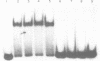
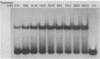
Abstract
The E. coli DNA photolyase is a flavoprotein that catalyzes the photoreversal of pyrimidine dimers. The enzyme binds to DNA containing pyrimidine dimers in a light-independent step and repairs the dimer upon absorbing a photon in the 300-600 nm range. The rate and equilibrium constants for the light-independent reaction were determined before, using randomly modified substrates that contained T mean value of T, T mean value of C and C mean value of C dimers in random sequence surrounding. In this paper we have determined these constants for a defined substrate (a 43 bp oligomer containing a T mean value of T dimer) using the gel retardation assay. We find that: the equilibrium constant and the off rate obtained with this substrate by this technique are similar to those obtained with randomly modified DNA using filter binding and flash photolysis techniques. the off rate with the defined substrate is heterogeneous indicating heterogeneity in the enzyme population or in the enzyme-substrate complexes, and the enzyme has 7.5 X 10(4)-fold higher affinity for pyrimidine dimer compared to non-dimer DNA nucleotides.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fried M. G., Crothers D. M. Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J Mol Biol. 1984 Jan 25;172(3):263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harm W. Analysis of photoenzymatic repair of UV lesions in DNA by single light flashes. V. Determination of the reaction-rate constants in E. coli cells. Mutat Res. 1970 Oct;10(4):277–290. doi: 10.1016/0027-5107(70)90043-6. [DOI] [PubMed] [Google Scholar]
- Harm W., Harm H., Rupert C. S. Analysis of photoenzymatic repair of UV lesions in DNA by single light flashes. II. In vivo studies with Escherichia coli cells and bacteriophage. Mutat Res. 1968 Nov-Dec;6(3):371–385. doi: 10.1016/0027-5107(68)90054-7. [DOI] [PubMed] [Google Scholar]
- Jorns M. S., Baldwin E. T., Sancar G. B., Sancar A. Action mechanism of Escherichia coli DNA photolyase. II. Role of the chromophores in catalysis. J Biol Chem. 1987 Jan 5;262(1):486–491. [PubMed] [Google Scholar]
- Jorns M. S., Sancar G. B., Sancar A. Identification of oligothymidylates as new simple substrates for Escherichia coli DNA photolyase and their use in a rapid spectrophotometric enzyme assay. Biochemistry. 1985 Apr 9;24(8):1856–1861. doi: 10.1021/bi00329a008. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac repressor binding to non-operator DNA: detailed studies and a comparison of eequilibrium and rate competition methods. J Mol Biol. 1972 Dec 30;72(3):671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- Madden J. J., Werbin H., Denson J. A rapid assay for DNA photolyase using a membrane-binding technique. Photochem Photobiol. 1973 Dec;18(6):441–445. doi: 10.1111/j.1751-1097.1973.tb06447.x. [DOI] [PubMed] [Google Scholar]
- RUPERT C. S. Photoenzymatic repair of ultraviolet damage in DNA. I. Kinetics of the reaction. J Gen Physiol. 1962 Mar;45:703–724. doi: 10.1085/jgp.45.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RUPERT C. S. Photoenzymatic repair of ultraviolet damage in DNA. II. Formation of an enzyme-substrate complex. J Gen Physiol. 1962 Mar;45:725–741. doi: 10.1085/jgp.45.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Smith F. W., Sancar G. B. Purification of Escherichia coli DNA photolyase. J Biol Chem. 1984 May 10;259(9):6028–6032. [PubMed] [Google Scholar]
- Sancar G. B., Jorns M. S., Payne G., Fluke D. J., Rupert C. S., Sancar A. Action mechanism of Escherichia coli DNA photolyase. III. Photolysis of the enzyme-substrate complex and the absolute action spectrum. J Biol Chem. 1987 Jan 5;262(1):492–498. [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Reid R., Payne G., Levy M., Sancar A. Action mechanism of Escherichia coli DNA photolyase. I. Formation of the enzyme-substrate complex. J Biol Chem. 1987 Jan 5;262(1):478–485. [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Sancar A. Binding of Escherichia coli DNA photolyase to UV-irradiated DNA. Biochemistry. 1985 Apr 9;24(8):1849–1855. doi: 10.1021/bi00329a007. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Berg O. G. On the specificity of DNA-protein interactions. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1608–1612. doi: 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]