Abstract
The present study searched for replicable risk genomic regions for alcohol and nicotine co-dependence using a genome-wide association strategy. The data contained a total of 3,143 subjects including 818 European-American (EA) cases with alcohol and nicotine co-dependence, 1,396 EA controls, 449 African-American (AA) cases and 480 AA controls. We performed separate genome-wide association analyses in EAs and AAs and a meta-analysis to derive combined p values, and calculated the genome-wide false discovery rate (FDR) for each SNP. Regions with p<5×10-7 together with FDR<0.05 in the meta-analysis were examined to detect all replicable risk SNPs across EAs, AAs and meta-analysis. These SNPs were followed with a series of functional expression quantitative trait locus (eQTL) analyses. We found a unique genome-wide significant gene region – SH3BP5-NR2C2 – that was enriched with 11 replicable risk SNPs for alcohol and nicotine co-dependence. The distributions of -log(p) values for all SNP-disease associations within this region were consistent across EAs, AAs, and meta-analysis (0.315≤r≤0.868; 8.1×10-52≤p≤3.6×10-5). In the meta-analysis, this region was the only association peak throughout chromosome 3 at p<0.0001. All replicable risk markers available for eQTL analysis had nominal cis- and trans-acting regulatory effects on gene expression. The transcript expression of the genes in this region was regulated partly by several nicotine dependence-related genes and significantly correlated with transcript expression of many alcohol and nicotine dependence-related genes. We concluded that the SH3BP5-NR2C2 region on Chromosome 3 might harbor causal loci for alcohol and nicotine co-dependence.
Keywords: GWAS, alcohol and nicotine co-dependence
Introduction
Alcohol dependence (AD) and nicotine dependence (ND) frequently co-occur in the same individuals. Alcohol and nicotine co-dependence may represent an independent phenotype. A large number of common risk genetic loci have been reported separately for AD and ND in the dopaminergic, serotoninergic, GABAergic, glutamatergic, cholinergic, opioid, and endocannabinoid systems by candidate gene approach; several genome-wide association studies (GWASs) on AD or ND reported other risk loci (summarized by [Zuo et al., 2011a; Zuo et al., In revision]). Only one study employed GWAS [Lind et al., 2010] to examine alcohol and nicotine co-dependence, and its results have yet to be replicated.
In the present study, we searched for replicable risk gene regions for alcohol and nicotine co-dependence in two distinct American populations using GWAS. In the association analysis, we separated European-Americans (EAs) and African-Americans (AAs) to increase population homogeneity, and controlled for admixture effects. The association findings from the EAs were replicated in the AAs and vice versa. Additionally, we used an independent sample with distinct tissues to detect expression quantitative trait locus (eQTL) signals, as a confirmation of the association findings. Furthermore, we applied a stringent definition of replication (see below). The primary target of investigation in the current study was not the top-ranked SNPs in the discovery sample as previous GWASs, but rather the replicable risk regions that might harbor the population-generalizable and functional variants. This strategy led to the discovery of novel risk loci for alcohol and nicotine co-dependence.
Materials and Methods
Subjects
The sample comprised of a total of 3,143 subjects for gene-disease association analysis, including 818 European-American (EA) cases with alcohol and nicotine co-dependence, 1,396 EA controls, 449 African-American (AA) cases and 480 AA controls. This sample was extracted from SAGE (dbGaP study accession phs000092.v1.p1)[Bierut et al., 2010]. SAGE subjects were recruited from 8 different study sites in 7 states and the District of Columbia; the majority of subjects were recruited in Missouri[Bierut et al., 2010]. All subjects were interviewed using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA)[Bucholz et al., 1994]. Affected subjects met lifetime DSM-IV criteria[American Psychiatric Association 1994] both for alcohol dependence and nicotine dependence, and were excluded if they had schizophrenia or other psychotic illnesses. Controls were defined as individuals who had been exposed to alcohol and nicotine in sufficient amounts for a sufficient time, but had never become dependent on or abused alcohol, nicotine or other illicit substances (see Supplemental Table S1). Additionally, controls were also screened to exclude individuals with major axis I disorders, including schizophrenia, mood disorders, and anxiety disorders. More demographic data were available in the Supplemental Table S1 or elsewhere [Bierut et al., 2010; Edenberg et al., 2005; Edenberg et al., 2010; Zuo et al., 2011a; Zuo et al., 2011b]. All subjects gave written informed consent to participating in protocols approved by the relevant institutional review boards and were de-identified in this study.
Genotyping
All samples were genotyped on the Illumina Human 1M beadchip at the Center for Inherited Disease Research (CIDR) at Johns Hopkins University (Baltimore, MD USA). Allele cluster definitions for each marker were determined using Illumina BeadStudio Genotyping Module version 3.1.14 and the combined intensity data from the samples.
Data cleaning
Before statistical analysis, we strictly cleaned the phenotype data and then the genotype data [Zuo et al., 2011a; Zuo et al., In revision]. After cleaning, 805,814 markers in EAs and 895,714 markers in AAs were included for association analysis. The cleaned data had high-quality, as evidenced by the following: (1) The homogeneity of the two samples was very high; that is, EAs and AAs were well differentiated. (2) The observed and expected p-values for the associations fit very well within EAs or AAs (see QQ plots in Supplemental Figure S1). (3) We also computed from these p-values a low genomic inflation factor (GIF) of 1.04 in EAs, 1.02 in AAs and 1.04 in meta-analysis.
Data analytic procedure
We performed genome-wide association analysis separately in EAs and AAs first, and then performed a meta-analysis of EAs and AAs. Genome-wide false discovery rates (FDRs)[Benjamini and Hochberg 1995] were calculated in EAs, AAs and meta-analysis. The replicable risk regions were identified, in which (1) many markers were associated with phenotype across EAs, AAs and meta-analysis, and (2) the distributions of -log(p) values for associations for all markers were consistent across EAs, AAs and meta-analysis; that is, the number of risk markers, the effect directions, the effect sizes and the significance strengths were congruent across three groups. These distributions were compared for similarity using Pearson correlations. Among these replicable risk regions, those with p<5×10-7 and FDR<0.05 in the meta-analysis were selected as “genome-wide significant and replicable regions” (screening step; see correction for multiple testing section below). And then, all SNPs in those significant and replicable regions were examined to identify all replicable risk SNPs (Table 1) (testing step). Also, around these significant and replicable regions, a region spanning 5Mb to whole chromosome was carefully studied, to know the width of the selected risk region where the putative causal loci may be located.
Table 1. P-values for the SNPs in SH3BP5-NR2C2 region with replicable association signals in EAs and AAs.
| SNP | Gene | Position (Build 36 Ref.) | Location | Minor Allele | EAs | AAs | Meta-analysis | eQTL▴ | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| OR | p | OR | p | Z score | p | p (Brain) | p (PBMC) | |||||
| rs17040623 | NR2C2 | 15003482 | Intron 1 | g | 0.87 | 0.025 | 0.69 | 3.4×10-3 | -3.46 | 5.3×10-4 | NA | NA |
| rs7635654 | NR2C2 | 15022966 | Intron 3 | c | 0.86 | 0.023 | 0.76 | 0.017 | -3.20 | 1.4×10-3 | NA | NA |
| rs28445844 | NR2C2 | 15049327 | Intron 11 | t | 0.87 | 0.026 | 0.75 | 9.5×10-3 | -3.27 | 1.1×10-3 | NA | NA |
| rs3773478 | ZFYVE20 | 15088046 | 3′ UTR | a | 0.85 | 0.033 | 0.72 | 0.036 | -2.93 | 3.4×10-3 | NA | NA |
| rs9851219 | ZFYVE20 | 15090727 | coding | c | 0.85 | 0.030 | 0.71 | 0.027 | -3.02 | 2.6×10-3 | NA | NA |
| rs9868848 | ZFYVE20 | 15090876 | coding | g | 0.86 | 0.038 | 0.72 | 0.037 | -2.87 | 4.1×10-3 | 0.004 | 0.016 |
| rs2306853 | ZFYVE20 | 15092129 | coding | a | 0.84 | 0.026 | 0.72 | 0.037 | -3.01 | 2.7×10-3 | NA | NA |
| rs735659 | intergenic | 15198998 | 24kb to 5′ of CAPN7 | c | 1.25 | 8.9×10-4 | 1.17 | 0.046 | 3.87 | 1.1×10-4 | 0.081 | 0.015 |
| rs1318937 | intergenic | 15270368 | 910bp to 3′ of SH3BP5 | g | 1.33 | 2.5×10-5 | 1.29 | 5.1×10-3 | 5.06 | 4.1×10-7 | 0.041 | 0.021 |
| rs3773471 | SH3BP5 | 15278051 | Intron 6 | c | 1.29 | 1.4×10-4 | 1.38 | 0.016 | 4.50 | 6.9×10-6 | NA | NA |
| rs8225 | SH3BP5 | 15280874 | Intron 5 | t | 1.29 | 1.4×10-4 | 1.36 | 0.026 | 4.41 | 1.0×10-5 | 0.040 | 0.103 |
All markers are in HWE. eQTL, expression quantitative trait locus analysis;
Exon-level expression changes in Brain and PBMC tissues (minimal p values are presented); ZFYVE20, CAPN7 and SH3BP5 have 15, 29 and 14 exons, respectively, and thus the corrected α for exon-level eQTL analysis was 0.003, 0.002 and 0.004, respectively. NA, not available.
In addition to the replication design described above, we also performed functional eQTL analysis on the replicable risk SNPs, which included (1) cis-eQTL analysis on exon-/transcript-level expression changes in peripheral blood mononucleated cells (PBMCs) (n=80) and cortical brain tissues (n=93), (2) transcriptome-wide trans-acting eQTL analysis on transcript expression, (3) genome-wide trans-acting eQTL analysis on the transcript expression of the replicable risk genes, and (4) transcriptome-wide expression correlation analysis [Heinzen et al., 2008; Zuo et al., 2011a; Zuo et al., 2011b]. Additionally, RNA secondary structure analysis [Zuker 2003; Zuo et al., 2011a; Zuo et al., 2011b] and a series of bioinformatic analysis (Supplemental Table S2) of the replicable risk markers were also performed.
Association analysis
Genome-wide association tests in EAs and AAs: The allele frequencies of the cleaned SNPs were compared between cases and controls using genome-wide logistic regression analysis implemented in the program Plink[Purcell et al., 2007], separately in EAs and AAs. Diagnosis served as the dependent variable, alleles or genotype served as the independent variables, and ancestry proportions (to control for admixture effects), sex, and age served as covariates (Supplementary Figure S2). Ancestry proportions of each individual were estimated from 3,172 completely independent ancestry-informative genetic markers [Zuo et al., 2011a]. The replicable regions between EAs and AAs were identified.
Meta-analysis of EAs and AAs: A meta-analysis of both populations was conducted to derive the effect directions of minor alleles, the Stouffer' Z scores, and the combined p values. The associations with opposite effects between EAs and AAs could become weaker in meta-analysis, and those with same direction of effects could become stronger in meta-analysis. Thus, to define replicable associations, we did not only require them to be positive in both EAs and AAs, but also required them to be stronger in meta-analysis, which increased the possibility that the replicable risk variants were causal. The associations in the genome-wide significant and replicable regions that were replicated across EAs, AAs and meta-analysis are shown in Table 1.
Cis-eQTL analysis: To examine relationships between genetic variants and local gene expression levels, we performed cis-eQTL analysis in the PBMC and brain tissues described above. Each of these associations was analyzed using a linear regression model by correcting for age, sex, source of tissues, and principle component scores. The rare homozygotes have been merged into the heterozygotes in the association analyses. P-values less than 0.05 were listed in Table 1 and the relationships between the exon expression levels (Y-axis) and the genotypes (X-axis) of the replicable risk markers were plotted in Figure S3. The genotype frequency distributions, the strength of associations (i.e., beta values from linear regression analysis) and the exon probe ID numbers are also shown on the plots and in their legends.
Correction for multiple testing on association and cis-eQTL analysis and power analysis
To mitigate false positive rates, genome-wide associations in the screening stage need to be corrected for multiple testing. Apparently, Bonferroni correction (α=5×10-8) is overly conservative because it treats all of the one million markers in the genome as independent (which is impossible). Alternatively, a WTCCC-defined α (=5×10-7) might be more appropriate to the present study[The Wellcome Trust Case Control Consortium 2007]. As a complementary approach, we also corrected the findings in screening stage by genome-wide false discovery rate (FDR)[Benjamini and Hochberg 1995], replicated the findings, and confirmed them by functional studies. Only when a region containing at least one association in meta-analysis that survived WTCCC-defined genome-wide correction (p<5×10-7) together with FDR<0.05, and was replicable across EAs, AAs, and meta-analysis and confirmed by functional studies, should it be taken as a “significant” region, which was conservative enough for statistical significance. In the testing step, (1) two independent samples were used to replicate each other, which significantly reduced the chance of false positive findings (i.e., false discovery rate). (2) We aimed to detect replicable regions, not individual markers. Thus, more than one risk marker were detected in the risk regions, which reduced the chance of false positive associations too. (3) Functional analysis in distinct tissues as confirmation of association finding further reduced the chance of false positive findings (including co-localization of association signals and eQTL signals randomly), although using different independent samples in one study might increase the false negative rates due to sample heterogeneity. (4) -log(P) value distributions across EAs, AAs and meta-analysis were compared using Pearson correlation analysis. The consistency between them would significantly reduce the chance of false positive findings. Therefore, in the testing step, when an association was replicable across EAs, AAs and meta-analysis, α could be set at 0.05 (except for the exon-level cis-eQTL findings that needed to be corrected for the number of exons and the types of tissues). Accordingly, the power of the discovery (α=5×10-7) and replication (α=0.05) samples to detect the significant genetic effects was analyzed using the power analysis package in R.
Results
There were a total of 29 SNPs in 15 genes in EAs, 9 SNPs in 9 genes in AAs and 22 SNPs in 16 genes in meta-analysis that were marginally to significantly (p<10-5) associated with alcohol and nicotine co-dependence (data available on request). The p values for the 10 top-ranked SNPs in EAs, AAs, and meta-analysis, respectively, are listed in Supplementary Table S3, and the p values in meta-analysis for the 10 top-ranked replicable SNPs are listed in Supplementary Table S4. After correction, 4 SNPs in EAs including rs7445832, rs4700575 and rs2169520 in IPO11-HTR1A region (7.0×10-9≤p≤3.0×10-7 and 6.3×10-4≤FDR≤0.031) and rs17427389 in PLEKHG1 (p=4.3×10-7 and FDR=0.019), rs4610908 in FAM47B in AAs (p=3.2×10-7 and FDR=0.032), and 2 SNPs in meta-analysis including rs9636470 in PLGLB2 (p=3.1×10-8 and FDR=0.003) and rs1318937 in SH3BP5 (p=4.1×10-7 and FDR=0.041) remained significant (p<5×10-7 together with FDR<0.05). Among these significant SNPs, only rs9636470 in PLGLB2 (p=2.4×10-6 in EAs and 0.004 in AAs) and rs1318937 in SH3BP5 (p=2.5×10-5 in EAs and 0.005 in AAs) were replicable between two populations. However, rs9636470 in PLGLB2 was the only replicable significant SNP in a 3Mb-wide region around this gene, and thus, this significant association could occur by chance. In contrast, the region around SH3BP5 was enriched with replicable risk SNPs and thus was the focus of interest in the present study.
Throughout the whole chromosome 3, SH3BP5-NR2C2 region was the only one that had gene-disease associations with p<10-4 in meta-analysis. In EAs, within the 8Mb-range around, SH3BP5-NR2C2 region was the only association peak with p<10-4.
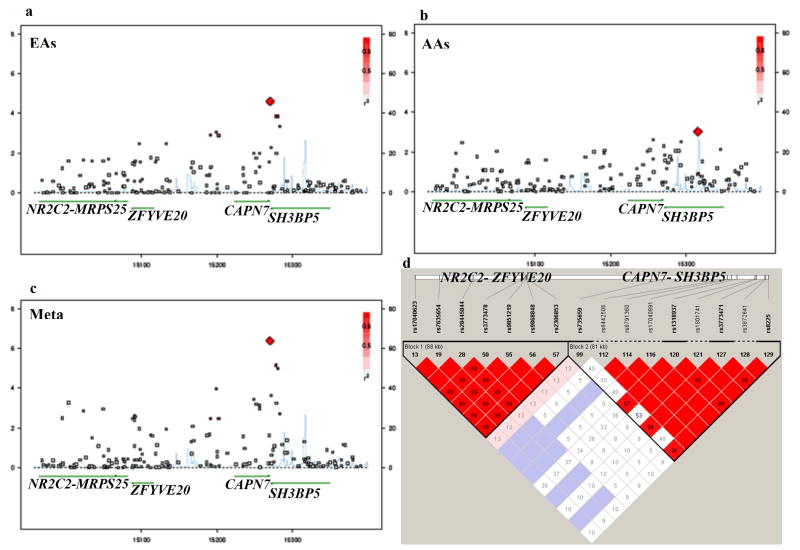
SH3BP5-NR2C2 region contains five known genes including SH3BP5, CAPN7, ZFYVE20, MRPS25 and NR2C2, all of which except MRPS25 were enriched with replicable risk SNPs. Thirty, 34 and 35 SNPs in this region were nominally associated with alcohol and nicotine co-dependence in EAs (2.5×10-5≤p≤0.038), AAs (9.3×10-4≤p≤0.046) and meta-analysis (4.1×10-7≤p≤0.049), respectively. Among them, 11 SNPs were replicable across EAs and AAs and became more significant in meta-analysis (4.1×10-7≤p≤4.1×10-3) (Table 1). Effects of all of these 11 SNPs were in the same direction between two populations. These 11 SNPs were located in two haplotype blocks, i.e., ZFYVE20-NR2C2 (block 1) and SH3BP6-CAPN7 (block 2) (Figure 1d). Minor alleles of all replicable SNPs in block 2 increased risk for disease (OR>1 in both EAs and AAs, and Z score > 0 in meta-analysis), but minor alleles in block 1 protected against disease (OR<1 in both EAs and AAs, and Z score < 0 in meta-analysis). The -log(p) values for all available SNPs across SH3BP5-NR2C2 region were plotted in Figure 1. The distributions of -log(p) values were consistent across EAs, AAs, and meta-analysis (0.315≤r≤0.868; 8.1×10-52≤p≤3.6×10-5; Table 2).
Figure 1. Regional association plots around SH3BP5-NR2C2.
region [X-axis: chromosome position; Y-axis: -log(p). (a) in EAs; (b) in AAs; (c) in meta-analysis; (d) LD map for the replicable risk markers in SH3BP5-NR2C2 region in EAs]
Table 2. Correlations of distributions of −log(p) values for gene-disease associations in SH3BP5-NR2C2 region between different populations.
| EA | AA | |||
|---|---|---|---|---|
|
|
|
|||
| r | p | r | p | |
| AA | 0.315 | 3.6×10-5 | ||
| EA | 0.315 | 3.6×10-5 | ||
| Meta | 0.868 | 8.1×10-52 | 0.544 | 3.5×10-14 |
r, Pearson correlation coefficient; p, p-values for pairwise correlations.
The LD structure (Figure 1d), the gene effect directions and the gene effect sizes shown in Table 1 indicated that SH3BP5-NR2C2 region could be represented by two independent risk SNPs from two LD blocks. After regressing out the effects of the two top-ranked SNPs in each block (i.e., rs17040623 at NR2C2 and rs1318937 at SH3BP5) by conditioning on them in the regression analysis, no other SNPs remained significantly associated with the phenotype (all p > 0.05; data not shown).
Our samples had a high power in detecting risk markers. For example, given that the risk allele of the most significant marker, i.e., rs7445832 (see Table S3), in the EA discovery sample had a frequency of 0.2892 in cases (n=818) and 0.2111 in controls (n=1396), our EA sample had a power of 90.1% to detect any risk marker that had a similar effect size to rs7445832 (α=5×10-7). Given that the risk allele of the most significant replicable marker, i.e., rs17040623 (see Table 1), in the AA replication sample had a frequency of 0.0.059 in cases (n=449) and 0.096 in controls (n=480), our AA sample had a power of 86.4% to detect any replicable risk marker that had a similar effect size to rs17040623 (α=0.05).
Among those 11 replicable risk markers, four SNPs were available for eQTL analysis, including rs1318937 and rs8225 in SH3BP5, rs735659 in CAPN7 and rs9868848 in ZFYVE20. eQTL analysis showed that all of them had nominal cis-acting regulatory effects on exon-level expression of local genes in Brain tissue and/or peripheral blood mononucleated cells (PBMCs) (0.004≤p≤0.041; Table 1 and Figure S3) and nominally regulated transcript expression of many genes across transcriptome (3.3×10-5≤p≤0.05; data not shown). After Bonferroni correction, none of these cis- and trans-acting regulatory effects remained significant.
Genome-wide trans-eQTL analysis showed that transcript expression of genes in SH3BP5-NR2C2 region was nominally regulated by multiple genes across the genome (data not shown). After Bonferroni correction (α=4.4×10-8), only rs7667919 in PET112L (Chromosome 4) showed significant regulatory effect on CAPN7 transcript expression in PBMC (p=7.8×10-9). Furthermore, 19 SNPs in 17 genes in brain and 5 SNPs in 5 genes in PBMC had nominal replicable trans-acting regulatory effects on all four genes in SH3BP5-NR2C2 region (5.0×10-5≤p≤4.5×10-3; data not shown).
Transcriptome-wide expression correlation analysis showed that expression of NR2C2, ZFYVE20, CAPN7 and SH3BP5 transcripts was significantly correlated with each other both in Brain and PBMC. Their expression was also significantly correlated with many alcohol or nicotine dependence-related genes (although some of these genes have not been widely replicated so far; data not shown). These genes were from the dopaminergic (DRD1, DRD2, NCAM1, TTC12, DRD3, DRD4, SLC6A3 and TH)[Batel et al., 2008; Stapleton et al., 2007], serotoninergic (HTR1B, HTR2A, HTR3B and SLC6A4)[Hasegawa et al., 2002], cholinergic (CHRM1, CHRNA1, CHRNA3, CHRNA7, CHRNB2, CHRND and CHRNG)[Lou et al., 2006; Saccone et al., 2009], GABAergic (GABARAP, GABBR1, GABRA1, GABRA2, GABRA4, GABRA6, GABRB1, GABRB2, GABRB3, GABRG1, GABRG2 and GABRG3) [Chang et al., 2002], glutamatergic (GAD1, GRIK2, GRIK3, GRIN2A, GRIN2B, GRIN2C, GRM5 and GRM7) [Edenberg et al., 2010], histaminergic (HNMT) [Oroszi et al., 2005], endocannabinoid (CNR1) [Zuo et al., 2007], opoid (OPRD1, OPRM1 and POMC)[Zhang et al., 2008], alcohol metabolic (ADH5 and ALDH3A2)[Luo et al., 2006; Uhl et al., 2008] and neuropeptide (NPY1R, NPY2R and NPY5R)[Wetherill et al., 2008] systems (α=4.2×10-7).
The main findings from the RNA secondary structure analysis included that (1) rs9868848 in ZFYVE20 significantly altered the RNA secondary structure; and (2) rs1318937 in SH3BP5, rs28445844 in NR2C2 and rs2306853 in ZFYVE20 slightly altered the RNA secondary structures (data not shown).
Discussion
Using a replication approach, we identified a unique genome-wide significant risk region - SH3BP5-NR2C2 - for alcohol and nicotine co-dependence. This region was enriched with replicable risk SNPs in two genetically distinct populations. Additionally, the effect directions and significance strengths of all available SNPs across this whole region matched between two populations; that is, the distributions of -log(p) values for these markers were consistent between two populations, and the associations became more significant in meta-analysis. These findings suggested that SH3BP5-NR2C2 region might harbor causal loci for alcohol and nicotine co-dependence.
Multiple lines of evidence support this conclusion. First, the SH3BP5-NR2C2 region was the only genome-wide significant (both p<5×10-7 and FDR<0.05) as well as replicable region across whole genome. Second, this region was the only association peak throughout Chromosome 3 at p<0.0001 in meta-analysis (in which the association signals were majorly attributable to the EA sample). It is thus highly likely that the putative causal loci for alcohol and nicotine co-dependence were located within this region. Third, RNA secondary structure may affect RNA stability, RNA 3D structure, intron splicing, exon recognition, transcription level and translation efficiency. Many replicable risk SNPs in SH3BP5-NR2C2 had potentials to slightly (in SH3BP5 and NR2C2) to significantly (in ZFYVE20) alter the RNA secondary structures (Supplemental Table S2), which might further influence the function of proteins and eventually affect the risk for disease, providing additional evidence in support of the hypothesis that SH3BP5-NR2C2 per se contributes to alcohol and nicotine co-dependence. Fourth, all SNPs in this region among those 11 replicable risk markers available for eQTL analysis had nominal cis- and trans-acting regulatory effects on gene expression.
Expression of NR2C2, ZFYVE20, CAPN7 and SH3BP5 transcripts was significantly correlated with expression of many alcohol and nicotine dependence-related genes, including those in the dopaminergic[Batel et al., 2008; Stapleton et al., 2007], serotoninergic[Hasegawa et al., 2002], cholinergic [Lou et al., 2006; Saccone et al., 2009], GABAergic [Chang et al., 2002], glutamatergic [Edenberg et al., 2010], histaminergic [Oroszi et al., 2005], endocannabinoid [Zuo et al., 2007], opoid [Zhang et al., 2008], alcohol metabolic[Luo et al., 2006; Uhl et al., 2008] and neuropeptide[Wetherill et al., 2008] systems. These findings suggested that SH3BP5-NR2C2 might also be implicated in alcohol dependence via the classical neurotransmission systems or metabolic pathways.
Additionally, the transcript expression of the genes in this region was nominally regulated by some nicotine dependence-related genes, including ITGA4, KCNN3, CCBE1, SGCZ, PARK2, PBK and CTNNA3. All of these genes have ever been associated with nicotine dependence or related traits before[Bierut et al., 2007; Uhl et al., 2010; Uhl et al., 2008], although some of them have not been well replicated yet. ITGA4 and KCNN3 regulated all four genes in SH3BP5-NR2C2 region in brain, and CCBE1 and SGCZ regulated these four genes in PBMC. PARK2 and PBK regulated SH3BP5 in PBMC and brain, respectively; and CTNNA3 regulated CAPN7 in brain. Although these regulatory effects were not significant after correction for multiple testing, those effects replicable across four genes might be robust. Biological functions of these nicotine dependence-related genes together with NR2C2, ZFYVE20, CAPN7 and SH3BP5 were summarized in Table S5. Most of these genes are integrin- or calcium-related, or encode proteases (Table S5), suggesting other possible molecular mechanisms that might be implicated in alcohol and nicotine dependence.
Taken together, these findings strongly support the hypothesis that SH3BP5-NR2C2 harbors causal loci for alcohol and nicotine co-dependence. This region can be represented by two independent markers located in two independent LD blocks, and it might harbor two independent causal loci that are in LD with these independent blocks. The first causal locus might be rs9868848 in ZFYVE20 in block 1 that is a non-synonymous variant (Leu591Pro) located in a coding region. It could significantly alter the RNA secondary structure. It was functional in brain and PBMC with nominal cis- and trans-acting regulatory effects (Table 1). Its minor allele protected against risk for alcohol and nicotine co-dependence. Two other SNPs (rs28445844 in NR2C2 and rs2306853 in ZFYVE20) in LD with this SNP could also slightly alter the RNA secondary structures, and rs2306853 is located in an exonic splicing silencer or enhancer (Table S2). Alternatively, SH3BP5-CAPN7 (block 2) might possibly harbor a second putative causal locus, because the most significant SNP in this region was rs1318937 in SH3BP5 and this SNP could also slightly alter the RNA secondary structures (Table S2). Its minor allele increased risk for alcohol and nicotine co-dependence. However, all risk markers in this region were predicted to most likely lack any phenotypic effect (by Polyphen-2 [Adzhubei et al., 2010]; Table S2), so that the causal loci might not be any one of these risk markers. It is warranted in future studies to identify the causal loci by sequencing the whole SH3BP5-NR2C2 region.
In the present study, although we studied alcohol and nicotine co-dependence, we obtained results that were similar to a previous GWAS that examined alcohol dependence using SAGE data[Bierut et al., 2010]. Many top-ranked risk SNPs (p<10-5) for alcohol dependence in that previous study (i.e., KIAA0040, HTR1A, PKNOX2, HAO2, CTTNBP2, TMEM47, SH3BP5 and PLEKHG1) were also listed as top-ranked genes in the present study (Table S3). Another study found that SH3BP5 was associated with polysubstance dependence in NIDA/MNB sample (rs9310472; p=0.008) and with cocaine dependence in SAGE sample (rs1318937; p=1.6×10-7) (both SNPs were claimed to be in CAPN7 in that study)[Drgon et al., 2010].
Finally, three genes or gene regions, i.e., GABRA2, CHRNA6-CHRNB3 and CHRNA5-CHRNA3-CHRNB4 that have previously been associated to both alcohol and nicotine dependence [Bierut et al., 2007; Edenberg et al., 2004; Liu et al., 2010; Ray et al., 2009; Saccone et al., 2007; Thorgeirsson et al., 2010] were also explored in the present study. They were only nominally associated with alcohol and nicotine co-dependence (Table S6); they were neither replicable between EAs and AAs, genome-wide significant, nor on the top-ranked gene list in the present study (Table S3), consistent with previous results using the same SAGE dataset [Bierut et al., 2010; Wang et al., 2011].
Supplementary Material
Supplemental Figure S1. QQ-plot for the p-values in EAs and AAs [X-axis: expected −log(P) values; Y-axis: observed −log(P) values; P-values correspond to associations between SNPs and alcohol and nicotine co-dependence]
Supplemental Figure S2. Manhattan plots for the p-values in EAs and AAs [Y-axis: -log0.05=1.3; -log10-5=5; -log(5×10-8)=7.3. X-axis: Chr1-22=Autosomes; Chr23=ChrX; Chr24=ChrY; SNPs were ordered by physical distance within each chromosome/region]
Supplemental Figure S3. Associations between replicable risk SNPs and exon expression levels
Supplemental Table S1. Demographics of the cleaned samples
Supplemental Table S2. Bioinformatics of replicable risk SNPs in SH3BP5-NR2C2 region [*,***These SNPs are predicted to slightly and significantly alter the RNA secondary structure, respectively. TFBS, transcription factor-binding site; ESS, exonic splicing silencer; ESE, exonic splicing enhancer. NA, not available]
Supplemental Table S3. The 10 top-ranked SNPs associated with alcohol and nicotine co-dependence in EAs, AAs and meta-analysis [“-”, p>0.05. ▸IPO11-HTR1A region is replicable only among European-origin populations, which was reported elsewhere [Zuo et al. In revision]]
Supplemental Table S4. P-values for the 10 top-ranked SNPs with replicable associations in meta-analysis
Supplemental Table S5. Biological functions of SH3BP5-NR2C2 and other nicotine dependence related genes
Supplemental Table S6. Associations between AD+ND and candidate genes
Acknowledgments
This work was supported in part by National Institute on Drug Abuse (NIDA) grants K01 DA029643, K24 DA017899, R01 DA016750 and K02 DA026990, National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants R01 AA016015, R21 AA020319, K24 AA013736, and P50 AA012870 and the National Alliance for Research on Schizophrenia and Depression (NARSAD) Award 17616 (L.Z.). It also received support from the Department of Veterans Affairs through its support of the VA Alcohol Research Center, the VA National Center for PTSD, and the Depression REAP. We thank NIH GWAS Data Repository, the Contributing Investigator(s) (Drs. Bierut and Edenberg) who contributed the phenotype and genotype data (SAGE and COGA) from his/her original study, and the primary funding organization that supported the contributing study. Funding support for SAGE and COGA was provided through the National Institutes of Health (NIH) Genes, Environment and Health Initiative (GEI) Grant U01 HG004422 and U01HG004438; the GENEVA Coordinating Center (U01 HG004446); the National Institute on Alcohol Abuse and Alcoholism (U10 AA008401); the National Institute on Drug Abuse (R01 DA013423); the National Cancer Institute (P01 CA089392); and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Genotyping was performed at the Johns Hopkins University Center for Inherited Disease Research or at deCODE.
Footnotes
Conflict of Interest: None.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. Washington, DC: American Psychiatric Press; 1994. [Google Scholar]
- Batel P, Houchi H, Daoust M, Ramoz N, Naassila M, Gorwood P. A haplotype of the DRD1 gene is associated with alcohol dependence. Alcohol Clin Exp Res. 2008;32(4):567–572. doi: 10.1111/j.1530-0277.2008.00618.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107(11):5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55(2):149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Chang YT, Sun HS, Fann CS, Chang CJ, Liao ZH, Huang JL, Loh EW, Yu WY, Cheng AT. Association of the gamma-aminobutyric acid A receptor gene cluster with alcohol dependence in Taiwanese Han. Mol Psychiatry. 2002;7(8):828–829. doi: 10.1038/sj.mp.4001110. [DOI] [PubMed] [Google Scholar]
- Drgon T, Johnson CA, Nino M, Drgonova J, Walther DM, Uhl GR. “Replicated” genome wide association for dependence on illegal substances: genomic regions identified by overlapping clusters of nominally positive SNPs. Am J Med Genet B Neuropsychiatr Genet. 2010;156(2):125–138. doi: 10.1002/ajmg.b.31143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Bierut LJ, Boyce P, Cao M, Cawley S, Chiles R, Doheny KF, Hansen M, Hinrichs T, Jones K, et al. Description of the data from the Collaborative Study on the Genetics of Alcoholism (COGA) and single-nucleotide polymorphism genotyping for Genetic Analysis Workshop 14. BMC Genet. 2005;6(1):S2. doi: 10.1186/1471-2156-6-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74(4):705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34(5):840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Higuchi S, Matsushita S, Miyaoka H. Association of a polymorphism of the serotonin 1B receptor gene and alcohol dependence with inactive aldehyde dehydrogenase-2. J Neural Transm. 2002;109(4):513–521. doi: 10.1007/s007020200042. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6(12):e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, de Moor MH, Smit AB, Hottenga JJ, Richter MM, Heath AC, et al. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res Hum Genet. 2010;13(1):10–29. doi: 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Ma JZ, Payne TJ, Beuten J, Crew KM, Li MD. Gene-based analysis suggests association of the nicotinic acetylcholine receptor beta1 subunit (CHRNB1) and M1 muscarinic acetylcholine receptor (CHRM1) with vulnerability for nicotine dependence. Hum Genet. 2006;120(3):381–389. doi: 10.1007/s00439-006-0229-7. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Diplotype trend regression analysis of the ADH gene cluster and the ALDH2 gene: multiple significant associations with alcohol dependence. Am J Hum Genet. 2006;78(6):973–987. doi: 10.1086/504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroszi G, Enoch MA, Chun J, Virkkunen M, Goldman D. Thr105Ile, a functional polymorphism of histamine N-methyltransferase, is associated with alcoholism in two independent populations. Alcohol Clin Exp Res. 2005;29(3):303–309. doi: 10.1097/01.alc.0000156128.28257.2e. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R, Tyndale RF, Lerman C. Nicotine dependence pharmacogenetics: role of genetic variation in nicotine-metabolizing enzymes. J Neurogenet. 2009;23(3):252–261. doi: 10.1080/01677060802572887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, Agrawal A, Breslau N, Grucza RA, Hatsukami D, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(4):453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton JA, Sutherland G, O'Gara C. Association between dopamine transporter genotypes and smoking cessation: a meta-analysis. Addict Biol. 2007;12(2):221–226. doi: 10.1111/j.1369-1600.2007.00058.x. [DOI] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Drgon T, Johnson C, Walther D, David SP, Aveyard P, Murphy M, Johnstone EC, Munafo MR. Genome-wide association for smoking cessation success: participants in the Patch in Practice trial of nicotine replacement. Pharmacogenomics. 2010;11(3):357–367. doi: 10.2217/pgs.09.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, David SP, Niaura R, Lerman C. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65(6):683–693. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. J Psychiatr Res. 2011;45(11):1419–1425. doi: 10.1016/j.jpsychires.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Wetherill L, Schuckit MA, Hesselbrock V, Xuei X, Liang T, Dick DM, Kramer J, Nurnberger JI, Jr, Tischfield JA, Porjesz B, et al. Neuropeptide Y receptor genes are associated with alcohol dependence, alcohol withdrawal phenotypes, and cocaine dependence. Alcohol Clin Exp Res. 2008;32(12):2031–2040. doi: 10.1111/j.1530-0277.2008.00790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2008;13(5):531–543. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CR, Wang F, Zhang XY, et al. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology. 2011a;37(2):557–566. doi: 10.1038/npp.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Kranzler HR, Luo X, Covault J, Gelernter J. CNR1 variation modulates risk for drug and alcohol dependence. Biol Psychiatry. 2007;62(6):616–626. doi: 10.1016/j.biopsych.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Zuo L, Zhang CK, Wang F, Li CS, Zhao H, Lu L, Zhang XY, Lu L, Zhang H, Zhang F, et al. A novel, functional and replicable risk gene region for alcohol dependence identified by genome-wide association study. PLoS One. 2011b;6(11):e26726. doi: 10.1371/journal.pone.0026726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang H, Winstead DK, Wang F, Tan Y, Cao Y, Li CSR, Lu L, Lu L, Krystal JH, et al. In revision. Genome-wide significant association signals for alcohol and nicotine co-dependence in European descent. Am J Psychiatry [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. QQ-plot for the p-values in EAs and AAs [X-axis: expected −log(P) values; Y-axis: observed −log(P) values; P-values correspond to associations between SNPs and alcohol and nicotine co-dependence]
Supplemental Figure S2. Manhattan plots for the p-values in EAs and AAs [Y-axis: -log0.05=1.3; -log10-5=5; -log(5×10-8)=7.3. X-axis: Chr1-22=Autosomes; Chr23=ChrX; Chr24=ChrY; SNPs were ordered by physical distance within each chromosome/region]
Supplemental Figure S3. Associations between replicable risk SNPs and exon expression levels
Supplemental Table S1. Demographics of the cleaned samples
Supplemental Table S2. Bioinformatics of replicable risk SNPs in SH3BP5-NR2C2 region [*,***These SNPs are predicted to slightly and significantly alter the RNA secondary structure, respectively. TFBS, transcription factor-binding site; ESS, exonic splicing silencer; ESE, exonic splicing enhancer. NA, not available]
Supplemental Table S3. The 10 top-ranked SNPs associated with alcohol and nicotine co-dependence in EAs, AAs and meta-analysis [“-”, p>0.05. ▸IPO11-HTR1A region is replicable only among European-origin populations, which was reported elsewhere [Zuo et al. In revision]]
Supplemental Table S4. P-values for the 10 top-ranked SNPs with replicable associations in meta-analysis
Supplemental Table S5. Biological functions of SH3BP5-NR2C2 and other nicotine dependence related genes
Supplemental Table S6. Associations between AD+ND and candidate genes