Abstract
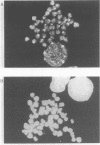
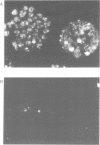
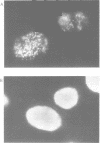
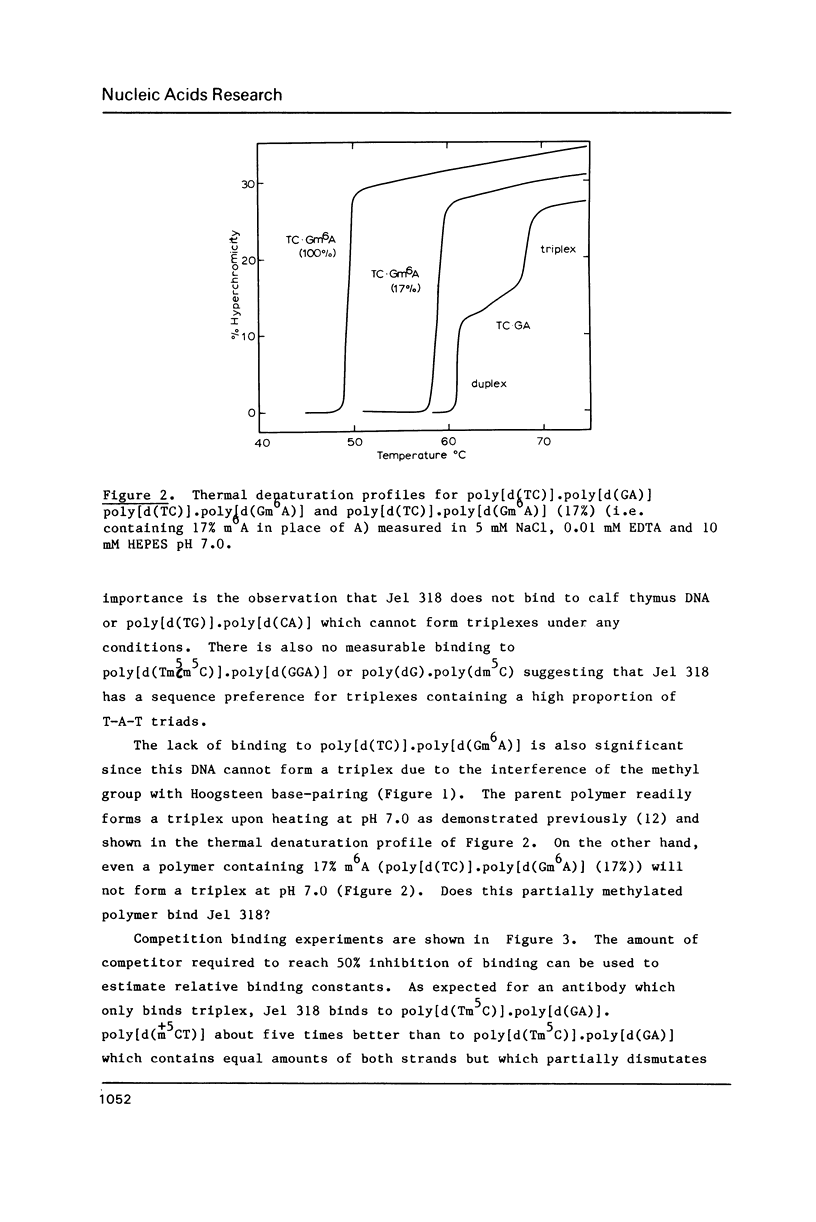
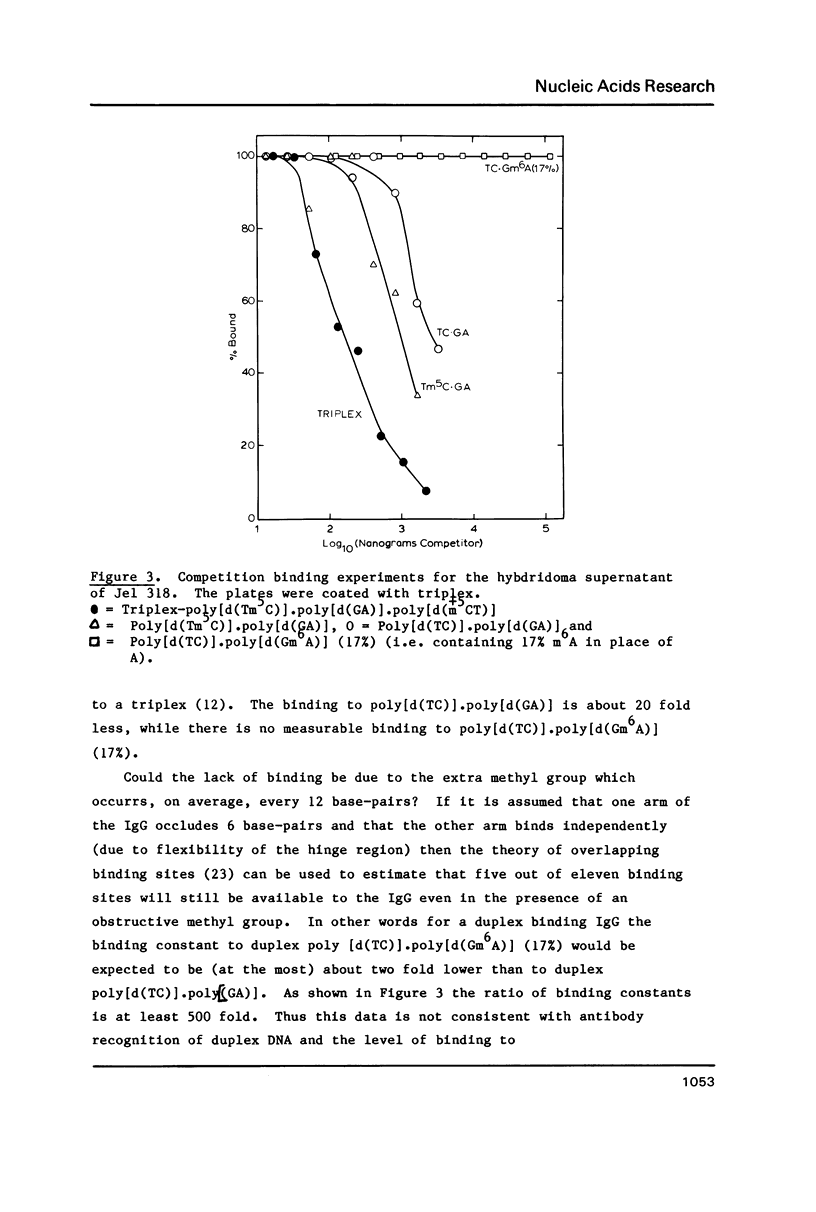
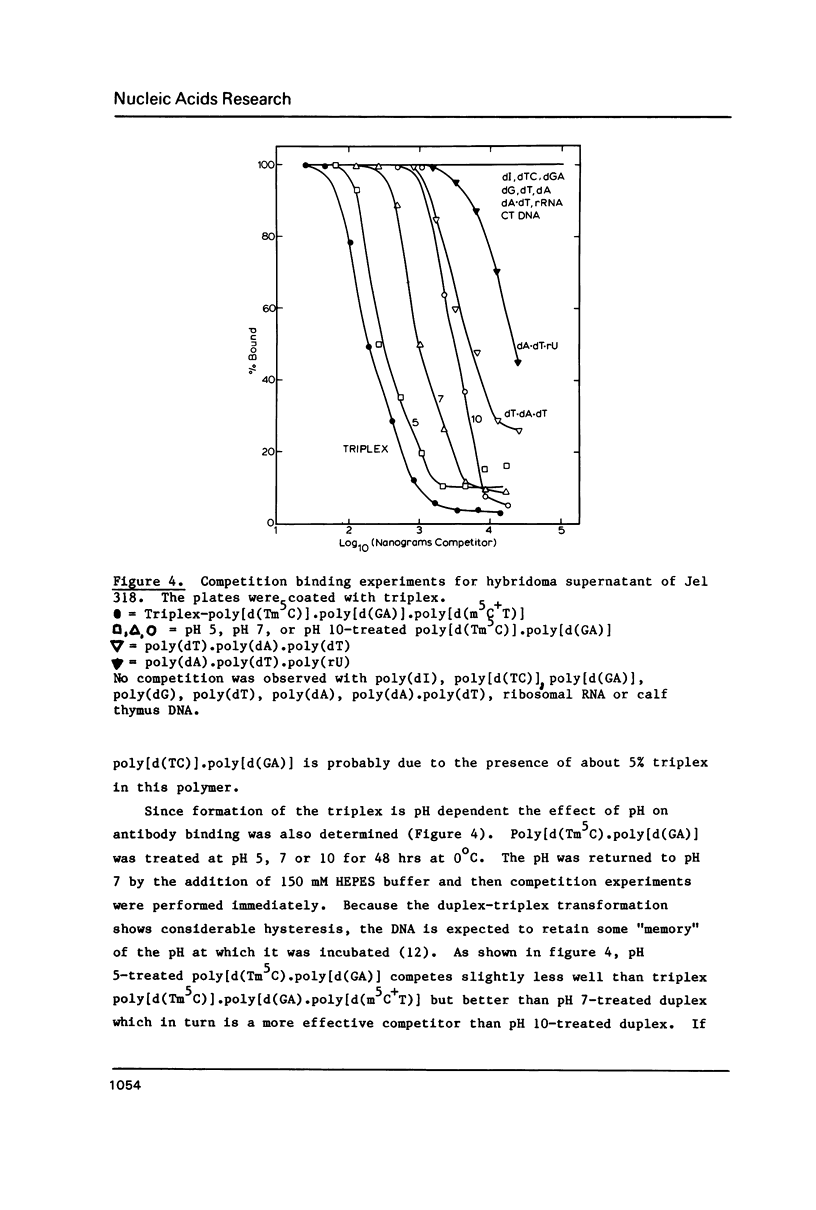
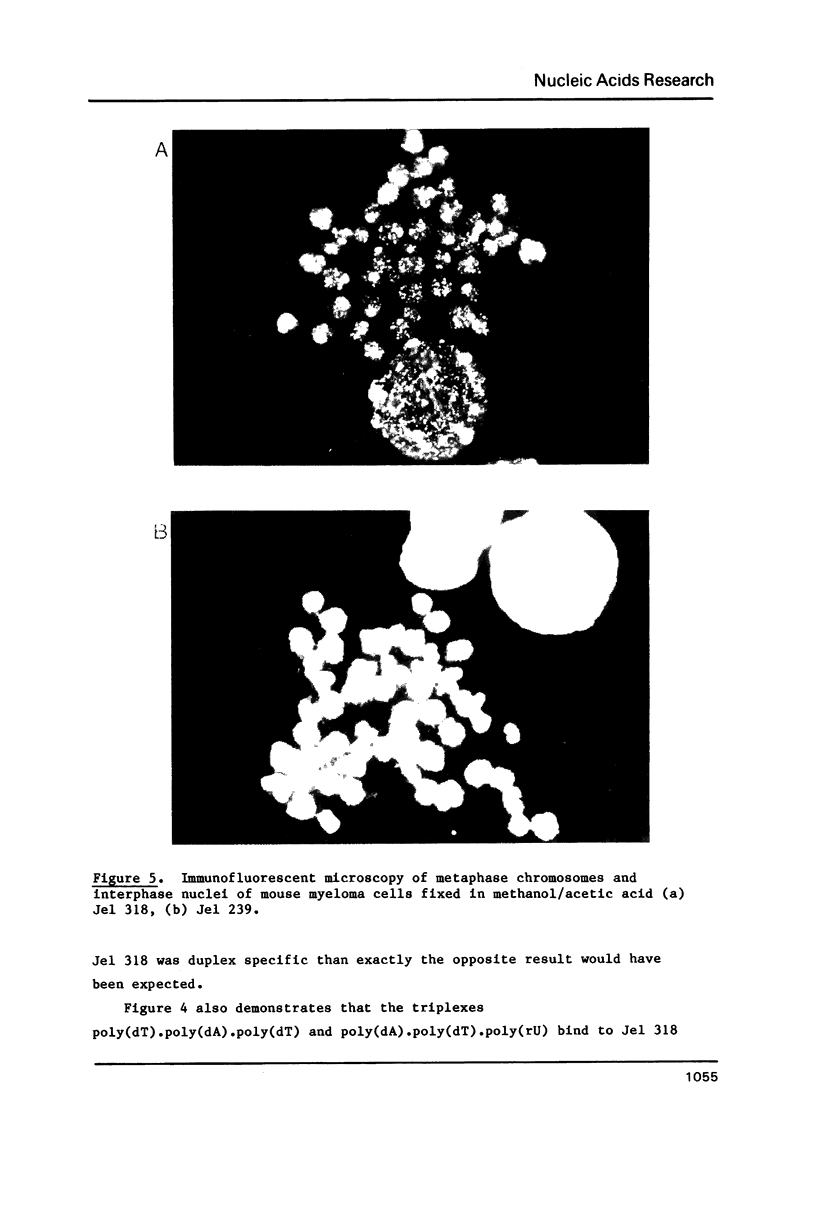
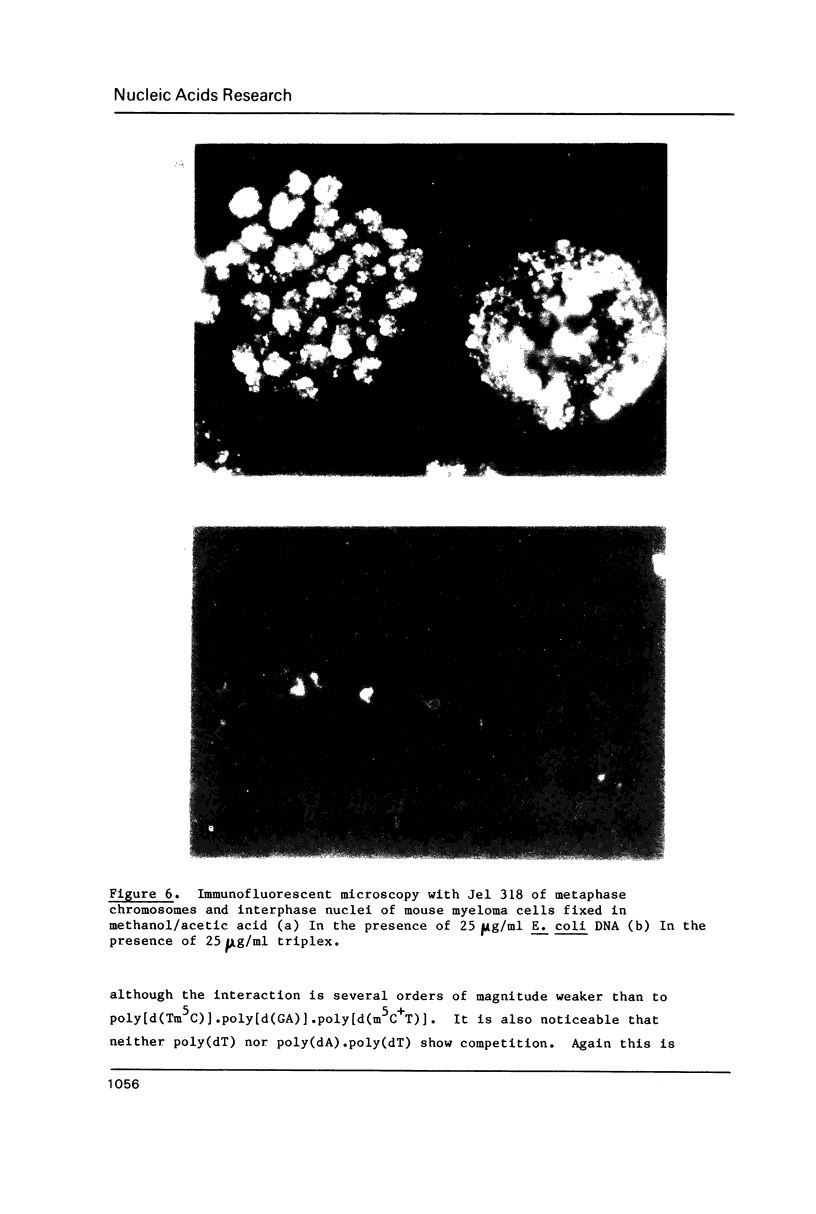
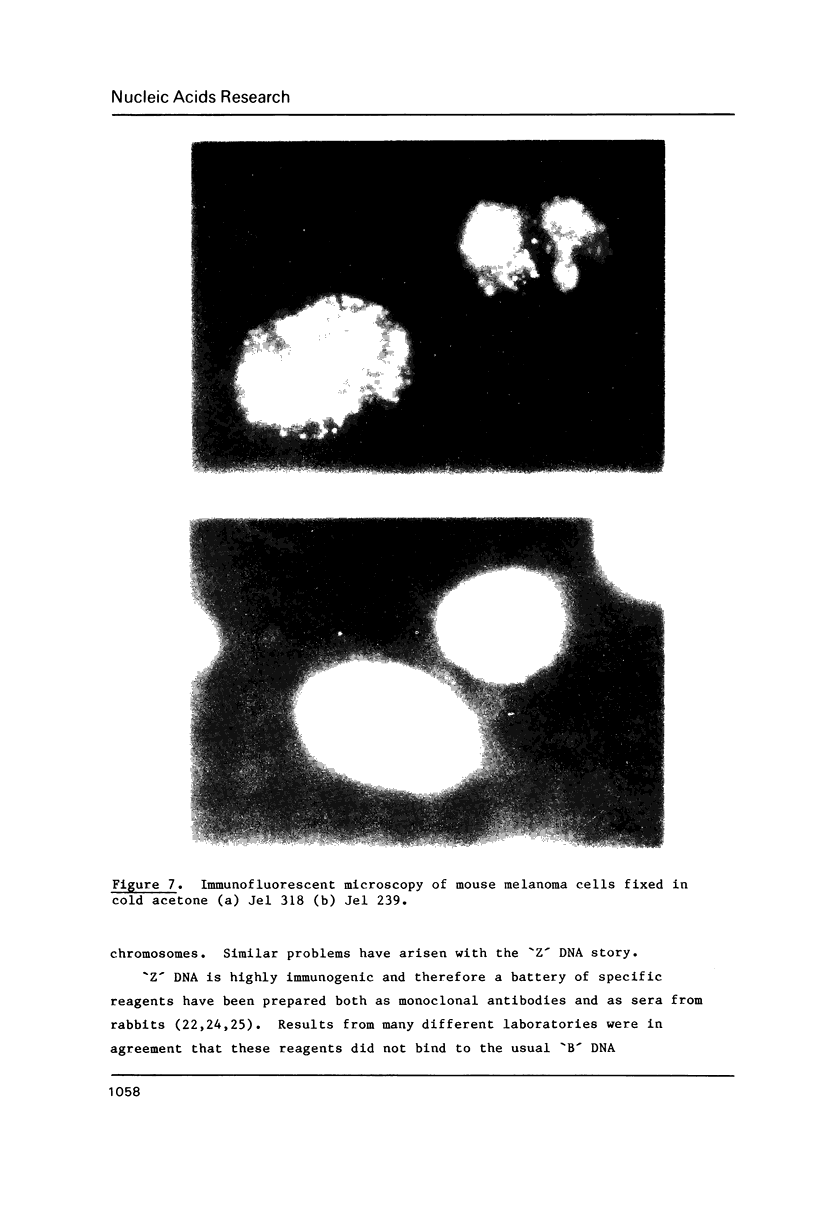
A monoclonal antibody (Jel 318) was produced by immunizing mice with poly[d(TmC)].poly[d(GA)].poly[d(mCT) which forms a stable triplex at neutral pH. Jel 318 did not bind to calf thymus DNA or other non pyrimidine.purine DNAs such as poly[d(TG)].poly[d(CA)]. In addition the antibody did not recognize pyrimidine.purine DNAs containing mA (e.g. poly[d(TC)].poly[d(GmA)]) which cannot form a triplex since the methyl group blocks Hoogsteen base-pairing. The binding of Jel 318 to chromosomes was assessed by immunofluorescent microscopy of mouse myeloma cells which had been fixed in methanol/acetic acid. An antibody specific for duplex DNA (Jel 239) served as a control. The fluorescence due to Jel 318 was much weaker than that of Jel 239 but binding to metaphase chromosomes and interphase nuclei was observed. The staining by Jel 318 was unaffected by addition of E. coli DNA but it was obliterated in the presence of triplex. Since an acid pH favours triplex formation, nuclei were also prepared from mouse melanoma cells by fixation in cold acetone. Again Jel 318 showed weak but consistent staining of the nuclei. Therefore it seems likely that triplexes are an inherent feature of the structure of eucaryotic DNA.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C. Spacing of polypyrimidine regions in mouse DNA as determined by poly(adenylate, guanylate) binding. J Mol Biol. 1978 Jun 5;121(4):541–559. doi: 10.1016/0022-2836(78)90399-6. [DOI] [PubMed] [Google Scholar]
- Braun R. P., Lee J. S. Variations in duplex DNA conformation detected by the binding of monoclonal autoimmune antibodies. Nucleic Acids Res. 1986 Jun 25;14(12):5049–5065. doi: 10.1093/nar/14.12.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe D., Cabrer B., Bacolla A., Targovnik H., Pohl V., Vassart G. An unusually long poly(purine)-poly(pyrimidine) sequence is located upstream from the human thyroglobulin gene. Nucleic Acids Res. 1985 Jul 25;13(14):5127–5144. doi: 10.1093/nar/13.14.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Spectrofluorometric measurement of the binding of ethidium to superhelical DNA from cell nuclei. Eur J Biochem. 1978 Mar 15;84(2):465–477. doi: 10.1111/j.1432-1033.1978.tb12188.x. [DOI] [PubMed] [Google Scholar]
- Doolittle W. F., Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980 Apr 17;284(5757):601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Engel J. D., von Hippel P. H. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J Biol Chem. 1978 Feb 10;253(3):927–934. [PubMed] [Google Scholar]
- Evans D. H., Lee J. S., Morgan A. R., Olsen R. K. A method for the specific inhibition of poly[d(A-T)] synthesis using the A-T specific quinoxaline antibiotic TANDEM. Can J Biochem. 1982 Feb;60(2):131–136. doi: 10.1139/o82-018. [DOI] [PubMed] [Google Scholar]
- Evans T., Efstratiadis A. Sequence-dependent S1 nuclease hypersensitivity of a heteronomous DNA duplex. J Biol Chem. 1986 Nov 5;261(31):14771–14780. [PubMed] [Google Scholar]
- Fowler R. F., Skinner D. M. Eukaryotic DNA diverges at a long and complex pyrimidine.purine tract that can adopt altered conformations. J Biol Chem. 1986 Jul 5;261(19):8994–9001. [PubMed] [Google Scholar]
- Gillies S. D., Folsom V., Tonegawa S. Cell type-specific enhancer element associated with a mouse MHC gene, E beta. Nature. 1984 Aug 16;310(5978):594–597. doi: 10.1038/310594a0. [DOI] [PubMed] [Google Scholar]
- Hill R. J., Stollar B. D. Dependence of Z-DNA antibody binding to polytene chromosomes on acid fixation and DNA torsional strain. Nature. 1983 Sep 22;305(5932):338–340. doi: 10.1038/305338a0. [DOI] [PubMed] [Google Scholar]
- Johnson G. D., Nogueira Araujo G. M. A simple method of reducing the fading of immunofluorescence during microscopy. J Immunol Methods. 1981;43(3):349–350. doi: 10.1016/0022-1759(81)90183-6. [DOI] [PubMed] [Google Scholar]
- Kanai Y., Tanuma S., Sugimura T. Immunofluorescent staining of poly(ADP-ribose) in situ in HeLa cell chromosomes in the M phase. Proc Natl Acad Sci U S A. 1981 May;78(5):2801–2804. doi: 10.1073/pnas.78.5.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Lewis J. R., Morgan A. R., Mosmann T. R., Singh B. Monoclonal antibodies showing sequence specificity in their interaction with single-stranded DNAs. Nucleic Acids Res. 1981 Apr 10;9(7):1707–1721. doi: 10.1093/nar/9.7.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Morgan A. R. Novel aspects of the structure of the Escherichia coli nucleoid investigated by a rapid sedimentation assay. Can J Biochem. 1982;60(10):952–961. doi: 10.1139/o82-122. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Woodsworth M. L., Latimer L. J. Monoclonal antibodies specific for poly(dG) X poly(dC) and poly(dG) X poly(dm5C). Biochemistry. 1984 Jul 3;23(14):3277–3281. doi: 10.1021/bi00309a024. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Woodsworth M. L., Latimer L. J., Morgan A. R. Poly(pyrimidine) . poly(purine) synthetic DNAs containing 5-methylcytosine form stable triplexes at neutral pH. Nucleic Acids Res. 1984 Aug 24;12(16):6603–6614. doi: 10.1093/nar/12.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Lee J. S., Pulleyblank D. E., Murray N. L., Evans D. H. Review: ethidium fluorescence assays. Part 1. Physicochemical studies. Nucleic Acids Res. 1979 Oct 10;7(3):547–569. doi: 10.1093/nar/7.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller A., Gabriels J. E., Lafer E. M., Nordheim A., Rich A., Stollar B. D. Monoclonal antibodies recognize different parts of Z-DNA. J Biol Chem. 1982 Oct 25;257(20):12081–12085. [PubMed] [Google Scholar]
- Möller A., Nordheim A., Kozlowski S. A., Patel D. J., Rich A. Bromination stabilizes poly(dG-dC) in the Z-DNA form under low-salt conditions. Biochemistry. 1984 Jan 3;23(1):54–62. doi: 10.1021/bi00296a009. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Pardue M. L., Lafer E. M., Möller A., Stollar B. D., Rich A. Antibodies to left-handed Z-DNA bind to interband regions of Drosophila polytene chromosomes. Nature. 1981 Dec 3;294(5840):417–422. doi: 10.1038/294417a0. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Larson J. E., Grant R. C., Shortle B. E., Cantor C. R. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J Mol Biol. 1970 Dec 28;54(3):465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]
- Zarling D. A., Arndt-Jovin D. J., Robert-Nicoud M., McIntosh L. P., Thomae R., Jovin T. M. Immunoglobulin recognition of synthetic and natural left-handed Z DNA conformations and sequences. J Mol Biol. 1984 Jul 5;176(3):369–415. doi: 10.1016/0022-2836(84)90495-9. [DOI] [PubMed] [Google Scholar]