Abstract
To mount an immune response, lymphocytes must re-circulate between the blood and lymph nodes, recognize antigens upon contact with specialized presenting cells, proliferate to expand a small number of clonally-relevant lymphocytes, differentiate to antibody-producing plasma cells or effector T cells, exit from lymph nodes, migrate to tissues, and engage in host-protective activities. All of these processes involve motility and cellular interactions – events that were hidden from view until recently. Introduced to immunology by three papers in this journal in 2002, in vivo live-cell imaging studies are revealing the behavior of cells mediating adaptive and innate immunity in diverse tissue environments, providing quantitative measurement of cellular motility, interactions, and response dynamics. Here, we review themes emerging from such studies and speculate on the future of immuno-imaging.
Introduction
During embryonic development of complex metazoans, rapid cell division, large-scale movement of cells, and inductive interactions result in further differentiation and specialization. These latter events depend greatly on cellular location and take account of both contact-dependent and soluble signals. But this panoply of highly dynamic processes is largely absent from adult organisms, replaced by relatively stable tissue architectures, and stereotypical spatial relocation of terminal cells in epithelial structures from basal progenitors. Neural networks undergo local modifications and pruning, but wide scale cell position changes and replacement are rare.
The cells of the immune system stand out against this general landscape in retaining many of the properties of the embryonic state. Aside from the initial seeding of some resident myeloid and lymphoid cells into specific tissues and organs, there is widespread movement throughout life of many cell types from bone marrow to the thymus and secondary lymphoid organs, entry into a variety of tissue sites in response to damage or microbial invasion, extensive signaling through transient contacts lasting minutes to hours, transient exchange of differentiation-inducing or viability-sustaining information, and rapid cell division that rivals the rates seen during embryogenesis.
Although the existence of circulating and tissue-invading immune cells has been recognized for half a century (1), and the dynamic process of leukocyte extravasation from blood to tissue studied using video imaging for nearly 20 years (2), it is only in the last decade that multiplex, high-resolution, dynamic, in situ examination of this complex choreography of immune cell motion, interaction, and function has been possible. Starting with a series of papers in 2002, three of which appeared together in this journal (3–5), our understanding of how cell movement, positioning, and interaction contribute to effective immune responses has undergone explosive growth using 1- and more commonly 2-photon (2P) microscopy to visualize living cells in vivo and in tissue explant preparations (Box 1). The observations made during this period have changed concepts of the relationship between tissue organization and the development of adaptive immunity, provided new insights into how innate immune effectors carry out their search and destroy missions, yielded quantitative data that have altered previous models of adaptive immune response development, and helped provide insight into the effect of gene mutations on immunity that could not have been gained by other means. Other studies have revealed in “Technicolor” detail how immune cells interact with a diverse arrays of pathogens, the basis for immunoregulation in secondary lymphoid tissues, and the effects of immunosuppressive drugs on immune cell behavior in vivo. In short, in situ imaging has proved a powerful tool to investigate the cellular dynamics of the immune response in lymphoid organs and in peripheral tissues (Fig. 1). Here we try to synthesize the key conceptual advances that have come from this research, not seeking a comprehensive review of the literature, but focusing on how the application of this technology has fundamentally changed our understanding of immune system organization and physiology. We end with some thoughts about the future.
Box 1.
2P basics
Although some important contributions have come from use of confocal (1P) imaging methods (as just two examples, (5, 85)), most studies now use two-photon (2P) microscopy as the technique of choice for relatively deep tissue imaging of living cells (86, 87). Two-photon microscopy uses incredibly bright pulses of near-infrared laser light, less than 1 picosecond in duration and focused to a spot by the objective lens of a microscope, to illuminate fluorescently labeled cells inside the tissue environment. When the light is on during the laser pulse, the photon density at the spot is such that two photons are absorbed almost simultaneously by a fluorescent dye or protein inside the cell, and a lower-wavelength photon is then emitted. Despite the intensity of light, less damage is produced than with other imaging methods, because the light is off most of the time in between laser pulses, and fluorescence excitation is confined to the diffraction-limited spot. Moreover, the near-infrared light used for excitation penetrates better through the tissue environment than lower wavelengths. The laser is scanned rapidly in the x-y plane to produce an image; volume images are obtained by repositioning the objective up and down in the z axis; emitted photons are detected by photomultiplier tubes. This process is then repeated to obtain a time-lapse ‘movie’ of cell behavior; volume sampling in less than 20 seconds is best to avoid blurring of rapidly migrating cells. Several detectors can be deployed to image differently labeled molecules or cell types simultaneously.
The first 2-photon imaging study of lymph nodes examined explanted lymph nodes that were superfused with warmed, oxygenated media (4). This study was followed shortly by studies in which lymph nodes were imaged in live, anesthetized mice (13, 24). The practical limit to tissue depth for most studies is around 300 microns, and thus some studies have used sliced tissue preparations to provide access to deep regions of tissue, such as the splenic white pulp (88, 89) and the thymic medulla (56, 58). Other studies have succeeded in imaging in liver (63, 64), lung (90, 91), bone marrow (92–94), pancreas (95), and the gastrointestinal mucosa (65, 96), as well as other sites. In general, the observations made with explanted and intravitally imaged tissues have been in close agreement. A crucial technical consideration for 2-photon tissue imaging is that care be given to maintain the health of the tissue during the experiment, regardless of what method is used to provide access for imaging.
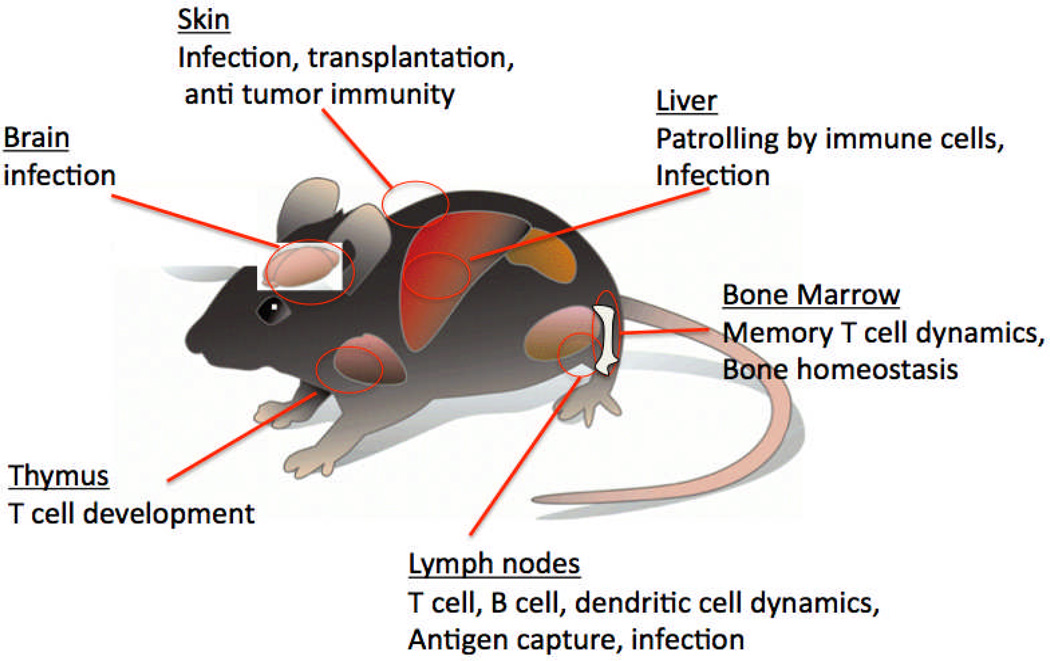
Figure 1. Two-photon imaging of different anatomical sites in the mouse.
Although early 2-photon studies of the immune system focused on T cell activation in the lymph node, in the past decade this approach has been extended to a variety of different tissues, using both intravital imaging approaches and tissue explants. Processes that have been examined include immune responses to infection, immune homeostasis, transplant immunology, anti-tumor immunology, and regulation of immune responses.
Lymph node cellular dynamics and the initiation of T cell adaptive immune responses
Among the most striking findings to emerge from dynamic imaging analyses is the seemingly random pattern of robust cellular migration exhibited by many cell types under basal conditions and the efficiency with which cells direct their attention to particular targets by short- or long-range migrations during active immune responses. T cells are able to crawl more rapidly than any other cell type in the body. Similar ameboid actin-based motility at a somewhat slower pace is the default status of B lymphocytes (4, 6), natural killer cells (7, 8), neutrophils (9, 10), and monocytes (11). Collectively, these cells can be regarded as the explorers, using cell surface receptors to sample the environment and responding with altered motility when signals are transmitted. On the other hand, antigen-presenting cells, such as dendritic cells (DCs) (5, 12–17) and Langerhans cells (18, 19), are generally sessile in tissue unless induced to migrate by microbial or inflammatory signals (16, 19, 20), actively waving their dendritic cell processes and displaying on their surface MHC-encoded proteins a short-term historical record of pathogen invasion. When antigen receptors are engaged by antigen or chemokines, lymphocytes often stop (5, 12, 13, 17, 21, 22) as signals begin to transform seemingly random motility into directed responses that reveal coordinated cellular behavior, including local swarming or directed migration from one region to another. Local motility can evolve into long-range migrations by cells leaving the tissue environment to migrate in blood or lymph to distant sites.
Lymph nodes are major sites of antigen capture, detection, and initial responses during an adaptive immune response (Fig. 2). After homing into lymph nodes from the blood, lymphocytes spend several hours to a day in a given lymph node (23). During this time, they sample the environment and most often leave the lymph node via efferent lymphatic vessels without finding antigen. Entry is regulated by the chemokine receptor CCR7, egress by the sphingosine-1-phosphate receptor 1 (S1P1); both G-protein coupled receptors ensure directed migration at the global level into and out of the lymph node (23). The dynamic nature of lymphocyte movement as revealed by the earliest imaging studies of these secondary lymphoid organs, first in explants (4, 12) and then intravital preparations (13, 24), demonstrated that lymphocytes actively migrate to pass from their entry sites at high endothelial venules to their exit at efferent lymphatics. The remarkably rapid pace of T cell movement while in the dense paracortical region of the node, however, and the way in which they scanned for antigen, were nonetheless unexpected. When viewed in time lapse, it looks chaotic – in fact, naïve T and B cell tracks are well described as a random walk (4, 13, 24). But when antigen is present, T and B cells respond by altering their ongoing random migration, initiating interactions that lead to antibody production, proliferation, differentiation to memory or effector cells, and exit from the lymph node.
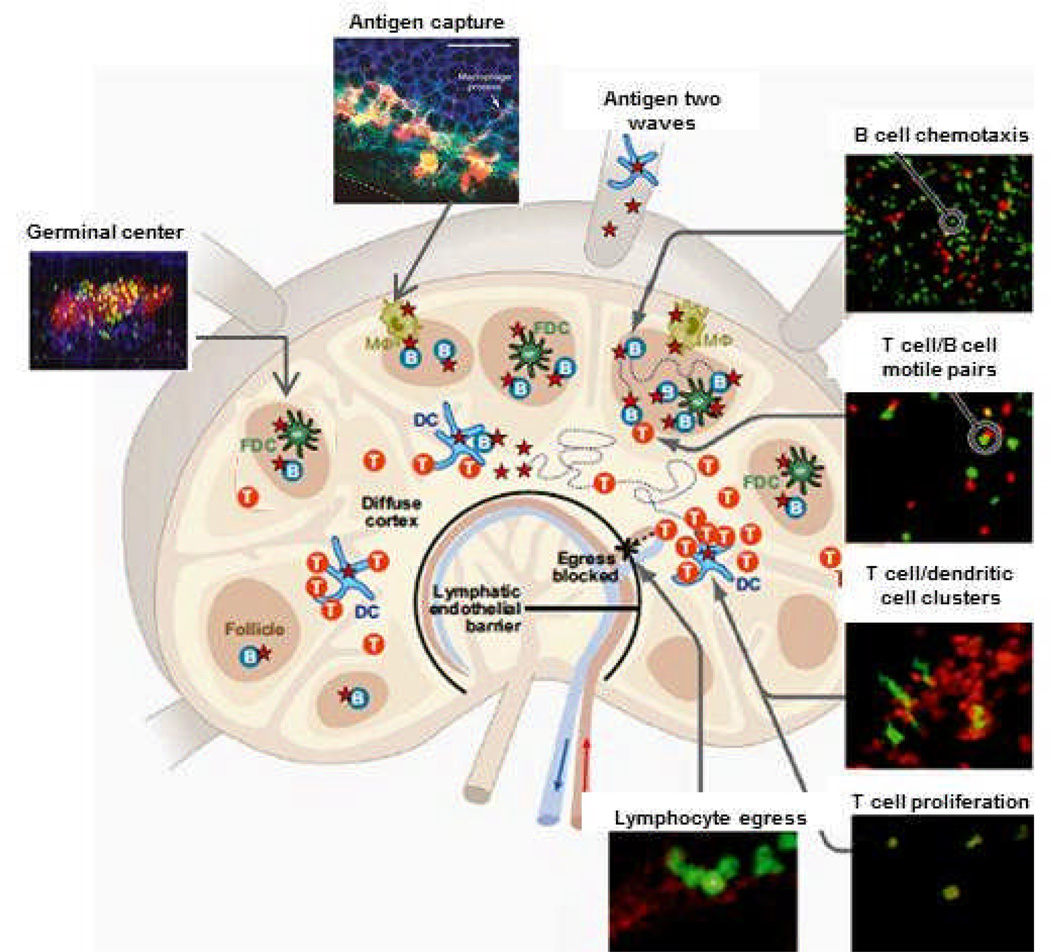
Figure 2. Lymph node cellular choreography in response to antigen.
T cells (T), B cells (B), dendritic cells (DC), follicular dendritic cells (FDC), and macrophages (Mφ) during the response to antigen, adapted from (6). Diagram depicts 2P images of antigen capture, T cell-dendritic cell interactions, T cell proliferation, chemotaxis of B cells to the follicle edge, motile T cell-B cell conjugates, germinal center dynamics, and lymphocyte egress as described in the text.
2P studies illuminated the remarkable process whereby cell types present in very small numbers (antigen presenting cells, specific T and B cells) find each other in the large volume of a lymph node to drive effective adaptive immune responses. The robust motility in lymph nodes initially suggested an antigen search strategy carried out by lymphocytes acting autonomously (24). Later, it became clear that T lymphocytes migrate in a random walk-like manner in contact with a network of fibroblastic reticular cells that are tightly associated with dendritic cells (FRCs; (25) and from which they acquire chemokinetic signals enabling more rapid migration (26). These imaging data refined earlier concepts based on static imaging that suggested a possible role of the FRC network in guiding intranodal lymphocyte movement (27). Inflammatory chemokine production by DC or DC-T cell combinations can also influence the migration of T cells within the LN, leading to more directed movement on this network (28, 29). On the antigen-presenting cell side of the equation, individual resident and migratory DCs extend agile dendrites and contact hundreds or thousands of motile T cells per hour to enable efficient repertoire scanning (14). Together these observations suggest that structural and chemical cues are used to enhance the likelihood that rare cells will co-localize and come into contact in the shortest possible time following antigen entry, driving effective adaptive responses.
Intravital imaging has also uncovered a previously unappreciated sequence of kinetic behaviors in the T cell response to antigen-bearing DCs (Figure 3a). This process evolves in three distinct phases for both CD4+ and CD8+ T cells (13, 15). Initially, T cells contact antigen-bearing DCs intermittently, briefly pausing and then migrating again to sample several DCs. During this time, T cell signaling is initiated, resulting in a series of Ca2+ spikes (30). The Ca2+ signal reduces motility acutely and also acts synergistically with other signaling pathways, resulting in enhanced gene expression, cytokine secretion, and cell proliferation. T cell-DC contact durations later increase, leading to prolonged interactions as several T cells cluster around individual DCs. After 16–24 hours, T cells resume their motility, swarm in the local vicinity, and undergo several rounds of proliferation. Activated CD4+ T cells then begin to interact with cognate B cells near the edge of the follicle.
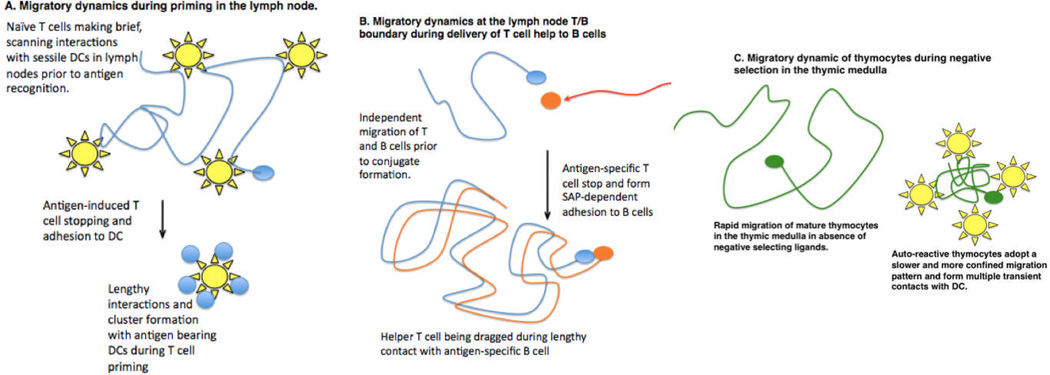
Figure 3. Cell migratory dynamics and intracellular communication.
Two-photon microscopy has revealed numerous examples in which T cells regulate their speed and cell adhesion to allow for efficient sampling of potential antigen presenting cells and effective intracellular communication. A. During T cell priming in lymph nodes, naive T cells (blue) migrate rapidly along FRC (not depicted here) making brief and frequent contact with dendritic cells (yellow). Upon antigen detection, T cells arrest and adhere to DC, leading to the formation of T cell swarms and clusters around individual DCs. In some cases, T cells undergo a phase of intermittent contacts prior to forming lengthy interactions with DC (not depicted). B. The independent migration of helper T cells and antigen-specific B cells is overcome when T cells arrest and form stable SAP-dependent conjugates with B cells. B cells continue to migrate, dragging the T cell behind. C. In the thymus, developing T cells (green) undergo rapid migration in the medulla while scanning thymic antigen presenting cells for self-antigens. Encounter with self-antigens can lead to slower and more confined migration, allowing for frequent, serial interactions with DC (yellow) and other potential APCs in the vicinity.
Imaging the induction of humoral immunity
If the scanning and motility data along with this newly revealed multistage progression of cell interaction dynamics first captured the field’s attention, a second wave of enthusiasm came with studies revealing how antigen accessed the lymph node and became available for lymphocyte recognition (31). Antigen can arrive in the form of molecules or microbial particles that travel passively via afferent lymphatic vessels, or they can arrive as peptide-MHC ligands on the surface of tissue-derived DCs and Langerhans cells that deliver a representative sample of peripheral material. Large antigen molecules or virus particles in lymph are taken up by subcapsular macrophages in draining lymph nodes and then handed off to B cells and, in turn, to follicular dendritic cells that provide a reservoir later sampled by B cells (32–35). Other pathways for delivery of soluble antigens include being conveyed directly to B cells in the follicle by conduits (36) or presented to B cells by DC (37).
Beyond revealing this first wave of antigen acquisition by the B cell, 2P imaging of T and B cell interaction provided additional insights. Activated B cells were known to take the initiative in seeking T cell help, moving by chemotaxis toward the follicle edge (38, 39), but live imaging provided a much richer appreciation of the choreography of these critical first steps in humoral immunity. Antigen-activated B cells migrate randomly within the follicle until they enter a zone about 100–200 µm from the follicle edge. Then, newly expressed CCR7 receptors detect a gradient of CCL19 and CCL21 that guides them toward the T cell zone. Near the follicle edge, antigen presentation-dependent motile B-T conjugates are formed, the B cells leading the way (40, 41). Both activated B and T cells then return to the deeper follicle to start the germinal center reaction.
The germinal center reaction, responsible for production of high affinity, isotype-switched antibodies, had been well studied using static imaging and elegant molecular tools. It had also been the subject an intensive efforts to quantitatively model immune function, specifically antibody affinity maturation. So it came as a surprise when the initial sets of results from live imaging of germinal centers did not fit easily into the established model of trafficking between germinal center subregions as implied by data from conventional histochemistry (42–44). Movement between these zones seemed more frequent and less regulated than expected and a clear division of proliferation vs. selective events in the two regions was less evident. However, recent studies using advances in tracking cells in vivo, especially employment of photo-activatable probes that permit cells to be tagged when present in one location and imaged as they move to another, have shown that the older model of selection based on T cell help in the ‘light’ zone and proliferation in the ‘dark’ zone was largely correct, while implicating the extent of interaction with T cells as a major determinant of inter-zonal migration and effective selection (45). Thus, intravital imaging combined with imaginative experimental design and new technology has substantially improved our understanding of a process at the heart of adaptive immunity.
Cell migratory dynamics place a threshold on cell-cell communication
The first 2-photon images of rapid T cell migration in lymph nodes (4, 12, 13, 24) necessitated a rethinking of intercellular communication and the impact of cell motility on this process. Although immunologists long appreciated that T cell responses require direct contact between a T cell and an antigen-bearing cell, these events had previously been examined using either end stage assays of in vivo events that occur over a period of days or weeks or in vitro studies in which the interacting cells are maintained in a constrained culture environment for days. Striking images of the distinctive migratory patterns of T cells and their potential partners forcefully pointed out that independent cellular movement must be overcome to prevent partner cells from moving outside of the range needed for effective molecular communication. This in turn points to the existence of “go-no go” thresholds for antigen signaling intensity – such signaling must elicit an adequate adhesive change or override the propensity for continued cell movement to ensure useful intercellular communication.
With this concept in mind, we can now better appreciate seminal observations showing that during the phase of T-B adhesion in lymph nodes, the B cells ‘drag’ the T cells behind them as they move (41) (Figure 3b). The early in situ imaging studies, as confirmed by many subsequent reports, helped resolve an existing controversy about whether T cells undergo a ‘stop’ signal when the T cell receptor (TCR) is adequately engaged by antigen (4, 5, 12, 13, 24), a phenomenon initially described in vitro (21, 46, 47). In contrast, B cells that have acquired antigen through the B cell receptor (BCR) and become activated antigen-presenting cells, do not get such a strong stop signal and continue to migrate. Therefore, to ensure a sufficient duration of cell-cell contact to permit upregulation of key mediators by the T cells (CD40L, cytokines, …) and effective sensing of these signals by the antigen-activated B cells, the T cell needs to depolarize, then adhere to and passively follow the moving B cell.
The significance of overcoming dispersive cell movement by regulated adhesion was acutely revealed while exploring the basis for immune defects produced by mutation of the small adaptor protein SAP. Functional loss of SAP in T cells results in X-linked lymphoproliferative disease in humans and a syndrome in mice characterized by the lack of germinal center responses (48). 2P imaging studies revealed that the immunodeficiency resulted from an insufficient duration of cell-cell contact between SAP-deficient activated helper T cells and activated B cells (49) The reduced time available for these interactions when the T cells lacked SAP prevented delivery of the molecular ‘help’ required for early B cell survival and clonal expansion. The critical role of adequate cell adhesion during developing T-dependent antibody responses was missed in vitro.
This theme of cell contact duration as a key regulatory checkpoint in immunity is further emphasized by data on how inhibitory receptors on effector T cells or how regulatory T cells mediate their suppressive effects. Operating in a cis fashion, CTLA-4 (50) and PD-1 (51) have both been reported in imaging analyses to limit the duration of T cell interaction with antigen-bearing DC. Other studies have implicated interference with stable T cell contact with antigen-presenting DC or B cells as one way Tregs interfere with CD4 T cell priming (52) or CD8 T cell effector activity (53). By reducing the duration of effective cell-cell contact and thus how long cellular receptors remain engaged, these immunoregulatory components amplify any inhibitory effects they have directly on TCR or costimulatory molecule signaling.
In contrast to examples where mature T cells adhere tightly to antigen-bearing cells during productive responses, 2P imaging has revealed other settings in which T cells remain relatively motile and collect signals from serial brief encounters with multiple antigen presenting cells. This mode of interaction may provide sufficient interactions to sustain TCR signaling under conditions where peptide-MHC ligands are broadly distributed on multiple APC, or when the directed release of effector molecules by T cells is not required. Behavior of this type has been reported for activation of CD8 T cells in lymph nodes under conditions of limiting antigen on DC (54) and in particular, developing T cells undergoing TCR repertoire selection in the thymus. Immature T cells in the thymus migrate relatively slowly via random walk through the cortex (55, 56), and encounters with positive selecting ligands lead to calcium-dependent pausing (56) and both dynamic and stable contacts with MHC-bearing stromal cells (3). These behaviors are consistent with the broadly distributed self-peptide MHC ligands that induce positive selection. On the other hand, the rapid and directional migration of positively selected thymocytes is incompatible with productive engagement of peptide-MHC ligands on immobile thymic epithelial cells (55, 57), and thus cessation of strong MHC recognition must occur as positively selected thymocytes relocate from the cortex to the medulla. Once in the medulla, thymocytes undergo further screening for recognition of self-antigen. 2P imaging of thymocytes undergoing negative selection in the medulla revealed a motile, but highly confined migration pattern (58) (Figure 3c) suggesting that some auto-reactive thymocytes sample multiple antigen-presenting cells in a local area of the medulla for some time before eventually being eliminated by clonal deletion.
The duration of T cell contact with antigen presenting cells has also been explored in peripheral tolerance induction. Some (59, 60) but not other (61) papers have described a striking difference in the length of T-DC contact under immunogenic vs. tolerogenic conditions, with the former being long-lived and the latter transient. Whether these different observations arise from the specific experimental systems employed remains to be determined, but such divergent results emphasize the need for further studies on how the length of cell-cell contact influences the quality and magnitude of T cell responses, not only with respect to events within secondary lymphoid tissues, but also in terms of effector T cell activity in peripheral tissues as we discuss in the next section.
Imaging host-pathogen interactions and understanding effector function in tissues
Besides the basic understanding of immune cell behavior revealed by these imaging studies, 2P imaging has also become a key tool to investigate the interplay between pathogens and the host immune system (62). The ability to directly visualize fluorescent pathogens as they move through the body and interact with immune cells has provided a new dimension to studies of host-pathogen interactions in diverse tissues including lymph nodes, brain, liver, gut, and skin (10, 63–69). Prior to 2P imaging, our understanding of pathogen-immune cell interactions relied largely on in vitro infection, which may miss the key role of specialized cells types that exist in vivo. Thus 2P imaging combined with in vivo infection models has proved a powerful approach to reveal when, where, and how pathogens are engaged by the immune system.
One example is the key role of lymph node subcapsular macrophages in the initial encounters with pathogens. Besides conveying antigen to B cells, these macrophages also trap lymph-borne pathogens and impede their dissemination through the body. A particularly fascinating example involves the neurotropic vesicular stomatitis virus (70). In a normal lymph node, subcapsular macrophages prevent the virus from gaining access to other cells. When these macrophages are removed, however, the virus invades neurons within the lymph node and can spread rapidly to the central nervous system. In another example, by allowing themselves to be invaded by intracellular pathogens, including viruses and the protozoan parasite, Toxoplasma gondii (10, 34, 68, 70, 71), subcapsular macrophages expose themselves to recognition and killing by CD8+ T cells (68, 71). These studies reveal that lymph node subcapsular macrophages provide an important battleground between host and pathogen during the initial phases of infection.
2P imaging has also been used to visualize the standoff between pathogens and T cell effectors at sites of chronic infection (63, 64, 66, 69). These studies reveal striking examples in which pathogens can remain undetected while surrounded by large numbers of actively migrating effector T cells. For example, effector CD8+ T cells ignore Toxoplasma containing cysts in the brains of chronically infected mice, in spite of the presence of abundant antigen, instead forming transient contacts with granuloma-like structures containing isolated parasites (66). Similarly, CD4+ effector T cells at sites of Leishmania major infection focused their attention on certain parasites, while ignoring others in the immediate vicinity (69). Limited T cell effector responses at sites of chronic infection, and the ability of some pathogens to avoid detection altogether, help to explain the ability of pathogens to persist in the face of a T cell response and the ability of T cells to contain pathogens while avoiding collateral damage to host tissues.
The question of whether transient contacts between T cell effectors and APC during chronic infection allow for delivery of effector functions remains controversial. Some studies suggested that short-lived interactions (‘kinapses’) mediated activation and function during anti-tumor or anti-viral CD8+ T cell responses (72, 73). A different view emerged from analysis of chronic mycobacterial granulomas. In such lesions, a clear correlation between a low rate of antigen-induced stopping by effector T cells and a low frequency of interferon-γ producing T cells was observed. Increasing the amount of antigen in the granuloma resulted in stopping by nearly all antigen-specific cells and cytokine production by the same large fraction of cells (64). These results reinforce the notion that strong stop signals are required for elicitation of and/or delivery of T cell effector molecules, at least under many circumstances, and also indicate that only a small number of potential effectors may do so at one time when antigen is limiting. This latter result that has important implications for assessing whether antigen-induced stopping is critical for effector function when analyzing data on large populations of effector cells in a tissue setting. If only a small fraction of pathogen-specific cells stops at any moment, measurements such as average velocity or average confinement calculated for all the specific cells being imaged will show little difference from those seen for control (antigen-unspecific) cells, obscuring the behavior of the functionally critical subpopulation of effectors and suggesting that activation without stopping had occurred.
Many other key issues related to tissue entry and in situ function of innate and adaptive effectors have recently been highlighted by dynamic imaging studies. For example, how directional are cell paths within a tissue – do neutrophils or effector T cells traffic directly to foci of infection or tissue damage or do they meander on the way to these end targets? Intravital imaging has shown that neutrophil migration from venules to sites of tissue damage is direct and linear, with little meandering and with essentially no neutrophils exiting an inflamed vessel on the side away from the damage (9, 10), documenting a precise control of cell migration directionality at both the vessel and tissue level. In tissues, what is the effect of tissue density and architecture on effector movement and how might this influence the search for pathogens and tumors? How long do T cells produce cytokines once in an antigen-rich tissue environment and do they do so locally around static antigen-presenting cells or in contrast, once activated by antigen within the tissue, do they move extensively, delivering effector cytokines to many distinct locations? These remain key questions for the future studies.
Tracking immunosuppression
Imaging has also been applied to investigate the action of drugs that interfere with immune processes. Two classes of immunosuppressants have been examined using this approach: egress blockers that resemble sphingosine and disrupt the normal trafficking of lymphocytes back into the circulation from the lymph node; and inhibitors of Kv1.3 potassium channels in T cells for specific suppression of effector T cells that are mediators of autoimmune disease and inflammatory responses. Several studies have imaged the egress step of lymphocytes traversing the lymphatic endothelial barrier in the medullary sinuses at particular sites or “portals” for egress to gain access to efferent lymphatic vessels (74–77). S1P1 is the target of FTY-720 (fingolimod), an agent that has shown efficacy in treatment of multiple sclerosis. After exposure to a metabolic product of this drug, lymphocytes fail to egress from lymph nodes, resulting in lymphopenia and a paucity of lymphocytes in the periphery. Intravital imaging showed that reversible agonists of S1P1 are able to prevent egress and, upon washout or addition of an S1P1 antagonist, lymphocytes were observed crossing into the medullary sinuses. Although some mechanistic aspects of egress are controversial (23), these studies documented the feasibility of 2P imaging to investigate drug action.
Imaging immunosuppression has also been accomplished in the periphery during chronic inflammatory immune responses. T effector memory cells recapitulate the events of antigen recognition in the lymph node, stopping in contact with tissue APCs and subsequently migrating on collagen as enlarged T cell blasts in dermal tissue during a delayed-type hypersensitivity response. Blockade of Kv1.3 channels selectively inhibits cell enlargement and motility of T effector memory cells in the tissue (78). Moreover, Kv1.3 channel blockade spares the motility of naïve T cells in the lymph node and, correspondingly, does not inhibit the acute immune response to bacterial or viral infection. These experiments provide important validation for selective immunosuppression based on Kv1.3 channels as a target to ameliorate chronic autoimmune and inflammatory conditions without disrupting an acute immune response.
Future Directions
Imaging has opened a new window to observe cells of the immune system in real time and in vivo. However, current immuno-imaging techniques are restricted in their ability to analyze the motility and interactions of cells over extended time and distance scales and to discriminate individual cells within a swarm of identically labeled cohorts. To address these limitations, novel approaches have been recently introduced utilizing photo-convertible genetic probes to unambiguously mark specific cells, and to image and track cells over long distances within intact tissue. This approach – “optical highlighting” – eliminates ambiguity when cells cross tracks with one another, and enables labeling of a subset of cells that have undergone specific behaviors, such as interactions with DC. The method has recently been used to clarify germinal center dynamics (45). Other technological advances in optical imaging promise to markedly improve our ability to image deeper and faster. These new methods include sheet illumination rather than point illumination (79) and a shift to far red probes whose emitted photons are better able to penetrate tissue without scattering to improve signals at depth. Beyond allowing for the tracking of individual cells over greater depths and distances, these improvements will also permit following cells for longer periods, allowing better linkage between early signaling events and subsequent differentiation / function of the imaged cells. But perhaps the most important frontier in intravital imaging of the immune system is that of combining molecular imaging with the cell-level dynamic measurements that have dominated to date. The goal is to monitor not just the behavior of cells, but to link cellular movement and positioning to changes in signaling and gene expression. Only by doing so can a robust and truly multidimensional picture of immune function in vivo be developed.
While it is still early days in this regard, progress is being made and there is an expectation of rapid advances in this arena. Existing fluorescent cytokine gene reporter animals can be used to follow the behavior of cells that are marked as committed to a specific effector fate, but because of the longevity of the reporter fluorescent proteins, these present indicator lines are not useful for real-time analysis of contemporaneous gene expression / cytokine production. The use of rapidly degraded reporter proteins or secreted rather than cytoplasmic reporters will likely help overcome this present limitation. Ca2+ imaging using dyes has already been used in several published studies (30, 37, 56) and improved FRET-based reporters (80) are likely to provide more robust systems for following this aspect of cellular signaling in the future. Fluorescent chimeric proteins with transcriptional factors whose nuclear translocation is important to their function have been described (81) as have adapters (73) or chimeric receptor proteins (82) that relocalize during TCR signaling, and techniques for deconvolving the complex data involved in measuring such molecular relocation in moving cells using intravital methods have been published, so we can anticipate new insights from application of these methods in the near future. Other studies will benefit from improved physiological preparations for imaging of tissues not well studied to date, including the gastrointestinal tract, pancreas, spleen, and lung. Portable imaging setups with miniaturized light delivery systems in endoscopes or implantable devices will bring this approach into the realm of clinical diagnosis.
As the number of different colors used for such imaging increases, as the tissue volume examined and number of cells imaged enlarges, as the duration of imaging sessions lengthens, and as the use of subcellular probes becomes commonplace, there will be a critical need for new analytic methods for distilling useful information from the resulting complex data sets. Analysis of the collected images is now a time-limiting feature of many intravital studies and this will only be an increasing bottleneck until more facile and robust ways of automated data processing are developed. Enhanced methods for tracking very large numbers of objects moving in three dimensions have been introduced in studies of embryogenesis (83) and certainly should be adapted for such work with immune cells, but many more computational tools for parsing the highly dynamic aspects of intravital data on immune cells will be needed to enable future studies to reach their full potential (84). In introducing more automated methods, it will be critical to avoid having investigators lose the intimate connection to their data that manual review now provides. The proper blend of computer assistance and direct viewing will be crucial so that unexpected behaviors that would not be automatically extracted from the data are not missed and so that artifacts that an algorithm would not notice are caught. In the end, the greatest value from imaging data comes from its integration with other modes of assessing the state and operation of the immune system. Imaging is just a tool, albeit a powerful one that has provided a new level of insight into the key dynamic aspects of immune system behavior. Systems biology methods for integrating diverse complex datasets will ultimately be a key element in extracting the greatest value from advanced imaging studies, helping to yield a more complete picture of immune function. Thus although a decade of imaging has given rise to a new appreciation of the importance of cell motility and interaction dynamics in producing immune responses, current studies have only scratched the surface. We look forward to even greater progress in the next decade of research in this rapidly developing field.
Acknowledgements
We would like to thank members of our laboratory groups for their contributions to the research described here and for comments on the manuscript. We apologize to the many authors who contributed importantly to the field of imaging the immune response, but whose work could not be included in the references for lack of space. This research was supported by the Intramural Research Program of NIAID, NIH (RNG); AI-065537 and AI-064227 (EAR); and NIH RO1 grant GM-41514 (MDC).
Contributor Information
Ronald N. Germain, Email: rgermain@nih.gov.
Ellen A. Robey, Email: erobey@berkeley.edu.
Michael D. Cahalan, Email: mcahalan@uci.edu.
References
- 1.Gowans JL. Int Rev Exp Pathol. 1966;5:1. [PubMed] [Google Scholar]
- 2.von Andrian UH. Microcirculation. 1996 Sep;3:287. doi: 10.3109/10739689609148303. [DOI] [PubMed] [Google Scholar]
- 3.Bousso P, Bhakta NR, Lewis RS, Robey E. Science. 2002 Jun 7;296:1876. doi: 10.1126/science.1070945. [DOI] [PubMed] [Google Scholar]
- 4.Miller MJ, Wei SH, Parker I, Cahalan MD. Science. 2002 Jun 7;296:1869. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 5.Stoll S, Delon J, Brotz TM, Germain RN. Science. 2002 Jun 7;296:1873. doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- 6.Cahalan MD, Parker I, Wei SH, Miller MJ. Nat Rev Immunol. 2002 Nov;2:872. doi: 10.1038/nri935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrod KR, Wei SH, Parker I, Cahalan MD. Proc Natl Acad Sci U S A. 2007 Jul 17;104:12081. doi: 10.1073/pnas.0702867104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celli S, Albert ML, Bousso P. Nat Med. 2011 Jun;17:744. doi: 10.1038/nm.2376. [DOI] [PubMed] [Google Scholar]
- 9.Peters NC, et al. Science. 2008 Aug 15;321:970. doi: 10.1126/science.1159194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chtanova T, et al. Immunity. 2008 Sep 19;29:487. doi: 10.1016/j.immuni.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auffray C, et al. Science. 2007 Aug 3;317:666. doi: 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- 12.Bousso P, Robey E. Nat Immunol. 2003 Jun;4:579. doi: 10.1038/ni928. [DOI] [PubMed] [Google Scholar]
- 13.Mempel TR, Henrickson SE, Von Andrian UH. Nature. 2004 Jan 8;427:154. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 14.Miller MJ, Hejazi AS, Wei SH, Cahalan MD, Parker I. Proc Natl Acad Sci U S A. 2004 Jan 27;101:998. doi: 10.1073/pnas.0306407101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller MJ, Safrina O, Parker I, Cahalan MD. J Exp Med. 2004 Oct 4;200:847. doi: 10.1084/jem.20041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindquist RL, et al. Nat Immunol. 2004 Dec;5:1243. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 17.Sen D, Deerinck TJ, Ellisman MH, Parker I, Cahalan MD. PLoS One. 2008;3:e3290. doi: 10.1371/journal.pone.0003290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kissenpfennig A, et al. Immunity. 2005 May;22:643. doi: 10.1016/j.immuni.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Sen D, Forrest L, Kepler TB, Parker I, Cahalan MD. Proc Natl Acad Sci U S A. 2010 May 4;107:8334. doi: 10.1073/pnas.0912817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tal O, et al. J Exp Med. 2011 Sep 26;208:2141. doi: 10.1084/jem.20102392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dustin ML, Bromley SK, Kan Z, Peterson DA, Unanue ER. Proc Natl Acad Sci U S A. 1997 Apr 15;94:3909. doi: 10.1073/pnas.94.8.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakami N, et al. J Exp Med. 2005 Jun 6;201:1805. doi: 10.1084/jem.20050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cyster JG, Schwab SR. Annu Rev Immunol. 2011 Mar 24; doi: 10.1146/annurev-immunol-020711-075011. [DOI] [PubMed] [Google Scholar]
- 24.Miller MJ, Wei SH, Cahalan MD, Parker I. Proc Natl Acad Sci U S A. 2003 Mar 4;100:2604. doi: 10.1073/pnas.2628040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajenoff M, et al. Immunity. 2006 Dec;25:989. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worbs T, Mempel TR, Bolter J, von Andrian UH, Forster R. J Exp Med. 2007 Mar 19;204:489. doi: 10.1084/jem.20061706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gretz JE, Anderson AO, Shaw S. Immunol Rev. 1997 Apr;156:11. doi: 10.1111/j.1600-065x.1997.tb00955.x. [DOI] [PubMed] [Google Scholar]
- 28.Castellino F, et al. Nature. 2006 Apr 13;440:890. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 29.Hugues S, et al. Nat Immunol. 2007 Sep;8:921. doi: 10.1038/ni1495. [DOI] [PubMed] [Google Scholar]
- 30.Wei SH, et al. J Immunol. 2007 Aug 1;179:1586. doi: 10.4049/jimmunol.179.3.1586. [DOI] [PubMed] [Google Scholar]
- 31.Cyster JG. Nat Immunol. 2010 Nov;11:989. doi: 10.1038/ni.1946. [DOI] [PubMed] [Google Scholar]
- 32.Phan TG, Grigorova I, Okada T, Cyster JG. Nat Immunol. 2007 Sep;8:992. doi: 10.1038/ni1494. [DOI] [PubMed] [Google Scholar]
- 33.Carrasco YR, Batista FD. Immunity. 2007 Jul;27:160. doi: 10.1016/j.immuni.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Junt T, et al. Nature. 2007 Nov 1;450:110. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- 35.Phan TG, Green JA, Gray EE, Xu Y, Cyster JG. Nat Immunol. 2009 Jul;10:786. doi: 10.1038/ni.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roozendaal R, et al. Immunity. 2009 Feb 20;30:264. doi: 10.1016/j.immuni.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi H, Egen JG, Huang AY, Germain RN. Science. 2006 Jun 16;312:1672. doi: 10.1126/science.1125703. [DOI] [PubMed] [Google Scholar]
- 38.Garside P, et al. Science. 1998 Jul 3;281:96. [Google Scholar]
- 39.Reif K, et al. Nature. 2002 Mar 7;416:94. doi: 10.1038/416094a. [DOI] [PubMed] [Google Scholar]
- 40.Gunzer M, et al. Blood. 2004 Nov 1;104:2801. doi: 10.1182/blood-2004-03-1193. [DOI] [PubMed] [Google Scholar]
- 41.Okada T, et al. PLoS Biol. 2005 Jun;3:1047. [Google Scholar]
- 42.Allen CD, Okada T, Tang HL, Cyster JG. Science. 2007 Jan 26;315:528. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 43.Schwickert TA, et al. Nature. 2007 Mar 1;446:83. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 44.Hauser AE, et al. Immunity. 2007 May;26:655. doi: 10.1016/j.immuni.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Victora GD, et al. Cell. 2010 Nov 12;143:592. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Negulescu PA, Krasieva TB, Khan A, Kerschbaum HH, Cahalan MD. Immunity. 1996 May;4:421. doi: 10.1016/s1074-7613(00)80409-4. [DOI] [PubMed] [Google Scholar]
- 47.Donnadieu E, Bismuth G, Trautmann A. Curr Biol. 1994 Jul 1;4:584. doi: 10.1016/s0960-9822(00)00130-5. [DOI] [PubMed] [Google Scholar]
- 48.Cannons JL, Tangye SG, Schwartzberg PL. Annu Rev Immunol. 2011 Apr 23;29:665. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 49.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. Nature. 2008 Oct 9;455:764. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider H, et al. Science. 2006 Sep 29;313:1972. [Google Scholar]
- 51.Fife BT, et al. Nat Immunol. 2009 Nov;10:1185. doi: 10.1038/ni.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang Q, et al. Nat Immunol. 2006 Jan;7:83. doi: 10.1038/ni1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mempel TR, et al. Immunity. 2006 Jul;25:129. doi: 10.1016/j.immuni.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 54.Henrickson SE, et al. Nat Immunol. 2008 Mar;9:282. doi: 10.1038/ni1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Witt CM, Raychaudhuri S, Schaefer B, Chakraborty AK, Robey EA. PLoS Biol. 2005 Jun;3:e160. doi: 10.1371/journal.pbio.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhakta NR, Oh DY, Lewis RS. Nat Immunol. 2005 Feb;6:143. doi: 10.1038/ni1161. [DOI] [PubMed] [Google Scholar]
- 57.Ehrlich LI, Oh DY, Weissman IL, Lewis RS. Immunity. 2009 Dec 18;31:986. doi: 10.1016/j.immuni.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le Borgne M, et al. Nat Immunol. 2009 Aug;10:823. doi: 10.1038/ni.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hugues S, et al. Nat Immunol. 2004 Dec;5:1235. doi: 10.1038/ni1134. [DOI] [PubMed] [Google Scholar]
- 60.Katzman SD, et al. Proc Natl Acad Sci U S A. 2010 Oct 19;107:18085. doi: 10.1073/pnas.1010560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shakhar G, et al. Nat Immunol. 2005 Jul;6:707. doi: 10.1038/ni1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coombes JL, Robey EA. Nat Rev Immunol. 2010 May;10:353. doi: 10.1038/nri2746. [DOI] [PubMed] [Google Scholar]
- 63.Egen JG, et al. Immunity. 2008 Feb;28:271. doi: 10.1016/j.immuni.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Egen JG, et al. Immunity. 2011 May 27;34:807. doi: 10.1016/j.immuni.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chieppa M, Rescigno M, Huang AY, Germain RN. J Exp Med. 2006 Dec 25;203:2841. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schaeffer M, et al. J Immunol. 2009 May 15;182:6379. doi: 10.4049/jimmunol.0804307. [DOI] [PubMed] [Google Scholar]
- 67.John B, et al. PLoS Pathog. 2009 Jul;5:e1000505. doi: 10.1371/journal.ppat.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hickman HD, et al. Nat Immunol. 2008 Feb;9:155. doi: 10.1038/ni1557. [DOI] [PubMed] [Google Scholar]
- 69.Filipe-Santos O, et al. Cell Host Microbe. 2009 Jul 23;6:23. doi: 10.1016/j.chom.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 70.Iannacone M, et al. Nature. 2010 Jun 24;465:1079. doi: 10.1038/nature09118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chtanova T, et al. Immunity. 2009 Aug 21;31:342. doi: 10.1016/j.immuni.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim JV, Kang SS, Dustin ML, McGavern DB. Nature. 2009 Jan 8;457:191. doi: 10.1038/nature07591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Azar GA, Lemaitre F, Robey EA, Bousso P. Proc Natl Acad Sci U S A. 2010 Feb 23;107:3675. doi: 10.1073/pnas.0905901107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei SH, et al. Nat Immunol. 2005 Dec;6:1228. doi: 10.1038/ni1269. [DOI] [PubMed] [Google Scholar]
- 75.Sanna MG, et al. Nat Chem Biol. 2006 Aug;2:434. doi: 10.1038/nchembio804. [DOI] [PubMed] [Google Scholar]
- 76.Grigorova IL, et al. Nat Immunol. 2009 Jan;10:58. doi: 10.1038/ni.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cahalan SM, et al. Nat Chem Biol. 2011 May;7:254. doi: 10.1038/nchembio.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matheu MP, et al. Immunity. 2008 Oct 17;29:602. doi: 10.1016/j.immuni.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Nat Methods. 2011 Sep;8:757. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- 80.Palmer AE, Tsien RY. Nat Protoc. 2006;1:1057. doi: 10.1038/nprot.2006.172. [DOI] [PubMed] [Google Scholar]
- 81.Melichar HJ, et al. Immunol Cell Biol. 2011 May;89:549. doi: 10.1038/icb.2010.122. [DOI] [PubMed] [Google Scholar]
- 82.Friedman RS, Beemiller P, Sorensen CM, Jacobelli J, Krummel MF. J Exp Med. 2010 Nov 22;207:2733. doi: 10.1084/jem.20091201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khairy K, Keller PJ. Genesis. 2011 Jul;49:488. doi: 10.1002/dvg.20698. [DOI] [PubMed] [Google Scholar]
- 84.Klauschen F, et al. Nat Protoc. 2009;4:1305. doi: 10.1038/nprot.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Geissmann F, et al. PLoS Biol. 2005 Apr;3:e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Germain RN, Miller MJ, Dustin ML, Nussenzweig MC. Nat Rev Immunol. 2006 Jul;6:497. doi: 10.1038/nri1884. [DOI] [PubMed] [Google Scholar]
- 87.Cahalan MD, Parker I. Annu Rev Immunol. 2008;26:585. doi: 10.1146/annurev.immunol.24.021605.090620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aoshi T, et al. Immunity. 2008 Sep 19;29:476. doi: 10.1016/j.immuni.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 89.Bajenoff M, Glaichenhaus N, Germain RN. J Immunol. 2008 Sep 15;181:3947. doi: 10.4049/jimmunol.181.6.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kreisel D, et al. Proc Natl Acad Sci U S A. 2010 Oct 19;107:18073. doi: 10.1073/pnas.1008737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Looney MR, et al. Nat Methods. 2011 Jan;8:91. doi: 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cavanagh LL, et al. Nat Immunol. 2005 Oct;6:1029. doi: 10.1038/ni1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ishii M, et al. Nature. 2009 Mar 26;458:524. doi: 10.1038/nature07713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kohler A, et al. Blood. 2011 Apr 21;117:4349. doi: 10.1182/blood-2010-09-308387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Coppieters K, Martinic MM, Kiosses WB, Amirian N, von Herrath M. PLoS One. 2010;5:e15732. doi: 10.1371/journal.pone.0015732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Toiyama Y, et al. J Gastroenterol. 2010 May;45:544. doi: 10.1007/s00535-009-0187-7. [DOI] [PubMed] [Google Scholar]