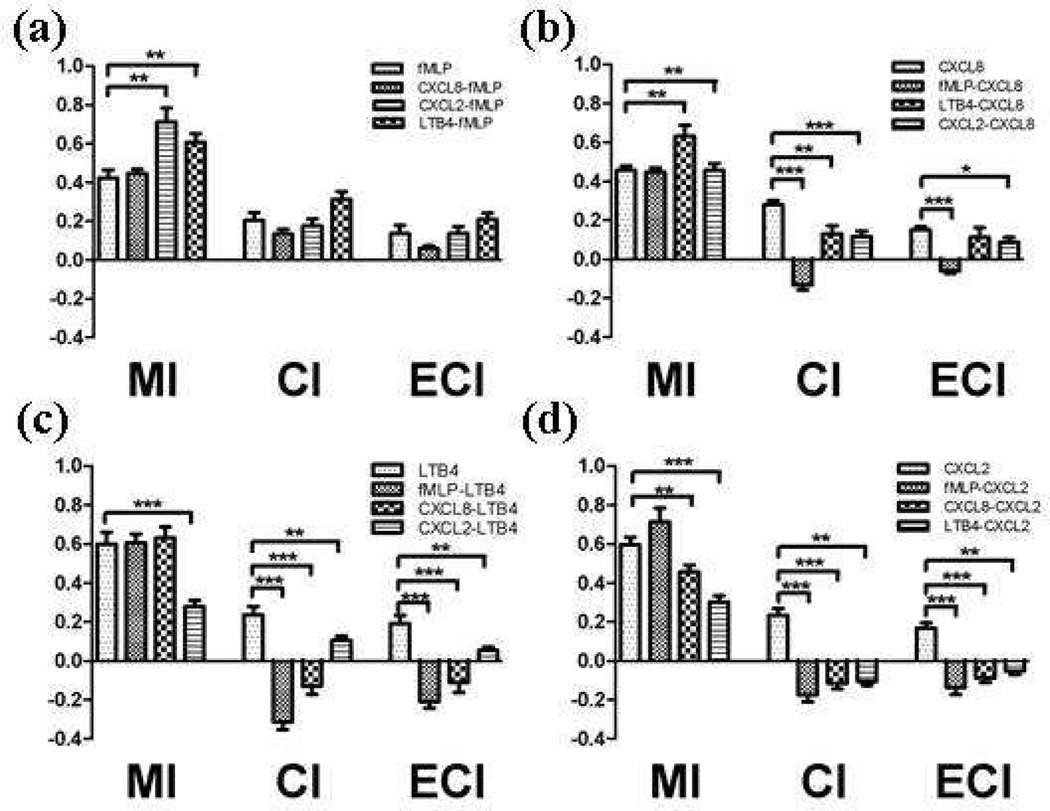
The results demonstrate a hierarchy among the employed chemoattractants in which neutrophils prioritize fMLP > CXCL8 > LTB4 > CXCL2 (Table 1 and Figure 5). More details about chemotaxis can be gained using the measure of MI to indicate motility of neutrophils’ migration, CI to quantify the orientation of neutrophils during migration, and ECI (the product of MI and CI) to quantify how effective chemotaxis is in the neutrophil migration.
Figure 5.
MI, CI, and ECI comparison summary (a) Comparison of MIs, CIs, and ECIs from fMLP-containing conditions, (b) Comparison of MIs, CIs, and ECIs from CXCL8-containing conditions, (c) Comparison of MIs, CIs, and ECIs from LTB4 containing conditions, and (d) Comparison of MIs, CIs and ECIs from CXCL2-containing conditions. MIs, CIs, and ECIs from competing gradient conditions were statistically tested using unpaired T-test against MI, CI, and ECI from single gradient conditions (*: p < 0.1, **: p < 0.01, ***: p < 0.0001).
An in vivo study reported that the PI3K-dependent pathway is not required for fMLP-induced chemotaxis but that it accelerates fMLP-induced chemotaxis.24 Comparison to this in vivo study reveals whether or not the results obtained using this in vitro platform mimic the in vivo milieu, while also providing some biophysical insight into neutrophil behavior. The results show that host-derived chemoattractant signals superimposed with fMLP signals result in no change in cellular orientation trend compared to the orientation trend achieved under the fMLP single gradient condition. While cells orient toward fMLP in complex conditions, the MIs, or motilities under the varied competing gradients, are not dominated by fMLP in the same way (Figure 5-a). That is, the MIs under CXCL2-fMLP and LTB4-fMLP conditions are 0.713 ± 0.0705 and 0.606 ± 0.0444, significantly different from the MI for fMLP-mediated chemotaxis (0.423 ± 0.0413). Higher motilities (higher MIs) indicate that neutrophils move, regardless of direction, better and faster, which can result in accelerated chemotaxis. Under most competing gradient conditions, the MI values measured are 1.5 - 2 times larger than those measured from fMLP single gradient conditions Further, the analyses show that the competing gradient MI values are actually comparable to those measured from single gradient CXCL2-, LTB4-, and CXCL8-mediated chemotaxis, 0.596 ± 0.0375 (p = 0.1792), 0.599 ± 0.0603 (p = 0.9285), and 0.447 ± 0.0224 (p = 0.7875), respectively (Figure 5-b, c, and d). These results suggest that while overall chemotaxis is dominated by one signal, the minor chemoattractant contributes to the character of chemotaxis, resulting in the accelerated fMLP-induced chemotaxis previously reported.24 MIs under fMLP-containing competing gradient conditions seem to originate from the secondary chemoattractant while CI values under the same conditions seem to originate from the dominant chemoattractant (Figure 5). Under these conditions, the dominant chemoattractant is fMLP and therefore overall orientation is toward fMLP while the MIs are comparable to the MIs from the weaker chemoattractant (CXCL8, CXCL2 or LTB4), as demonstrated by the trends found in Figure 4. MI histograms of CXCL8-fMLP, CXCL2-fMLP, and LTB4-fMLP conditions are more similar to MI histograms of CXCL8, CXCL2, and LTB4 than the histogram for fMLP-mediated chemotaxis (Figure S-7 to S-10). Combined, these data suggest that neutrophils prioritize a certain signal while they integrate multiple signals, and both the dominant and the secondary chemoattractants contribute to the cellular migration behavior (orientation and motility).
Neutrophil chemotaxis under competing gradient conditions: Against only host-derived chemoattractants
Experiments under the CXCL2-CXCL8, CXCL2-LTB4, and LTB4-CXCL8 investigate how neutrophils respond to signals that trigger the same signaling cascade (in this case, the PI3K-dependant pathway). CXCL2 and CXCL8 interact with neutrophils via CXCR1 and CXCR2 receptors while LTB4 interacts with neutrophils via BLT1 and BLT2 receptors.10–16 Competing gradients of chemoattractants that initiate signaling cascades via either CXCR receptors or BLT receptors (or both) generally suppressed chemotaxis toward the competing signal. For example, a LTB4 signal (via BLT receptors) suppressed CXCL2-induced chemotaxis (via CXCR receptors), resulting in significantly lower MI, CI, and ECI values than those from CXCL2-mediated chemotaxis (Figure S-6). CXCL2 (via CXCR receptors) also suppressed the effectiveness of CXCL8-induced chemotaxis (Figure S-5). The only exceptions to this trend were the LTB4 signal boosting motility of neutrophils during CXCL8-mediated chemotaxis and the CXCL2 signal resulting in no change in the motility of neutrophils during CXCL8- mediated chemotaxis (Figure 4, S-5). These results suggest that, even when competing chemoattractants trigger the same signaling cascade, there is still a dominant signal mediating neutrophil chemotaxis and both signals contribute to the migration characteristics.
Interestingly, depending on whether or not the p38 MAP kinase-dependent pathway is triggered with PI3K pathway, a different hierarchy among the three host-derived chemoattractants is obtained (Table 1). When only the PI3K pathway is triggered (host-derived chemoattractants only), CXCL8 is the strongest among the three host-derived chemoattractants, and LTB4 showed stronger effects on neutrophil chemotaxis when it is introduced in competition with CXCL2. The overall population of cells that moved toward LTB4 under the LTB4-CXCL8 gradient was more than those that moved toward CXCL2 under the CXCL2-CXCL8 condition. Experiments under the CXCL2-LTB4 condition confirmed the hierarchy of CXCL8 > LTB4 > CXCL2 (Figure 4, S-5, S-6). However, under fMLP-containing competing gradient conditions, when the PI3K pathway is triggered along with the p38 MAP kinase-dependent pathway, the hierarchy was fMLP > CXCL8 > CXCL2 > LTB4, showing stronger effects of CXCL2 than LTB4. This alteration in the hierarchy might be caused by the p38 MAP kinase-dependent pathway triggered with PI3K pathway. Alternatively, the alteration might be caused by saturation/desensitization of surface receptors for a certain chemoattractant under the conditions used in this study (10 ng/mL concentration), which likely result in suppressed chemotaxis toward the overdosed chemoattractant.
Considering all results, it can be concluded that neutrophils 1) sense and integrate multiple signals during chemotaxis and 2) use a hierarchy to prioritize among signals. With regards to the order of a chemoattractant in the hierarchy, the results suggest that, of the few considered here, CXCL8 is the most potent, LTB4 is second, and CXCL2 is the least potent among the host-derived chemoattractants; if a bacterially derived chemoattractant (fMLP) is introduced, it acts as the most potent chemoattractant and CXCL8, CXCL2 and LTB4 follow (CXCL2 becomes stronger than LTB4).
Conclusions
This work provides detailed information about neutrophil chemotaxis under single and competing gradients of multiple chemoattractants (fMLP, CXCL8, CXCL2, and LTB4). The microfluidic platform successfully incorporates two chemoattractants simultaneously to which the cells sense and respond. The single gradient experiments confirmed that the type of chemoattractants, mean concentration, and gradient itself are important aspects to be considered when studying neutrophil chemotaxis, and the proper gradient for each chemoattractant was selected for competing gradient studies. Through competing gradient experiments, we found that neutrophils respond to multiple chemoattractants by integrating and prioritizing the signals. A hierarchy of chemoattractants is found, and the hierarchy among the chemoattractants (fMLP > CXCL8 > CXCL2 > LTB4) is dynamic depending on which signaling cascades are triggered. Importantly, this research reveals that a dynamic in vitro platform can provide results that correspond well with in vivo studies; specifically herein, each chemoattractant signal contributes to the cellular migration (motility from the weaker chemoattractant and orientation from the dominant chemoattractant). The distinct behavior of each chemoattractant within the heterogeneous population of primary culture neutrophils is confirmed by histogram analysis of the MI, CI, and ECI parameters (see Supporting Information). This tool for screening neutrophil response to a variety of chemoattractants will lend significant insight into clinical research on neutrophil-related diseases such as asthma, chronic obstructive pulmonary disease or chronic granulocytic leukaemia.1–4, 7, 9, 32, 34, 35
Supplementary Material
ACKNOWLEDGMENT
This work was funded by grants from the National Institutes of Health New Innovator Award (DP2 OD004258-01). Device fabrication was done in Nanofabrication Center (NFC), and COMSOL Multiphysics modeling was performed with the support of Minnesota Supercomputing Institute (MSI) at the University of Minnesota. We thank to Ozlem Ersin, an associate professor at Manchester College School of Pharmacy (Fort Wayne, IN, U.S.A.) for valuable discussions regarding neutrophil chemotaxis, and Leah Laux, a NNIN REU awardee from Washington University in St. Louis, MO, for her efforts in fabricating preliminary microfluidic devices.
REFERENCES
- 1.Cooper JAD, Jr, Carcelen R, Culbreth R. J. Infect. Dis. 1996;173:279–284. doi: 10.1093/infdis/173.2.279. [DOI] [PubMed] [Google Scholar]
- 2.Braciale TJ, Sun J, Kim TS. Nat. Rev. Immunol. 2012;12:295–305. doi: 10.1038/nri3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CCM, Beck PL, Muruve DA, Kubes P. Science. 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- 4.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Nat. Rev. Immunol. 2011;11:519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 5.Fantone JC, Kunkel SL, Ward PA. Annu. Rev. of Physiol. 1982;44:283–293. doi: 10.1146/annurev.ph.44.030182.001435. [DOI] [PubMed] [Google Scholar]
- 6.Ng LG, Qin JS, Roediger B, Wang Y, Jain R, Cavanagh LL, Smith AL, Jones CA, de Veer M, Grimbaldeston MA, Meeusen EN, Weninger W. J. Invest. Dermatol. 2011;131:2058–2068. doi: 10.1038/jid.2011.179. [DOI] [PubMed] [Google Scholar]
- 7.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 8.Mendonça M, Cunha F, Murta E, Tavares-Murta B. Cancer Chemoth. Pharm. 2006;57:663–670. doi: 10.1007/s00280-005-0086-4. [DOI] [PubMed] [Google Scholar]
- 9.Carpagnano GE, Palladino GP, Lacedonia D, Koutelou A, Orlando S, Foschino-Barbaro MP. BMC cancer. 2011;11:226. doi: 10.1186/1471-2407-11-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luster AD. New Engl. J. Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 11.Luster AD. Chemotaxis: Role in Immune Response. John Wiley & Sons, Ltd; 2001. eLS. [Google Scholar]
- 12.Gillitzer R, Ritter U, Spandau U, Goebeler M, Brocker E-B. J. Invest. Dermatol. 1996;107:778–782. doi: 10.1111/1523-1747.ep12371803. [DOI] [PubMed] [Google Scholar]
- 13.Matsushima K, Terashima Y, Toda E, Shand F, Ueha S. Inflammation and Regeneration. 2011;31:11–22. [Google Scholar]
- 14.Li J, Gyorffy S, Lee S, Kwok CS. Inflammation. 1996;20:361–372. doi: 10.1007/BF01486739. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs AA, Huber JL, Ward RA, Klein JB, McLeish KR. J. Leukocyte Biol. 1995;57:679–686. doi: 10.1002/jlb.57.4.679. [DOI] [PubMed] [Google Scholar]
- 16.Cotter TG, Keeling PJ, Henson PM. J. Immunol. 1981;127:2241–2245. [PubMed] [Google Scholar]
- 17.Boyden S. J. Exp. Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson RD, Quie PG, Simmons RL. J. Immunol. 1975;115:1650–1656. [PubMed] [Google Scholar]
- 19.Brown AF. J. Cell Sci. 1982;58:455–467. doi: 10.1242/jcs.58.1.455. [DOI] [PubMed] [Google Scholar]
- 20.Zigmond SH. J. Cell Biol. 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zicha D, Dunn GA, Brown AF. J. Cell Sci. 1991;99:769–775. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- 22.Hopkins NK, Schaub RG, Gorman RR. BBA-Mol. Cell Res. 1984;805:30–36. doi: 10.1016/0167-4889(84)90033-8. [DOI] [PubMed] [Google Scholar]
- 23.Inoue T, Meyer T. PLoS ONE. 2008;3:e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heit B, Liu L, Colarusso P, Puri KD, Kubes P. J. Cell Sci. 2008;121:205–214. doi: 10.1242/jcs.020412. [DOI] [PubMed] [Google Scholar]
- 25.Heit B, Robbins SM, Downey CM, Guan Z, Colarusso P, Miller BJ, Jirik FR, Kubes P. Nat. Immunol. 2008;9:743–752. doi: 10.1038/ni.1623. [DOI] [PubMed] [Google Scholar]
- 26.Jeon NL, Baskaran H, Dertinger SKW, Whitesides GM, Van De Water L, Toner M. Nat. Biotech. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 27.Atencia J, Morrow J, Locascio LE. Lab Chip. 2009;9:2707–2714. doi: 10.1039/b902113b. [DOI] [PubMed] [Google Scholar]
- 28.Toetsch S, Olwell P, Prina-Mello A, Volkov Y. Integr. Biol. 2009;1:170–181. doi: 10.1039/b814567a. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell P. Nat. Biotech. 2001;19:717–721. doi: 10.1038/90754. [DOI] [PubMed] [Google Scholar]
- 30.Blow N. Nat. Meth. 2009;6:683–686. [Google Scholar]
- 31.Jablons D, Bolton E, Mertins S, Rubin M, Pizzo P, Rosenberg SA, Lotze MT. J. Immunol. 1990;144:3630–3636. [PubMed] [Google Scholar]
- 32.Reilly IA, Knapp HR, Fitzgerald GA. J. Clin. Pathol. 1988;41:1163–1167. doi: 10.1136/jcp.41.11.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokomizo T, Izumi T, Chang K, Takuwa Y, Shimizu T. Nature. 1997;387:620–624. doi: 10.1038/42506. [DOI] [PubMed] [Google Scholar]
- 34.Randis TM, Puri KD, Zhou H, Diacovo TG. Eur. J. Immunol. 2008;38:1215–1224. doi: 10.1002/eji.200838266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathis SP, Jala VR, Lee DM, Haribabu B. J. Immunol. 2010;185:3049–3056. doi: 10.4049/jimmunol.1001031. [DOI] [PubMed] [Google Scholar]
- 36.Barnett ML, Lamb KA, Costello KM, Pike MC. BBA-Mol. Cell Res. 1993;1177:275–282. doi: 10.1016/0167-4889(93)90123-7. [DOI] [PubMed] [Google Scholar]
- 37.Kreisle RA, Parker CW. J. Exp. Med. 1983;157:628–641. doi: 10.1084/jem.157.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cavicchioni G, Fraulini A, Falzarano S, Spisani S. Eur. J. Med. Chem. 2009;44:4926–4930. doi: 10.1016/j.ejmech.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Foxman EF, Kunkel EJ, Butcher EC. J. Cell Biol. 1999;147:577–588. doi: 10.1083/jcb.147.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin F, Nguyen CM-C, Wang S-J, Saadi W, Gross SP, Jeon NL. Ann. Biomed. Eng. 2005;33:475–482. doi: 10.1007/s10439-005-2503-6. [DOI] [PubMed] [Google Scholar]
- 41.Heit B, Tavener S, Raharjo E, Kubes P. J. Cell Biol. 2002;159:91–102. doi: 10.1083/jcb.200202114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh H, Siano B, Diamond S. J. Vis. Exp. 2008;17:e745. doi: 10.3791/745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brune D, Kim S. Proc. Natl. Acad. Sci. USA. 1993;90:3835–3839. doi: 10.1073/pnas.90.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delgado JMPQ. J. Phase Equilib. Diff. 2007;28:427–432. [Google Scholar]
- 45.Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Anal. Chem. 2001;73:1240–1246. [Google Scholar]
- 46.Papaioannou TG, Stefanadis C. Hellenic. J. Cardiol. 2005;46:9–15. [PubMed] [Google Scholar]
- 47.Souto FO, Zarpelon AC, Staurengo-Ferrari L, Fattori V, Casagrande R, Fonseca MJV, Cunha TM, Ferreira SH, Cunha FQ, Verri WA. J. Nat. Prod. 2011;74:113–118. doi: 10.1021/np1003017. [DOI] [PubMed] [Google Scholar]
- 48.Moghe PV, Nelson RD, Tranquillo RT. J. Immunol. Methods. 1995;180:193–211. doi: 10.1016/0022-1759(94)00314-m. [DOI] [PubMed] [Google Scholar]
- 49.Foxman EF, Campbell JJ, Butcher EC. J. Cell Biol. 1997;139:1349–1360. doi: 10.1083/jcb.139.5.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiu DT, Jeon NL, Huang S, Kane RS, Wargo CJ, Choi IS, Ingber DE, Whitesides GM. Proc. Natl. Acad. Sci. USA. 2000;97:2408–2413. doi: 10.1073/pnas.040562297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Billadeau DD. Nat. Immunol. 2008;9:716–718. doi: 10.1038/ni0708-716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.