Abstract
Between June and November 2010, a concerning rise in the number of cases of puerperal sepsis, a postpartum pelvic bacterial infection contracted by women after childbirth, was observed in the New South Wales, Australia, hospital system. Group A streptococcus (GAS; Streptococcus pyogenes) isolates PS001 to PS011 were recovered from nine patients. Pulsed-field gel electrophoresis and emm sequence typing revealed that GAS of emm1.40, emm75.0, emm77.0, emm89.0, and emm89.9 were each recovered from a single patient, ruling out a single source of infection. However, emm28.8 GAS were recovered from four different patients. To investigate the relatedness of these emm28 isolates, whole-genome sequencing was undertaken and the genome sequences were compared to the genome sequence of the emm28.4 reference strain, MGAS6180. A total of 186 single nucleotide polymorphisms were identified, for which the phylogenetic reconstruction indicated an outbreak of a polyclonal nature. While two isolates collected from different hospitals were not closely related, isolates from two puerperal sepsis patients from the same hospital were indistinguishable, suggesting patient-to-patient transmission or infection from a common source. The results of this study indicate that traditional typing protocols, such as pulsed-field gel electrophoresis, may not be sensitive enough to allow fine epidemiological discrimination of closely related bacterial isolates. Whole-genome sequencing presents a valid alternative that allows accurate fine-scale epidemiological investigation of bacterial infectious disease.
INTRODUCTION
Puerperal sepsis (PS) is a postpartum pelvic bacterial infection contracted by women after vaginal or abdominal delivery. The condition is identified by fever at 1 day postpartum, although more rapid and severe infection leading to death may occur. Puerperal sepsis has been recognized as a major contributor to maternal and newborn morbidity since ancient times. The introduction of lying-in hospitals in the 1600s triggered a steep rise in puerperal sepsis cases and deaths, which remained unchanged until the late 1800s. Epidemics of PS were common in the 1600s to 1800s as the by-product of hospital practices in the days before infection control and antimicrobial therapy (12).
Nosocomial bacterial infections are a worldwide problem requiring constant and targeted surveillance. Antiseptic control measures must be implemented to minimize the occurrence and spread of such infections, a notion accepted only relatively recently in the history of health care (12, 25, 27, 33). While death was the outcome for most infected mothers in the preantibiotic era, PS deaths are less common today, with most instances occurring in developing countries (17). The WHO ranks puerperal sepsis as the 6th-highest cause of maternal mortality worldwide (34). Risk factors that contribute to infection include breakdown of hygiene standards during delivery and postdelivery care, prolonged manipulation of patients during delivery, prolonged labor or rupture of membranes, as well as poor sanitary conditions and inadequate services within health facilities (23).
Group A beta-hemolytic streptococcus (GAS; Streptococcus pyogenes) is the infectious cause of puerperal fever (13). S. pyogenes is a strictly human pathogen usually found in the skin and throat and less frequently in the rectum and the female genital tract (19). GAS of serotype M28 has been associated with recent PS outbreaks (8, 14, 29). Pulsed-field gel electrophoresis (PFGE) and random amplified polymorphic DNA analysis have been used to determine the clonal relatedness of strains isolated in clusters of PS infection (10, 24, 29). However, advances in whole-genome sequencing technology provide an opportunity to overcome the limitations inherent in these techniques, offering highly sensitive and unequivocal sequence comparison at the single nucleotide level (3, 9, 28).
CASE REPORTS
In Australia, deaths due to GAS puerperal sepsis are rare, but outbreaks of infection still occur (26). In New South Wales, nine cases of GAS puerperal sepsis were identified between June and November 2010 from five different hospitals in the greater Sydney area. The mothers presented high temperatures on the day after childbirth, and all made a full recovery upon treatment. Given the close temporal distribution of multiple PS cases, an investigation of this potential outbreak was initiated. A total of 11 GAS isolates were recovered from vaginal swabs and urine and blood samples and subjected to molecular epidemiological characterization (Table 1).
Table 1.
GAS strains used in this study
| Identifier | Hospital of origin | Date of collection (day/mo/yr) | Specimen or site | Patient | emm type | Source (reference) |
|---|---|---|---|---|---|---|
| MGAS6180 | NAa | NA | NA | NA | emm28.4 | CP000056 (15) |
| PS001 | A | 26/06/10 | Vaginal swab | 1 | emm28.8 | This study |
| PS002 | A | 26/06/10 | Vaginal swab | 2 | emm1.40 | This study |
| PS003 | A | 5/07/10 | Vaginal swab | 3 | emm77.0 | This study |
| PS004 | A | 6/07/10 | Vaginal swab | 3 | emm77.0 | This study |
| PS005 | A | 27/06/10 | Urine | 4 | emm28.8 | This study |
| PS006 | A | 26/06/10 | Urine | 1 | emm28.8 | This study |
| PS007 | B | 21/09/10 | Blood | 5 | emm28.8 | This study |
| PS008 | C | 11/11/10 | Vaginal swab | 6 | emm28.8 | This study |
| PS009 | C | 7/11/10 | Vaginal swab | 7 | emm89.0 | This study |
| PS010 | D | 20/11/10 | Blood | 8 | emm75.0 | This study |
| PS011 | C | 25/11/10 | Vaginal swab | 9 | emm89.9 | This study |
NA, not applicable.
MATERIALS AND METHODS
Isolation and typing.
The 11 GAS isolates were emm typed by sequencing of the emm gene using the PCR amplification method of Beall et al. (4). Bidirectional Sanger sequencing was performed, and consensus sequences were submitted to the Centers for Disease Control and Prevention (http://www.cdc.gov/ncidod/biotech/strep/M-ProteinGene_typing.htm). Five isolates that were found to be emm28.8 were further characterized by PFGE (refer to the supplemental material for details of PFGE method).
Whole-genome sequencing and comparative analysis.
To investigate the genetic diversity of the emm28.8 isolates within the infection cluster, whole-genome sequencing was carried out using an Illumina HiSeq 2000 apparatus (Illumina, San Diego, CA). The previously sequenced emm28.4 strain S. pyogenes MGAS6180 (15) was also sequenced under the same conditions to provide an internal reference for subsequent comparative analyses. For each isolate, 1 million read pairs were mapped to the S. pyogenes MGAS6180 reference genome (GenBank accession number CP000056) using Burrows-Wheeler Aligner (BWA) (21). Putative single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) were called using bcftools (22) with settings that retained only high-quality variants. Whole-genome phylogenetic reconstruction using maximum likelihood was carried out with 186 high-quality SNPs. To identify any large-scale differences, such as phage or genomic islands, we carried out a de novo assembly of each genome using the Velvet program (35). This was done using the 1 million read pair subsets previously selected for the read-mapping step. Automated annotation was performed using the RAST annotation server (2), and subsequent genomic comparisons were undertaken using a combination of the software Mauve (11), Artemis (7), and BRIG (1). Further details of the DNA preparation, sequencing, phylogenetic, and comparative genome analysis methods are available in the supplemental material.
RESULTS
Initial typing of GAS puerperal sepsis isolates.
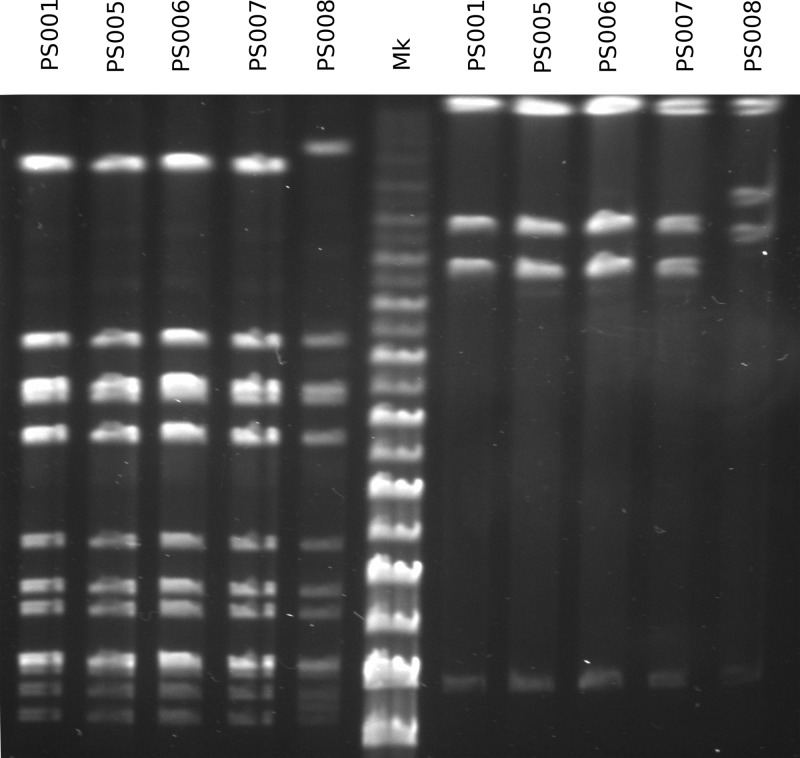
To characterize the relationship between the 11 GAS isolates in this cluster, we initially employed emm PCR typing and PFGE. Isolates with different emm types (emm1.40, emm75.0, emm77.0, emm89.0, and emm89.9) were each recovered from single patients (Table 1), ruling out a single source of infection. The five remaining emm28.8 isolates were further characterized by PFGE. Four of five emm28.8 isolates presented an identical PFGE profile (Fig. 1).
Fig 1.
PFGE profiles of the five emm28.8 isolates examined in this study, using SmaI on the left of the marker lane (Mk) and AscI on the right.
Whole-genome sequencing to identify the relationship between emm28.8 isolates.
For each GAS emm28 isolate, between 4.6 million and 15.8 million sequence read pairs were obtained, corresponding to 920 and 3,160 mega-base pairs of sequence data, respectively. As this amount of data exceeds the required sequence coverage, we carried out subsequent analysis steps using 1 million read pairs randomly sampled from each data set (approximately 100× read coverage relative to the reference emm28.4 strain S. pyogenes MGAS6180 [15]). Reads were mapped to the reference genome (Fig. 2). High-quality variants, including 186 SNPs and 33 small insertions/deletions (indels), were identified from the read-mapping data. Out of a total of 186 SNPs identified compared to the sequence of MGAS6180, only 72 are common to all emm28.8 isolates, ruling out the scenario of the emm28.8 strains having diverged from a recent common ancestor and spread from one hospital to another (see Table S1 in the supplemental material).
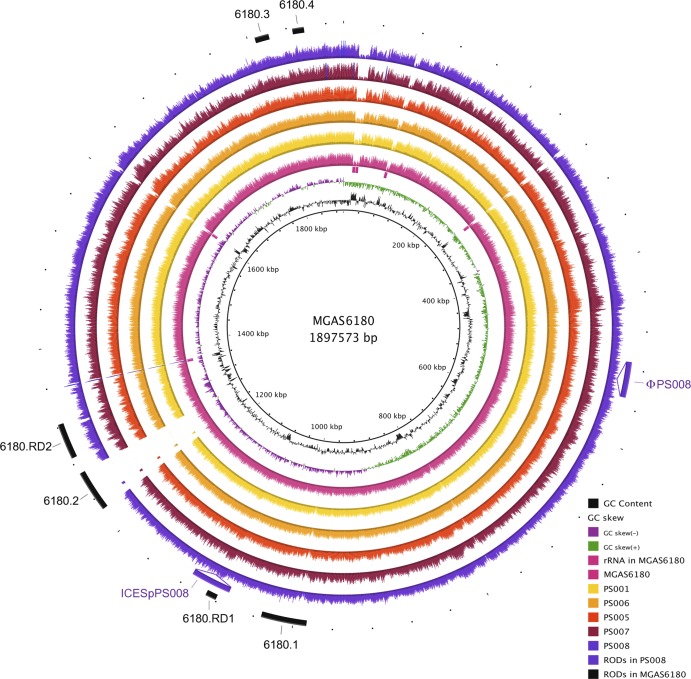
Fig 2.
Visualization of the reads selected for each strain mapped onto the S. pyogenes MGAS6180 reference genome. The innermost circles represent the GC content (black), GC skew (purple/green), and rRNA operons of MGAS6180 (pink boxes). BRIG (1) shows the distribution of the number of reads for each individual strain mapped onto the central reference using a window size of 500, arranged from inner to outer colored circles as follows: resequenced reference MGAS6180 (pink), PS001 (yellow), PS006 (orange), PS005 (red), PS007 (maroon), and PS008 (purple). Additional strain-specific regions of difference (RODs) (ϕPS008 and ICESpPS008) are represented as insertions. The outermost circle represents previously reported regions of difference in MGAS6180, namely, prophage elements 6180.1 and 6180.2, prophage remnants 6180.3 and 6180.4, and regions of difference 6180.RD1 and 6180.RD2 (15) (black).
Three isolates are virtually identical according to our SNP analysis, with only two SNPs identified in PS001 compared to the sequences of PS005 and PS006. To rule out the possibility of an error in sequencing or read mapping, the nucleotide sequence covering these SNPs was independently confirmed by Sanger sequencing. Notably, strains PS001, PS005, and PS006 were isolated from two patients from the same hospital within a 2-day window. In contrast, the emm28.8 GAS strains PS007 and PS008 were isolated from different hospitals 3 and 5 months after PS001, PS005, and PS006, respectively. Whereas PS007 and PS008 harbor a similar number of SNPs compared to the sequence of MGAS6180 (107 and 119 SNPs), these isolates have a substantially larger number of unique SNPs (32 and 47 SNPs) and therefore are not clonal.
The relationships between all emm28.8 isolates are best visualized as a phylogeny of the emm28.8 GAS strains based on the core 186 SNPs relative to the reference strain MGAS6180 (Fig. 3; see methods in the supplemental material). Indel analysis also indicates that PS001, PS005, and PS006 are closely related in comparison to the other emm 28.8 GAS strains (see Table S2 in the supplemental material). Thus, we hypothesize that PS001, PS005, and PS006 are clonal and are likely the result of either patient-to-patient- or staff-to-patient-mediated transmission.
Fig 3.
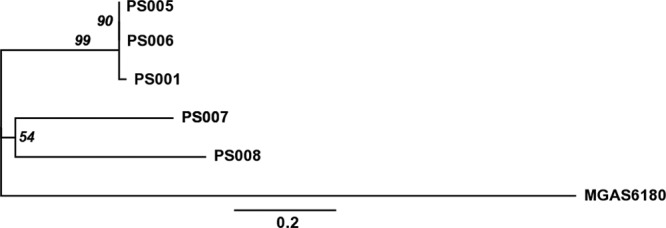
Phylogeny of the emm28 strains based on SNPs. An unrooted maximum-likelihood tree including the five emm28.8 strains and the reference strain MGAS6180 was constructed using the 172 core SNPs out of the 186 total SNPs identified compared to the sequence of MGAS6180, which were obtained by excluding 14 SNPs associated with recombination events, as described in the supplemental material. Numbers on internal nodes represent the percentage of replicated trees (>50%) in which the associated samples clustered together in the bootstrap test (based on 1,000 replicates). The scale bar represents the number of base substitutions per site.
Identification of mobile genetic elements.
Read-mapping visualization revealed that all M28-specific islands previously identified in MGAS6180, with the exception of the prophage 6180.2, were also present in the five PS emm28.8 isolates. Of note, this includes the 6180.RD2 region, which is known to carry several virulence factors and surface proteins, including the R28 protein that may play a role in vaginal carriage (15, 30, 32) (Fig. 2).
Identifying strain-specific accessory genomes and macrovariations, such as new mobile genetic elements, provides insight into horizontal gene transfer and evolution of GAS (5, 31). In order to identify regions that are not found in the reference genome, we produced a de novo assembly for each isolate. The whole-genome comparison of de novo assemblies also revealed the presence of two putative mobile genetic elements in PS008 (Fig. 2). The novel 37-kb prophage ϕPS008 does not share significant similarity to any previously described GAS prophage and does not carry any characterized virulence factors. The 41-kb integrative and conjugative element ICESpPS008 is nearly identical to ICESp2905 (6), although it harbors neither the erm(TR) nor the tet(O) antibiotic resistance gene and instead carries putative virulence factors, including genes encoding multidrug efflux proteins and a lipoprotein. Of note, ICESpPS008 is integrated at the 5′ end of the RNA uracil-methyltransferase rum gene, a well-known integration hot spot for several mobile streptococcal elements (6).
DISCUSSION
Despite the persistence of puerperal fever as a primary cause of maternal death, most of the predisposing factors leading to this disease are preventable. Control of PS incidence depends on the implementation of established techniques, including high standards of hygiene and cleanliness, strict adherence to asepsis, as well as preventive antibiotic therapy (17, 18, 33). In this study, we have demonstrated, using whole-genome sequence analysis, that in-ward transmission of infection may still be implicated in the spread of disease even within hospitals where hygiene standards are of a high level.
Whole-genome sequencing and SNP analysis have proven useful in discriminating between closely related isolates and allow analysis of the epidemiology of small infection clusters (9, 16, 20). This study shows that the 11 GAS strains recovered from a temporal cluster of PS infection in New South Wales hospitals did not represent a single clone and that most of the strains isolated from different hospitals were nonclonal. However, three strains isolated in the same hospital from two different patients were found to be clonal. PFGE was unable to discriminate these GAS emm28.8 isolates, whereas high-resolution SNP analysis allowed the deduction of fine-scale epidemiological links.
Analysis of the genome sequence of the five emm28 strains demonstrates variation in the accessory components of the genome compared to the sequence of the reference MGAS6180 strain. Although all emm28.8 strains carry the RD2 region, implicated in the development of PS (15, 32), of MGAS6180, they lack the 6180.2 phage of MGAS6180 that encodes two streptococcal virulence factors, the superantigen SpeK and the phospholipase SlaA. However, strain PS008 has acquired two novel mobile genetic elements, the phage ϕPS008 and the integrative chromosomal element ICESpPS008, carrying uncharacterized putative virulence factors. It has been established that horizontal gene transfer provides the main means for the evolution of new invasive clones via rearrangement of GAS genomes (5). Prophage sequences make up 10% of the GAS genome and account for most of the gene variation among different M types (5). Assessing the impact of such genome variation on the ability of these strains to cause disease will likely allow better understanding of the virulence potential of emm28 GAS.
Rapid, high-resolution genetic analysis of bacterial isolates is important for determining the epidemiology of hospital infection clusters. This study demonstrates the utility of whole-genome sequencing technology in the context of bacterial infection and transmission. Platforms for rapid whole-genome typing of microbial pathogens are becoming invaluable tools in epidemiological investigations (9). While there are still some limitations to the full integration of whole-genome sequencing as part of a standard typing pipeline in clinical settings, such as data analysis and cost, current efforts in implementing automated data analysis pipelines will allow whole-genome sequencing to be widely used as a routine diagnostic tool. Ongoing advances in sequencing technology, such as the Illumina platform, allow rapid sequencing of whole bacterial genomes with high coverage and will soon obviate the limitations of PFGE and other traditional epidemiological tools. This study provides an exemplar for real-time surveillance and management of suspected infection outbreaks.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the technical assistance of Mitchell Stanton-Cook and Nabil Alikhan.
This work is supported by the National Health and Medical Research Council of Australia (511224, 573401, and 565526). S. A. Beatson is the recipient of an Australian Research Council Australian research fellowship (DP0881347). M. J. Walker is the recipient of a National Health and Medical Research Council of Australia principal research fellowship (631386).
The sponsors of this study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
We declare that they have no competing interests.
Footnotes
Published ahead of print 18 April 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402 doi:10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aziz RK, et al. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75 doi:10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aziz RK, Nizet V. 2010. Pathogen microevolution in high resolution. Sci. Transl. Med. 2:16ps4. [DOI] [PubMed] [Google Scholar]
- 4. Beall B, Facklam R, Thompson T. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beres SB, Musser JM. 2007. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One 2:e800 doi:10.1371/journal.pone.0000800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brenciani A, et al. 2011. Two distinct genetic elements are responsible for erm(TR)-mediated erythromycin resistance in tetracycline-susceptible and tetracycline-resistant strains of Streptococcus pyogenes. Antimicrob. Agents Chemother. 55:2106–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carver TJ, et al. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423 doi:10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- 8. Chuang I, Van Beneden C, Beall B, Schuchat A, and the Active Bacterial Core Surveillance/Emerging Infections Program Network 2002. Population-based surveillance for postpartum invasive group A Streptococcus infections 1995-2000. Clin. Infect. Dis. 35:665–670 [DOI] [PubMed] [Google Scholar]
- 9. Croucher NJ, et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Darenberg J, et al. 2007. Molecular and clinical characteristics of invasive group A streptococcal infection in Sweden. Clin. Infect. Dis. 45:450–458 [DOI] [PubMed] [Google Scholar]
- 11. Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147 doi:10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Costa CM. 2002. “The contagiousness of childbed fever”: a short history of puerperal sepsis and its treatment. Med. J. Aust. 177:668–671 [DOI] [PubMed] [Google Scholar]
- 13. Gilbert GL. 1985. Group A streptococci and puerperal fever. Then and now. Med. J. Aust. 143:271–272 [DOI] [PubMed] [Google Scholar]
- 14. Green NM, et al. 2005. Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J. Clin. Microbiol. 43:4083–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Green NM, et al. 2005. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J. Infect. Dis. 192:760–770 [DOI] [PubMed] [Google Scholar]
- 16. Harris S, et al. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hussein J, Mavalankar DV, Sharma S, D'Ambruoso L. 2011. A review of health system infection control measures in developing countries: what can be learned to reduce maternal mortality. Global. Health 7:14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kwast BE. 1991. Puerperal sepsis: its contribution to maternal mortality. Midwifery 7:102–106 [DOI] [PubMed] [Google Scholar]
- 19. Larsen B, Monif GRG. 2001. Understanding the bacterial flora of the female genital tract. Clin. Infect. Dis. 32:e69–e77 [DOI] [PubMed] [Google Scholar]
- 20. Lewis T, et al. 2010. High-throughput whole-genome sequencing to dissect the epidemiology of Acinetobacter baumannii isolates from a hospital outbreak. J. Hosp. Infect. 75:37–41 [DOI] [PubMed] [Google Scholar]
- 21. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maharaj D. 2007. Puerperal pyrexia: a review. Part I. Obstet. Gynecol. Surv. 62:393–399 [DOI] [PubMed] [Google Scholar]
- 24. Mihaila-Amrouche L, Bouvet A, Loubinoux J. 2004. Clonal spread of emm type 28 isolates of Streptococcus pyogenes that are multiresistant to antibiotics. J. Clin. Microbiol. 42:3844–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Noakes TD, Borrensen J, Hew-Butler T, Lambert MI, Joordan E. 2008. Semmelweis and the aetiology of puerperal sepsis 160 years on: an historical review. Epidemiol. Infect. 136:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. NSWHealth 2010. Safety Alert Broadcast System. Safety notice SN:017/10. NSW Government, Sydney, Australia: http://www.health.nsw.gov.au/quality/sabs Accessed October 2011 [Google Scholar]
- 27. Pittet D, Mourouga P, Perneger TV, and the members of the Infection Control Program 1999. Compliance with handwashing in a teaching hospital. Ann. Intern. Med. 130:126–130 [DOI] [PubMed] [Google Scholar]
- 28. Rasko DA, et al. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365:709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raymond J, Schlegel L, Garnier F, Bouvet A. 2005. Molecular characterization of Streptococcus pyogenes isolates to investigate an outbreak of puerperal sepsis. Infect. Control Hosp. Epidemiol. 26:455–461 [DOI] [PubMed] [Google Scholar]
- 30. Sitkiewicz I, et al. 2010. Adaptation of group A Streptococcus to human amniotic fluid. PLoS One 5:e9785 doi:10.1371/journal.pone.0009785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sitkiewicz I, Green NM, Guo N, Mereghetti L, Musser JM. 2011. Lateral gene transfer of streptococcal ICE element RD2 (region of difference 2) encoding secreted proteins. BMC Microbiol. 11:65–77 doi:10.1186/1471-2180-11-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stalhammar-Carlemalm MT, Areschoug T, Larsson C, Lindahl G. 1999. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol. Microbiol. 33:208–219 [DOI] [PubMed] [Google Scholar]
- 33. WHO 2005. WHO guidelines on hand hygiene in health care (advanced draft): summary. Clean hands are safe hands. World Alliance for Patient Safety, World Health Organization, Geneva, Switzerland [Google Scholar]
- 34. WHO 2008. The global burden of disease: 2004 update. World Health Organization, Geneva, Switzerland [Google Scholar]
- 35. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.