Abstract
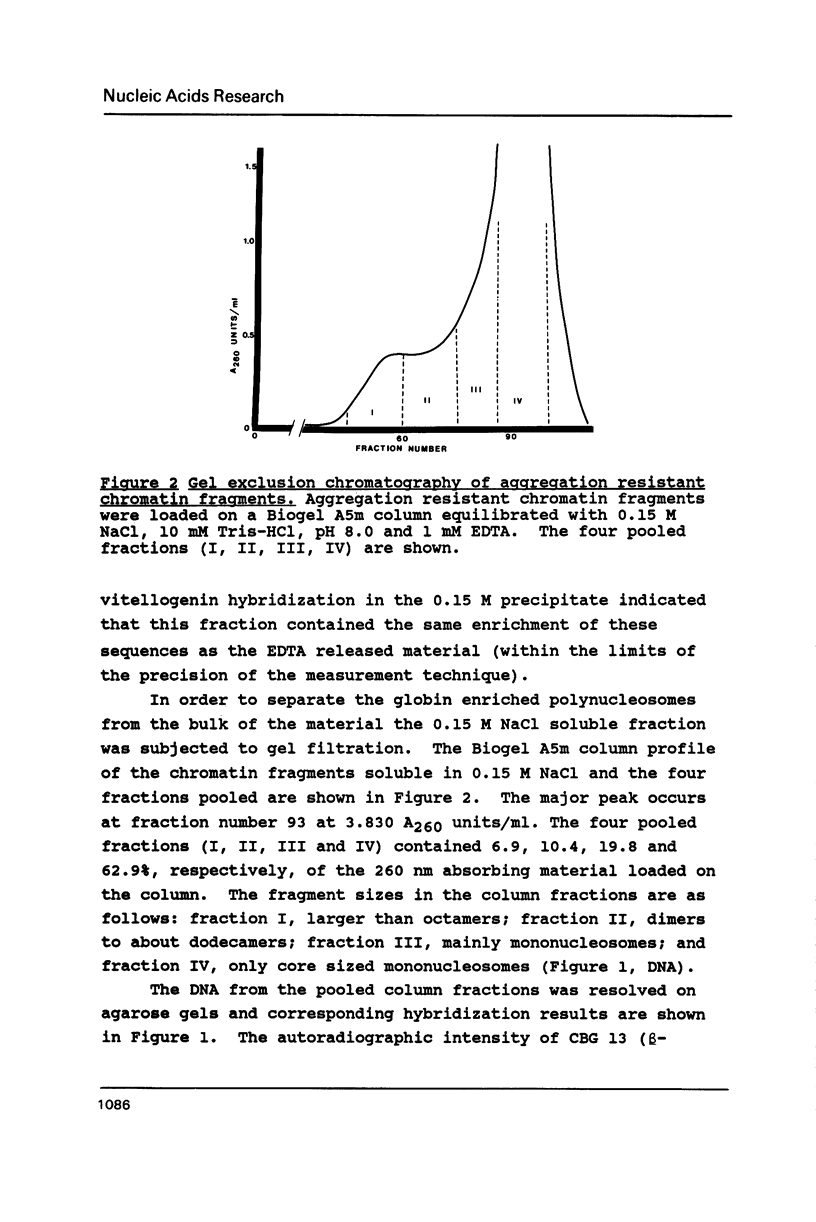
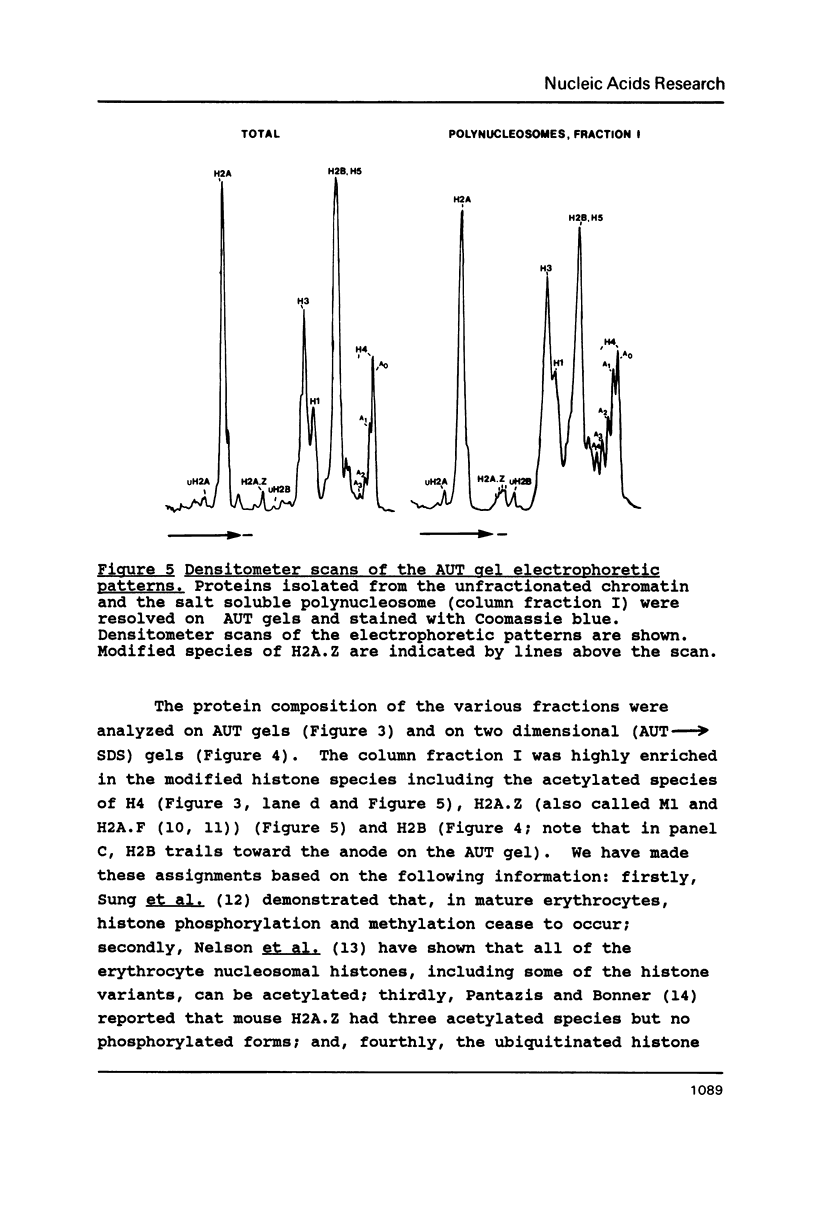
Mature chicken erythrocyte polynucleosomes which are soluble at physiological ionic strength are enriched in beta-globin DNA sequences. Vitellogenin chromatin, which is not expressed in this tissue, is found in aggregation prone, salt insoluble chromatin. There is a direct correlation between the size of soluble fragments and the degree of globin gene enrichment, with the largest fragments being most highly enriched. The highly globin enriched (about 50 fold) polynucleosomes contain significantly elevated levels of acetylated histones H4, H2A.Z, and H2B, and ubiquitinated (prefix "u") histones H2A and H2B (with a significant relative increase of uH2B over uH2A). These polynucleosomes were complexed with histones H1 and H5 but at a lower level than that found in unfractionated chromatin.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ausio J., van Holde K. E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986 Mar 25;25(6):1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- Brotherton T. W., Covault J., Shires A., Chalkley R. Only a small fraction of avian erythrocyte histone is involved in ongoing acetylation. Nucleic Acids Res. 1981 Oct 10;9(19):5061–5073. doi: 10.1093/nar/9.19.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Davie J. R., Numerow L., Delcuve G. P. The nonhistone chromosomal protein, H2A-specific protease, is selectively associated with nucleosomes containing histone H1. J Biol Chem. 1986 Aug 5;261(22):10410–10416. [PubMed] [Google Scholar]
- Davie J. R. Two-dimensional gel systems for rapid histone analysis for use in minislab polyacrylamide gel electrophoresis. Anal Biochem. 1982 Mar 1;120(2):276–281. doi: 10.1016/0003-2697(82)90348-7. [DOI] [PubMed] [Google Scholar]
- Ferenz C. R., Nelson D. A. N-Butyrate incubation of immature chicken erythrocytes preferentially enhances the solubility of beta A chromatin. Nucleic Acids Res. 1985 Mar 25;13(6):1977–1995. doi: 10.1093/nar/13.6.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. C., Cole R. D. The distribution of H1 histone is nonuniform in chromatin and correlates with different degrees of condensation. J Biol Chem. 1984 Nov 25;259(22):14237–14242. [PubMed] [Google Scholar]
- Jin Y. J., Cole R. D. H1 histone exchange is limited to particular regions of chromatin that differ in aggregation properties. J Biol Chem. 1986 Mar 5;261(7):3420–3427. [PubMed] [Google Scholar]
- Komaiko W., Felsenfeld G. Solubility and structure of domains of chicken erythrocyte chromatin containing transcriptionally competent and inactive genes. Biochemistry. 1985 Feb 26;24(5):1186–1193. doi: 10.1021/bi00326a020. [DOI] [PubMed] [Google Scholar]
- Nelson D. A., Ferris R. C., Zhang D. E., Ferenz C. R. The beta-globin domain in immature chicken erythrocytes: enhanced solubility is coincident with histone hyperacetylation. Nucleic Acids Res. 1986 Feb 25;14(4):1667–1682. doi: 10.1093/nar/14.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazis P., Bonner W. M. Quantitative determination of histone modification. H2A acetylation and phosphorylation. J Biol Chem. 1981 May 10;256(9):4669–4675. [PubMed] [Google Scholar]
- Perry M., Chalkley R. The effect of histone hyperacetylation on the nuclease sensitivity and the solubility of chromatin. J Biol Chem. 1981 Apr 10;256(7):3313–3318. [PubMed] [Google Scholar]
- Razin S. V., Yarovaya O. V., Georgiev G. P. Low ionic strength extraction of nuclease-treated nuclei destroys the attachment of transcriptionally active DNA to the nuclear skeleton. Nucleic Acids Res. 1985 Oct 25;13(20):7427–7444. doi: 10.1093/nar/13.20.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Rocha E., Davie J. R., van Holde K. E., Weintraub H. Differential salt fractionation of active and inactive genomic domains in chicken erythrocyte. J Biol Chem. 1984 Jul 10;259(13):8558–8563. [PubMed] [Google Scholar]
- Seale R. L. Rapid turnover of the histone-ubiquitin conjugate, protein A24. Nucleic Acids Res. 1981 Jul 10;9(13):3151–3158. doi: 10.1093/nar/9.13.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung M. T., Harford J., Bundman M., Vidalakas G. Metabolism of histones in avian erythroid cells. Biochemistry. 1977 Jan 25;16(2):279–285. doi: 10.1021/bi00621a019. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Rees C. Exchange of histones H1 and H5 between chromatin fragments. A preference of H5 for higher-order structures. Eur J Biochem. 1983 Jul 15;134(1):109–115. doi: 10.1111/j.1432-1033.1983.tb07538.x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Rees C., Pearson E. C. Histone H5 promotes the association of condensed chromatin fragments to give pseudo-higher-order structures. Eur J Biochem. 1985 Feb 15;147(1):143–151. doi: 10.1111/j.1432-1033.1985.tb08730.x. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban M. K., Franklin S. G., Zweidler A. Isolation and characterization of the histone variants in chicken erythrocytes. Biochemistry. 1979 Sep 4;18(18):3952–3960. doi: 10.1021/bi00585a017. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Landes G. M., Pankratz M. J., Martinson H. G. The chicken beta globin gene region. Delineation of transcription units and developmental regulation of interspersed DNA repeats. J Biol Chem. 1982 Sep 25;257(18):11015–11023. [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Kohn K. W., Bonner W. M. Metabolism of ubiquitinated histones. J Biol Chem. 1981 Jun 10;256(11):5916–5920. [PubMed] [Google Scholar]
- Wu R. S., Panusz H. T., Hatch C. L., Bonner W. M. Histones and their modifications. CRC Crit Rev Biochem. 1986;20(2):201–263. doi: 10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]