Abstract
The Atg4 cysteine proteases play crucial roles in the processing of Atg8 proteins during autophagy, but their regulation during cellular stress and differentiation remains poorly understood. We have found that two Atg4 family members—Atg4C and Atg4D—contain cryptic mitochondrial targeting sequences immediately downstream of their canonical (DEVD) caspase cleavage sites. Consequently, caspase-cleaved Atg4D (ΔN63 Atg4D) localizes to the mitochondrial matrix when expressed in mammalian cells, where it undergoes further processing to a ~42 kDa mitochondrial form. Interestingly, caspase cleavage is not needed for Atg4D mitochondrial import, because ~42 kDa mitochondrial Atg4D is observed in cells treated with caspase inhibitors and in cells expressing caspase-resistant Atg4D (DEVA63). Using HeLa cell lines stably expressing ΔN63 Atg4D, we showed that mitochondrial Atg4D sensitizes cells to cell death in the presence of the mitochondrial uncoupler, CCCP, and that mitochondrial cristae are less extensive in these cells. We further showed that the organization of mitochondrial cristae is altered during the mitochondrial clearance phase in differentiating primary human erythroblasts stably expressing ΔN63 Atg4D, and that these cells have elevated levels of mitochondrial reactive oxygen species (ROS) during late stages of erythropoiesis. Together these data suggest that the import of Atg4D during cellular stress and differentiation may play important roles in the regulation of mitochondrial physiology, ROS, mitophagy and cell viability.
Keywords: Atg4C, Atg4D, caspase, erythropoiesis, mitochondria, mitophagy
Introduction
Autophagosome biogenesis requires the dual actions of the Atg4 class of endopeptidases to first cleave newly synthesized Atg8 at conserved C-terminal glycine residues (a process known as priming), and then later to delipidate autophagosome-bound Atg8 (through hydrolysis of the glycine-phosphatidyl ethanolamine (PE) bond that is formed during autophagosome biogenesis).1,2 Studies in yeast, which contain only one copy each of Atg4 and Atg8, have demonstrated the indispensible role of Atg8 priming during autophagy,1 and have also highlighted the importance of the delipidation step during late stages of autophagosome maturation when material is delivered to the vacuole for degradation.3 The situation in mammalian cells is more complex, because these contain four Atg4 family members (Atg4A, B, C, D; also known as “autophagins”4) and seven Atg8 family members (LC3A, B, C and 4 GABARAPs; e.g., ref. 5). Understanding the individual functions of these family members has proved challenging, although a picture is now emerging of cooperative/sequential actions for LC3 and GABARAP family members during early stages of autophagosome assembly,6 in turn regulated by Atg4 proteins with broad Atg8 specificity (e.g., Atg4B7), or with a preference for individual Atg8 proteins (Atg4A for GATE-168; Atg4D for GABARAP-L19). It is conceivable, then, that assembly of autophagosomes in response to disparate autophagic stimuli in mammalian cells will involve tight spatiotemporal regulation of the actions of specific Atg4 family members, and corresponding fine-tuning of Atg8 recruitment to the nascent autophagosome. By this model, different autophagosomal properties including their size,5,6 their preferred cargoes (e.g.10), and perhaps also their directed transport to lysosomes could be programmed by the balance of Atg8 classes, affording cells with considerable functional flexibility in their autophagosomal populations.
Given their important roles as regulators of autophagosome biogenesis, and the need to carefully control their activities locally within the cytoplasm, it can be assumed that Atg4 family members will be subject to post-translational control. We know, for example, that Atg4A and Atg4B contain oxidation-prone cysteine residues in their active sites which render these proteins sensitive to reactive oxygen species (ROS).11 Indeed, it has been proposed that elevated ROS is needed to facilitate the productive assembly of autophagosomes by suppressing Atg8 delipidation at the autophagosome assembly site (phagophore).11,12 A requirement for the local modulation of Atg4 delipidation activity to allow sustained Atg8 incorporation into the nascent autophagosome is supported by data from a number of groups showing that overexpression of Atg4B inhibits autophagosome formation throughout the cytoplasm (e.g., see refs. 7 and 9). Hence, the tight spatial control of Atg4 action is an essential facet of the autophagosome biogenesis pathway: cells need Atg4 for Atg8 priming, but too much Atg4 activity blocks autophagosome formation at the expansion stage via unregulated Atg8 delipidation. Once assembled, autophagosomes retain Atg8 on their outer surface throughout their maturation.13 In yeast, removal of Atg8 via delipidation may be coupled with autophagosome/vacuole fusion, because in cells in which delipidation is prevented (ΔAtg4 cells) delivery of cargo to the vacuole is dramatically delayed.3 Although the comparable scenario has not been tested in mammalian cells, these data nevertheless point once more to a requirement for tight spatial control of Atg4 activities during autophagosome assembly, maturation and fusion with degradative compartments.
As further evidence for post-translational control of Atg4 function, we have demonstrated that Atg4D is subject to caspase cleavage at DEVD63 during apoptosis, and that cleavage stimulates Atg4D priming and delipidation activities toward GABARAP-L1 in vitro.9 Exactly how caspase cleavage of Atg4D contributes to cellular stress responses remains unclear. Importantly, it has recently been documented that several other human autophagy proteins—including all Atg4 family members—can be subject to caspase and/or calpain cleavage:14 Atg4A is cleaved in vitro at its C-terminus by recombinant caspase-3, and centrally by calpain 1, while Atg4B is cleaved at its C-terminus by calpain 1 (the partial cleavage at the N-terminus of Atg4B reported in the presence of recombinant caspase-3 was most likely due to cleavage of the epitope tag14). Alignment of the N-terminal sequences of Atg4C and Atg4D reveals the presence of canonical DEVD caspase sites in both, and we predicted that Atg4C would undergo a similar cleavage event at its corresponding site, DEVD.9,10 The recent data from Norman et al. are consistent with this prediction, and support a model in which both Atg4D and Atg4C undergo cleavage at identical N-terminal DEVD sites during apoptosis.14
Accepting that various members of the mammalian Atg4 family are subject to regulation by apoptotic proteases,9,14 what might be the consequence of Atg4 cleavage in relation to endopeptidase action, autophagy and ultimately, cell fate? Based on published crystal structures of Atg4B in free form and in complex with LC3, the N-termini of Atg4 family members are predicted to partially occlude access/exit of Atg8 substrates at the active site.15 Notably, recombinant Atg4B lacking its N-terminus was demonstrated to have enhanced priming activity against LC3,15 leading us to speculate that caspase cleavage of Atg4D stimulates GABARAPL1 priming through removal of the auto-inhibitory N-terminal domain of Atg4D.16 Whether Atg4C is subject to the same regulatory influences is less likely because its cleavage is predicted to only remove 10 amino acids from the N-terminus,9 possibly indicating different roles/consequences of caspase cleavage of these related Atg4 family members. Significantly, the caspase-cleaved form of Atg4D (ΔN63 Atg4D) was found to be highly toxic when overexpressed in HeLa cells; with cell death associated with the enrichment of Atg4D at the mitochondria and at least partly mediated by a BH3-type domain at the C-terminus of this protein.9
Here, we provide data suggesting that caspase action may control the subcellular distribution of Atg4C and Atg4D. Immediately downstream of the caspase cleavage sites in these molecules are stretches of amino acids that constitute strong mitochondrial-targeting motifs. We show that these sequences are sufficient to target the caspase-cleaved forms of these proteins into mitochondria, and that HeLa cells expressing mitochondrial Atg4D are more susceptible to cell death mediated by prolonged mitochondrial uncoupling. Furthermore, we show that expression of mitochondrial-targeted Atg4D in differentiating human erythroblasts influences mitochondrial structure and density, leading to elevated ROS and advanced mitochondrial clearance (mitophagy) in this physiologically relevant system.
Results
Caspase cleavage of Atg4C and Atg4D reveals latent mitochondrial targeting sequences
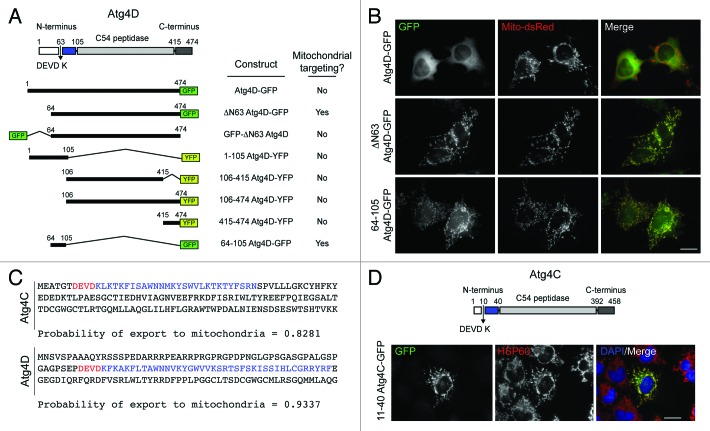
One unexpected observation arising from our previous characterization of Atg4D was the tendency of this enzyme to associate with mitochondria during oxidative stress and cell death.9 To determine the domain of Atg4D responsible for its dynamic mitochondrial targeting, we made truncation/deletion constructs with N- or C-terminal GFP or YFP fusions (Fig. 1A) and transiently expressed these in HeLa cells (Fig. 1B; Fig. S1). Most of these constructs gave similar staining patterns in viable cells, with strong cytosolic distribution super-imposed upon weaker reticular/ER patterns (Fig. 1B; Fig. S1). The exception was caspase truncated ΔN63 Atg4D-GFP which was exclusively mitochondrial (Fig. 1B; Fig. S1). Atg4D contains a proposed BH3 domain within its C-terminus9 that has the potential to engage Bcl-2 family members on the mitochondria or at the ER. A construct consisting of the C-terminus of Atg4D had the same subcellular distribution as full-length Atg4D, suggesting that the BH3 domain does not influence subcellular targeting of Atg4D (Fig. S1). Notably, placing the GFP tag at the new N-terminus of ΔN63 Atg4D (i.e., GFP-ΔN63 Atg4D) blocked its mitochondrial association, suggesting that determinants immediately downstream of the caspase site might be responsible for targeting truncated Atg4D to the mitochondria (Fig. S1). Consistent with this observation, subcellular targeting algorithms (e.g., Mitoprot, http://ihg.gsf.de/ihg/mitoprot.html) predicted that the sequence of the caspase-cleaved form of Atg4D (ΔN63 Atg4D) would be targeted to mitochondria (Fig. 1C). Additional processing to remove a putative mitochondrial-targeting domain at amino acid position RYRF104 was also predicted by the software (Fig. 1C), and consistent with this, a construct containing this domain was an excellent mitochondrial-targeting sequence for GFP (64-105 Atg4D-GFP; Fig. 1B; Fig. S1). Significantly, when we repeated this process using the comparable sequences in Atg4C, we observed the same phenomenon: amino acids 11–40 were predicted to form a mitochondrial-targeting domain (Fig. 1C), and a 11–40 Atg4C-GFP construct was strongly targeted to mitochondria in HeLa cells (Fig. 1D).
Figure 1. Human Atg4C and Atg4D contain latent mitochondrial targeting motifs preceded by caspase sites. (A) Domain structure of Atg4D and cartoon of some of the constructs used in this study. Example images of the domain localization for each of these constructs can be found in Figure S1. (B) Example images of HeLa cells co-transfected with GFP-tagged Atg4D domain constructs and Mito-dsRed. The domain spanning from the caspase site to the C54 peptidase domain of Atg4D (64-105) is a strong mitochondrial targeting sequence. (C) Comparison of the N-terminal amino acid sequences of human Atg4C and Atg4D showing the predicted probability of mitochondrial import (Mitoprot: http://ihg.gsf.de/ihg/mitoprot.html). Caspase sites are shown in red; mitochondrial import sequences are shown in blue. (D) Domain structure of human Atg4C and activity of the predicted Atg4C mitochondrial-targeting motif. HeLa cells were transiently transfected with 11–40 Atg4C-GFP, then fixed and stained with anti-HSP60 antibodies. Bars = 20 µm.
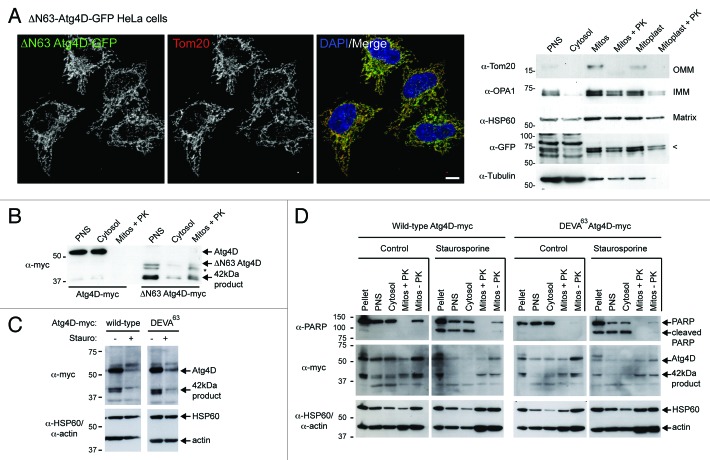
The mitochondrial recruitment of caspase-cleaved Atg4C and Atg4D raises important questions about the roles of these enzymes during autophagy, and about their potential influence upon mitochondrial function during cellular stress. To confirm that ΔN63 Atg4D was imported into mitochondria, and not simply associated with the outer mitochondrial membrane (OMM), we used lentivirus to generate HeLa cells stably expressing ΔN63 Atg4D-GFP (Fig. 2A). As expected, in these cells, ΔN63 Atg4D was exclusively mitochondrial (Fig. 2A). We then used cellular fractionation to isolate mitochondria and mitoplasts (mitochondria lacking their outer membrane), which we then used in protease protection assays in the absence or presence of proteinase K (PK) (Fig. 2A). Blotting for the OMM protein Tom20, the inner membrane (IMM) protein OPA1 (optic atrophy 1), and the matrix resident HSP60 confirmed that the isolation process had been successful (Fig. 2A). Importantly, ΔN63 Atg4D-GFP was retained in the PK-treated mitoplast fraction, suggesting that ΔN63 Atg4D is probably imported into the mitochondrial matrix (Fig. 2A).
Figure 2. Import of ΔN63 Atg4D into the mitochondrial matrix does not depend upon caspase action. (A) Mitochondrial targeting of ΔN63 Atg4D-GFP. To the left, example field of HeLa cells stably expressing ΔN63 Atg4D-GFP and stained with Tom20 antibodies (Bar = 10 µm). To the right, immunoblots of ΔN63 Atg4D-GFP stable HeLa cell extracts fractionated into post nuclear supernatant (PNS), cytosol, mitochondria (mitos) and mitoplasts (PK: proteinase K). Arrow on the anti-GFP blot indicates the position of ΔN63 Atg4D-GFP. (OMM: outer mitochondrial membrane; IMM: inner mitochondrial membrane). (B) Immunoblot of PNS, cytosol, and PK-treated mitochondrial fractions of HeLa cells transiently expressing Atg4D-myc or ΔN63 Atg4D-myc. The ~42 kDa mitochondrial form of Atg4D is indicated (* denotes an intermediate band resolving between ΔN63 Atg4D-myc and the ~42 kDa mitochondrial product). (C) Immunoblots of lysates of HeLa cells transiently expressing wild-type or caspase-resistant (DEVA63) Atg4D-myc, incubated in the absence or presence of staurosporine (6 h). (C) Immunoblots of fractionated HeLa cells expressing Atg4D-myc or ΔN63 Atg4D-myc, incubated in the absence or presence of staurosporine (6 h).
Bona fide mitochondrial proteins are normally parted from their targeting sequences following import by the actions of mitochondrial peptidases.17 To test whether Atg4D conforms to this paradigm, we placed a myc tag at the C-terminus of full-length and caspase-cleaved Atg4D (Atg4D-myc; ΔN63 Atg4D-myc, respectively) and transiently transfected these into HeLa cells (an myc tag was used for this experiment because the gel shift in ΔN63 Atg4D due to protease action within mitochondria was more pronounced). Immunoblotting confirmed that full-length Atg4D did not associate with mitochondria (~55 kDa form) (Fig. 2B). By contrast, the majority of ΔN63 Atg4D-myc resolved at ~42 kDa, with two additional species observed: one at ~47 kDa (the expected size of ΔN63 Atg4D), and another at ~45 kDa, which may be an intermediate product of mitochondrial metalloproteinase action (Fig. 2B). Most significantly, we found that the 42 kDa form of Atg4D was retained in the proteinase K-treated mitochondrial fraction (Fig. 2B). This product resolved at the size predicted for Atg4D lacking both its N-terminal extension and mitochondrial-targeting domain, and is likely to represent the short form of Atg4D that we had previously described.9 Together, these data suggest that following caspase cleavage, Atg4D can be imported into mitochondrial matrix where it is processed to a ~42 kDa product. Clearly, it is unlikely that from this location 42 kDa Atg4D influences autophagy directly through processing of Atg8 family members, and it is therefore possible that Atg4D (and Atg4C) can regulate mitochondrial function via endopeptidase activity against unidentified mitochondrial substrates.
Using live-cell imaging, we previously observed the enrichment of Atg4D at or within mitochondria as a prelude to apoptosis.9 Whether this was due to the binding of Atg4D to the mitochondrial surface or to the uptake of Atg4D into mitochondria was not formally tested; however, because we observed this phenomenon using GFP-Atg4D (i.e., a construct with blocked mitochondrial import sequence), in the absence or presence of caspase inhibitors, the former possibility is much more likely. To examine this further, we expressed wild-type and caspase-resistant (DEVA63) Atg4D-myc in HeLa cells and analyzed the pattern of Atg4D expression in the absence or presence of staurosporine (to induce apoptosis) (Fig. 2C). In crude lysates, wild-type Atg4D-myc resolved as a 55 kDa full-length protein with evidence of a truncated 42kDa product (Fig. 2C). Surprisingly, this pattern was similar in cells expressing caspase-resistant DEVA63 Atg4D-myc, implying that caspase cleavage may not be necessary for import into mitochondria and subsequent processing (Fig. 2C). This is consistent with our previous detection of 42 kDa Atg4D in HeLa cells treated with the proteasome inhibitor, MG132, in the presence of the caspase inhibitor, zVAD.fmk.9 In cells treated with staurosporine, we observed a general decrease in the amount of full-length and 42 kDa Atg4D in both wild-type and DEVA63 expressing cells, which might indicate degradation of Atg4D by apoptotic proteases (Fig. 2C). Notably, we did not observe any differences in the relative amounts of cytosolic or mitochondrial Atg4D between cells expressing wild-type or caspase-resistant Atg4D-myc in the presence of staurosporine (Fig. 2C). To extend these findings, we compared the mitochondrial association of wild-type vs. caspase-resistant Atg4D in cells treated with staurosporine (Fig. 2D). Cell fractionation and PK treatment revealed that a fraction of 42 kDa Atg4D could always be found within mitochondria at steady-state, and that this fraction is not affected markedly either by caspase activation during staurosporine-induced apoptosis, or by the expression of a caspase-resistant form of Atg4D in viable or apoptotic cells (Fig. 2D). The presence of a small but significant fraction of 42 kDa Atg4D within mitochondria at steady-state raises interesting questions about the roles of this protein within cells, so we set out to examine the possible roles of mitochondrial Atg4D in viable and stressed cells.
Atg4D sensitizes cells to mitochondrial uncoupling
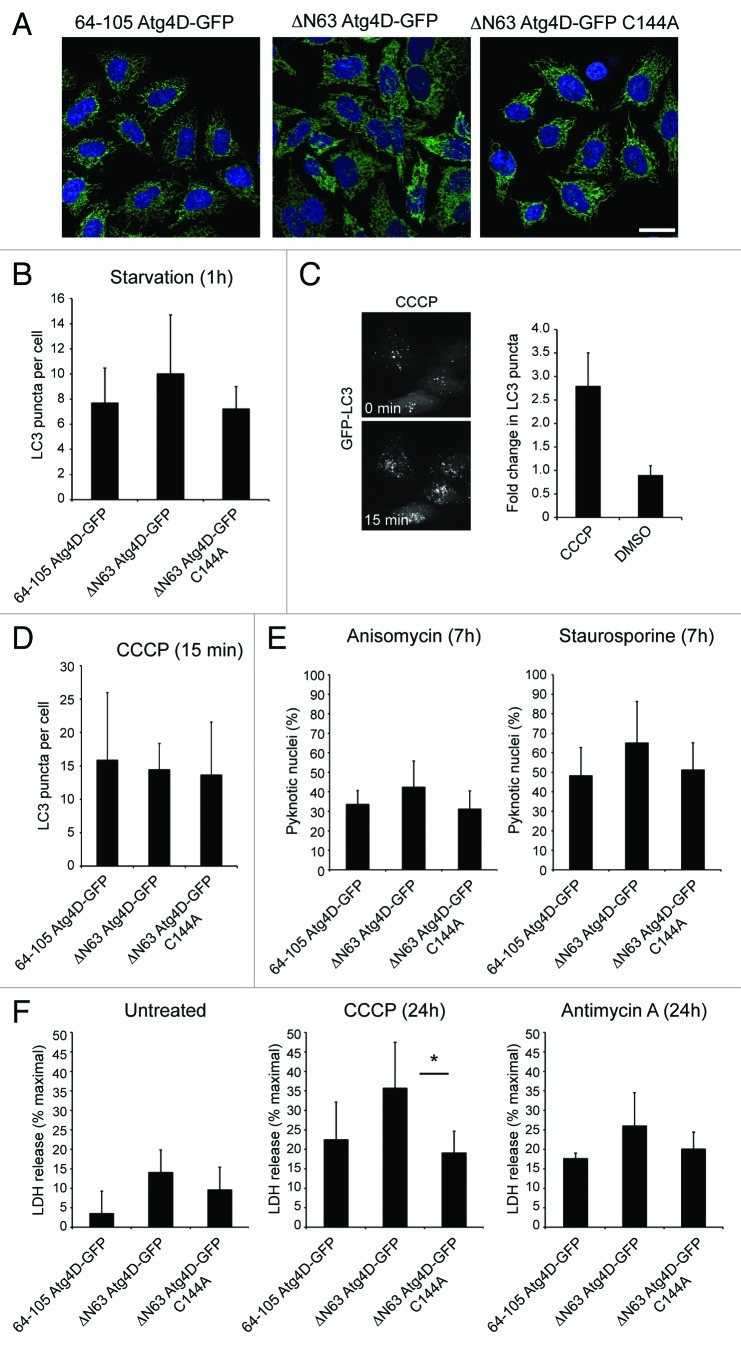
To gain insight into the possible influence of mitochondrial Atg4D upon cell viability, we used lentiviruses to generate HeLa cell-lines stably expressing active site mutant (C144A) ΔN63 Atg4D-GFP,9 or mitochondrial GFP [using the mitochondrial targeting sequence of Atg4D (64-105 Atg4D-GFP)] to compliment the wild-type ΔN63 Atg4D-GFP expressing cells previously introduced (Fig. 2A). As shown in Figure 3A, each of these constructs was expressed exclusively in mitochondria, and we observed no differences in mitochondrial density, distribution or dynamics between these cell lines (data not shown).
Figure 3. Effect of mitochondrial Atg4D on cell viability and autophagy. (A) Fields of HeLa cells stably expressing 64-105 Atg4D-GFP, ΔN63 Atg4D-GFP or ΔN63 Atg4D-GFP (C144A) showing mitochondrial labeling in all cases. DAPI staining is shown in blue (Bar = 20 µm). (B–D) Autophagy response in HeLa cell lines stably expressing wild-type or mutant ΔN63 Atg4D-GFP under amino acid/growth factor starvation (B), or following treatment with the mitochondrial uncoupler, CCCP (C and D). (E) Cell death response of HeLa cell lines stably expressing wild-type or mutant ΔN63 Atg4D-GFP following treatment with staurosporine or anisomycin (7 h). (F) Cell death response of HeLa cell lines stably expressing wild-type or mutant ΔN63 Atg4D-GFP following prolonged treatment with CCCP or antimycin A (24 h). *p < 0.01.
We next used these cell lines to compare the impact of mitochondrial Atg4D expression upon autophagy responses (amino acid starvation; CCCP) and the susceptibility of cells to inducers of stress and apoptosis (Fig. 3B–F). Each of the cell lines responded similarly to amino acid starvation when analyzed for LC3 puncta numbers (Fig. 3B). We also tested the responses of these cells to the induction of autophagy by short (15 min) treatment with the mitochondrial uncoupler, CCCP (Fig. 3C and D). No differences in autophagosome numbers were observed between the cell lines (Fig. 3D), suggesting that the presence of Atg4D within mitochondria has minimal impact upon amino acid starvation and mitochondrial dysfunction-induced autophagy. We next subjected these cell lines to the apoptosis inducers anisomycin and staurosporine (Fig. 3E), or to prolonged exposure to the mitochondrial poisons CCCP and antimycin A (an inhibitor of mitochondrial complex III which increased cellular ROS) (Fig. 3F), and measured cell death by assessment of pyknotic chromatin and LDH release respectively. When treated with either anisomycin or staurosporine, the ΔN63 Atg4D-GFP cells consistently showed a trend toward greater susceptibility to apoptosis (Fig. 3E); however, this was not statistically significant leading us to conclude that mitochondrial-targeted Atg4D does not contribute to apoptosis under these conditions. Under basal conditions and in the presence of antimycin A, there were small but not significant increases in cell death in cells expressing wild-type ΔN63 Atg4D-GFP (Fig. 3F). By contrast, cell death was significantly greater in wild-type ΔN63 Atg4D-GFP cells treated with CCCP (Fig. 3F). This suggests that mitochondrial Atg4D sensitizes cells to cell death in the presence of mitochondrial uncouplers. Interestingly, cytoplasmic ΔN63 Atg4D is potently cytotoxic when overexpressed in HeLa cells,9 suggesting that the proposed import/sequestration of Atg4D within mitochondria may have a protective role. Furthermore, siRNA suppression of Atg4D expression sensitized HeLa cells to starvation and staurosporine,9 suggesting that Atg4D is cytoprotective until caspase cleavage renders it cytotoxic, possibly via exposure or activation of its putative BH3 domain.9
Altered mitochondrial physiology in primary human erythroblasts expressing mitochondrial ΔN63 Atg4D
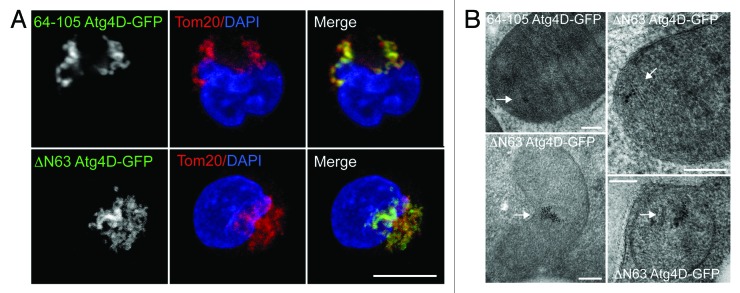
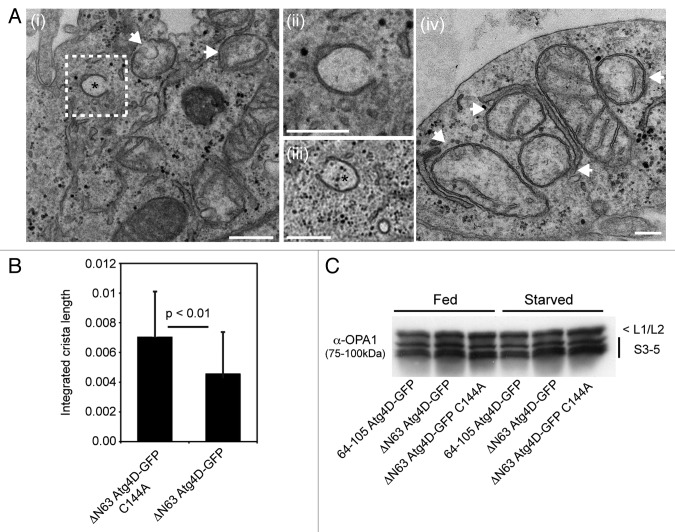
Autophagy is dramatically upregulated during erythropoiesis, when it contributes to the removal of mitochondria and other organelles from the cytoplasm of the nascent erythrocyte.18 Recent studies in mice genetically depleted of key autophagy/mitophagy genes (e.g., Ulk1, Atg7, Nix) suggest that the failure to remove mitochondria at the correct stage of erythroid differentiation can cause elevated oxidative stress, reduced reticulocyte viability and chronic anemia.18-25 Interestingly, sublethal caspase action has been shown to contribute to erythroblast terminal differentiation (e.g., see refs. 26 and 27), prompting us to investigate the potential involvement of caspase-cleaved Atg4D during erythroid mitophagy. We used lentiviruses to generate primary human erythroid precursors stably expressing 64-105 Atg4D-GFP or caspase-cleaved wild-type ΔN63 Atg4D-GFP. To determine whether these constructs influence mitochondrial clearance during late erythropoiesis, we then differentiated these cells along the erythroid lineage in vitro using established culture conditions.28,29 As with the HeLa cell lines that we described above (Fig. 3A), 64-105 Atg4D-GFP and ΔN63-Atg4D-GFP were expressed exclusively in the mitochondria of erythroblasts (Fig. 4A), and the ultrastructural localization of ΔN63-Atg4D-GFP was confirmed as being in the mitochondrial matrix by immuno EM (Fig. 4B).
Figure 4. Expression of ΔN63 Atg4D-GFP in primary human erythroid cells. (A) Confocal images of primary human erythroblasts stably expressing 64-105 Atg4D-GFP or ΔN63 Atg4D-GFP, stained with anti-Tom20 antibodies. Bar = 10 µm. (B) ΔN63 Atg4D-GFP is targeted to the mitochondrial matrix in human erythroid cells. Immuno EM images of mitochondria in human PCE cells stably expressing 64-105 Atg4D-GFP or ΔN63 Atg4D-GFP, stained with anti-GFP antibodies and 6 nm gold secondary antibodies. Arrows indicate clusters of gold particles. Bar = 100 nm.
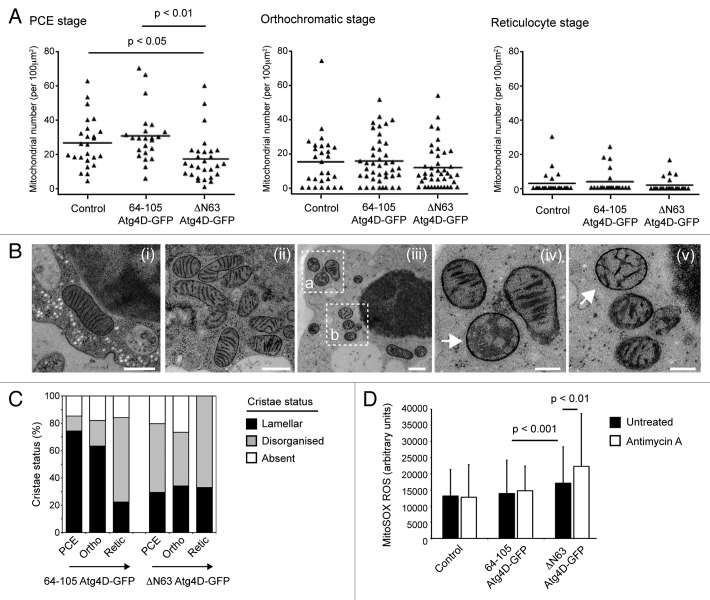
In parallel, extensive characterization of autophagy and organelle clearance in the human in vitro erythroid differentiation system which we are currently preparing for publication, we have found that the bulk of mitochondria are eliminated between the polychromatic erythroblast (PCE) and reticulocyte stages in human erythroid cells differentiated in vitro (Betin VMS, Parsons SF, Anstee DJ, Lane JD, manuscript in preparation). This is temporally associated with elevated expression of Atg4D and several other autophagy genes (Betin VMS, Parsons SF, Anstee DJ, Lane JD, manuscript in preparation). To determine whether mitochondrial Atg4D influences the rate and timing of mitophagy during human erythropoiesis, we used EM to count mitochondrial profiles at late stages of erythroid maturation using cells differentiated from the stably transformed erythroid progenitor cells (Fig. 5A). We found that mitochondrial numbers were already significantly reduced in PCE cells expressing ΔN63 Atg4D-GFP (PCE control vs. PCE ΔN63 Atg4D-GFP: p < 0.05; PCE 64-105-GFP vs. PCE ΔN63 Atg4D-GFP: p < 0.01) (Fig. 5A), suggesting that mitophagy may have been initiated sooner in these cells. Thereafter the rates of mitochondrial clearance were very similar between erythroid cells expressing ΔN63 Atg4D-GFP, those expressing 64-105 Atg4D-GFP, and untransfected (control) erythroblasts (Fig. 5A). Importantly, no significant differences in the rates of differentiation were observed between control, 64-105 Atg4D-GFP and ΔN63 Atg4D-GFP expressing erythroid cells (Fig. S2), although there was a notable increase in cell death in the ΔN63 Atg4D-GFP expressing populations (data not shown). These observations suggest that the expression of ΔN63 Atg4D-GFP in differentiating human erythroblasts causes a reduction in mitochondrial content at the PCE stage, without influencing the rate of erythroid differentiation in vitro.
Figure 5. Influence of mitochondrial ΔN63 Atg4D-GFP on mitochondrial structure and mitophagy in differentiating human erythroid cells. (A) Mitophagy kinetics in human erythroid cells differentiated in vitro. At the PCE stage, there are significantly fewer mitochondria in erythroblasts expressing ΔN63 Atg4D-GFP; however, the numbers of mitochondria at the orthochromatic-reticulocyte stages are comparable with controls. (B and C) Mitochondrial morphology in differentiating erythroid cells. (B) EM images of erythroid cells showing examples of mitochondria with damaged or missing cristae (arrows). (iv) and (v) are zoomed images of (iii), area (a and b) respectively. Bars, (i–iii) = 500 nm; (iv and v) = 200 nm. (C) Quantitation of mitochondrial morphology in differentiating erythroid cells stably expressing 64-105 Atg4D-GFP or ΔN63 Atg4D-GFP. (C) Measurements of mitochondrial ROS (mitoSOX fluorescence) in human erythroid cells (population consists of: 30% reticulocytes; 70% orthochromatic cells) stably expressing 64-105 Atg4D-GFP or ΔN63 Atg4D-GFP.
Analysis of the status of mitochondria in erythroblasts expressing mitochondrial GFP (64-105 Atg4D-GFP) suggested a trend toward less organized cristae concomitant with the transition to reticulocyte stage (Fig. 5B and C). Mitochondria were often observed having reduced numbers of misshapen/disorganized cristae, or lacking discernable cristae (Fig. 5B and C), at the time when mitochondria were being actively removed by mitophagy (Fig. 5A). Altered mitochondrial profiles have previously been described in cultured rat reticulocytes30 and in mouse fibroblasts deficient in prohibitin-mediated OPA1 processing.31 Importantly, the density and organization of mitochondrial cristae is directly related to the dimeric status of ATP synthase, and thus to the respiratory efficiency of the organelle.32,33 Meanwhile, disrupted mitochondrial ultrastructure has been linked with reduced mitochondrial inner membrane fusion and subsequent cell death signaling in pathology.34,35 Aberrant cristae ultrastructure can therefore be viewed as a common feature of dysfunctional mitochondria, and may correlate with those mitochondria destined for autophagic removal in the erythroid lineage. Interestingly, in ΔN63 Atg4D-GFP expressing erythroblasts, changes to crista structure were advanced, with a greater proportion of mitochondria having disorganized cristae at the PCE and orthochromatic stages (Fig. 5C). Significantly, this is the stage during which we had recorded reduced mitochondrial numbers in ΔN63 Atg4D-GFP expressing erythroblasts (Fig. 5A), suggesting that expression of ΔN63 Atg4D-GFP alters mitochondrial cristae organization and may be linked to advanced differentiation triggered mitophagy in this system.
One possible consequence of the presence of increased numbers of abnormal mitochondria in late erythroid cells expressing ΔN63 Atg4D-GFP is an increase in oxidative stress. Reactive oxygen species are generated in mitochondria as a normal consequence of oxidative phosphorylation, but their production is greatly enhanced in mitochondria that are inefficient at ATP generation and therefore hold a high protonmotive force (Δp).36 Mitochondria with reduced densities of cristae are predicted to have a less efficient ATP synthase population,33 and may consequently generate more ROS.36 To test whether this is a feature of erythroid cells stably expressing ΔN63 Atg4D-GFP, we used the mitochondrial ROS reporter, MitoSOX. Late stage erythroblasts (day 8 of differentiation: typically 30% reticulocyte, 70% orthochromatic for each culture condition; Fig. S2) were stained with MitoSOX in the absence or presence of antimycin A. At least 400 erythroblasts from two separate cultures (eight individual platelet donors) were imaged using the same exposure settings, with ROS assessed by measuring fluorescence levels in individual cells (Fig. 5D shows data from one culture). Basal MitoSOX fluorescence was significantly increased in cells expressing ΔN63 Atg4D-GFP (Fig. 5D), perhaps linked with the disruption of cristae architecture in these cells. Meanwhile, although there was very little change in MitoSOX fluorescence when control or 64-105 Atg4D-GFP expressing erythrocytes were treated with antimycin A, ΔN63 Atg4D-GFP erythroblasts showed a significant increase in ROS upon antimycin A treatment (Fig. 5D). It is unclear why the addition of antimycin A failed to increase ROS levels in control and 64-105 Atg4D-GFP expressing erythroblasts, but this perhaps reflects the differing physiological status of these cells. Overall, these data suggest that overexpression of ΔN63 Atg4D-GFP within the mitochondria of differentiating human erythroid cells influences mitochondrial structure and physiology, rendering cells more prone to oxidative stress.
HeLa cells expressing ΔN63 Atg4D-GFP display evidence of reduced mitochondrial crista density
The observation of increased numbers of mitochondria with disordered cristae in differentiating human erythroid cells expressing ΔN63 Atg4D-GFP (Fig. 5C), prompted us to assess the status of mitochondria in HeLa cells stably expressing wild-type or C144A ΔN63 Atg4D-GFP at the ultrastructural level. We found that wild-type ΔN63 Atg4D-GFP expressing HeLa cells appeared morphologically normal (when compared with cells expressing 64-105 Atg4D-GFP or C144A ΔN63 Atg4D-GFP; data not shown): membranes of rough ER, Golgi and autophagosomal origin were observed (including autophagic phagophores), and numerous mitochondria could be clearly identified (Fig. 6A). The majority of mitochondria in wild-type ΔN63 Atg4D-GFP expressing cells contained extensive cristae; however, evidence of reduced density of cristae was apparent in many (Fig. 6A). We therefore measured the total length of cristae in each mitochondrion relative to mitochondrial area, and observed a significant reduction in mitochondrial crista density in wild-type ΔN63 Atg4D-GFP expressing cells when compared with cells expressing C144A ΔN63 Atg4D-GFP (Fig. 6B). These data suggest that in both primary human erythroid cells and cultured HeLa cells, ΔN63 Atg4D-GFP expression disrupts mitochondrial crista organization (dependent upon Atg4D endopeptidase activity), altering mitochondrial function and rendering cells more susceptible to oxidative stress. One possible reason for changes in the organization/density of mitochondrial cristae is altered process/activity of the inner membrane fusion factor, OPA1.31 We checked whether OPA1 processing was deficient in ΔN63 Atg4D-GFP expressing cells by immunoblotting cell lysates with anti-OPA1 antibodies; however, no differences in OPA1 band profiles were observed between these cell lines (Fig. 5C).
Figure 6. Mitochondrial crista density is reduced in HeLa cells stably expressing ΔN63 Atg4D-GFP. (A) Electron micrographs of areas of HeLa cells expressing wild-type ΔN63 Atg4D-GFP, showing mitochondria and autophagosomal phagophores (*) (i–iii). Examples of mitochondria with reduced crista density are highlighted by arrows in micrographs (i and iv). The image shown in micrograph (ii) is a zoom of the area highlighted in micrograph (i). Bars, (i–iii) = 500 nm; (iv) 200 nm. (B) Mitochondria in HeLa cells expressing wild-type ΔN63 Atg4D-GFP have reduced crista length when compared with those stably expressing C144A ΔN63 Atg4D-GFP. (C) HeLa cell lysates immunoblotted with an antibody against OPA1, showing no differences in OPA1 processing following expression of ΔN63 Atg4D-GFP.
Discussion
The Atg4 endopeptidase family plays essential, but partially redundant roles in mammalian autophagy.4,37-39 Why the diversification in Atg4 family members—and indeed, also the diversification in their recognized autophagic substrates, the Atg8 proteins—has evolved remains unclear. In addition to functional redundancy, one plausible explanation for multiple Atg4 family members is that some or all have gained additional functions within cells that may or may not be related to autophagy.39 This idea is supported by the data that we have reported here, because our results show that Atg4C and Atg4D both contain strong, cryptic mitochondrial import sequences that can change the localization of these proteins from cytosol to mitochondria under certain conditions. Interestingly, each of these Atg4 family members is a caspase substrate,9,14 and we have mapped the mitochondrial import sequences to stretches of amino acids immediately downstream of the caspase sites in these molecules. This suggests that caspase cleavage may be one mechanism to trigger mitochondrial import of Atg4C and Atg4D (through exposure of import sequences) therefore implicating proteolysis in the regulated import of these Atg4 family members into mitochondria during cellular stress. Importantly, though, our data also show that caspase action is not necessary for mitochondrial Atg4D import, because a subfraction of Atg4D is present within mitochondria at steady-state, and that mitochondrial Atg4D is detected in cells expressing uncleavable Atg4D or those treated with caspase inhibitors. The key questions are: What are the roles of Atg4D and Atg4C within mitochondria, and should these Atg4 family members continue to be considered as true regulators of autophagy? Although we do not attempt to address the latter question in this article, we have made some progress toward understanding the potential mitochondrial roles of Atg4D in cultured cells and primary, differentiating human erythroblasts.
Using cell fractionation and immunoblotting, we have found that in common with other nuclear encoded mitochondrial proteins, the import sequence of Atg4D is removed upon import into mitochondria. The resulting ~42 kDa product—which we had reported in our earlier characterization of Atg4D9—resides in the mitochondrial matrix (as determined by biochemical fractionation and immuno EM). Interestingly, we also showed previously that the amounts of both full-length Atg4D and its ~42 kDa mitochondrial species increase on addition of proteasome inhibitors.9 Atg4D contains a strong PEST sequence within its caspase cleavable N-terminus,9 suggesting that Atg4D is subject to tight proteasomal regulation. Whether removal of this domain by caspases also effectively stabilizes the truncated product remains to be tested.
We have taken parallel approaches to test the potential roles of Atg4D in mitochondria. We generated HeLa cell lines stably expressing wild-type or inactive (C144A) mitochondrial Atg4D which we have used to investigate cell viability and autophagy. We also derived primary human CD34+ erythroid precursors stably expressing wild-type mitochondrial Atg4D to explore the potential role of Atg4D during physiological mitophagy. Although HeLa cells expressing mitochondrial Atg4D had apparently normal mitochondrial contents and organization/dynamics, when treated with the mitochondrial uncoupler CCCP, those expressing wild-type ΔN63 Atg4D were more prone to cell death. This provided some clues as to the possible influence of mitochondrial Atg4D upon mitochondrial function, although expression of ΔN63 Atg4D did not enhance the susceptibility of HeLa cells to antimycin A, or to the apoptotic inducers anisomycin or staurosporine. Like many transformed cells grown in glucose-rich media, HeLa cells are highly glycolytic, meaning that they rely less on oxidative phosphorylation for generation of ATP. Upon addition of CCCP, the mitochondrial inner membrane becomes highly permeable to protons, effectively disconnecting the electron transport chain from oxidative phosphorylation. Under these conditions, the mitochondrial electron transport chain continues to operate, but heat is generated instead of ATP. Why the presence of mitochondrial ΔN63 Atg4D influences cell viability in cells treated with CCCP but not with antimycin A (an inhibitor of electron transport and potent ROS inducer) remains unclear. Interestingly, evidence suggests that the activity of several mitochondrial proteases is controlled by mitochondrial bioenergetics. For example, the proteolytic processing and therefore the activities of OPA1 and Pink1 depend on mitochondrial membrane potential,40-42 demonstrating how mitochondrial quality pathways are coupled to ATP generation, apoptosis and mitophagy. It is possible, then, that altered mitochondria bioenergetics might influence the activity of mitochondrial Atg4D; however, until any mitochondrial targets for Atg4D are identified, this aspect of Atg4D biology cannot readily be determined. Of note, reduced mitochondrial crista density was observed in HeLa cells expressing wild-type ΔN63 Atg4D-GFP; although this phenomenon was not associated with altered OPA1 processing.
Erythropoiesis is characterized by a dramatic upregulation of autophagy that facilitates the clearance of mitochondria and all other organelles from the cytoplasm of the nascent erythrocyte. We have exploited a human ex vivo erythroid differentiation protocol to test whether expression of mitochondrial Atg4D influences erythroid mitochondrial quality control and cellular differentiation. We were motivated by the suggestion that limited, sublethal caspase action is needed for erythroid differentiation,26 and by the dramatic upregulation of Atg4D expression observed during erythropoiesis (Betin VMS, Parsons SF, Anstee DJ, Lane JD, manuscript in preparation). Although we have not been able to formally demonstrate the presence of Atg4D within erythroid mitochondria, due to the lack of a suitable antibody for the detection of processed Atg4D,9 we set out to determine the potential roles of mitochondrial Atg4D during erythropoiesis by lentiviral expression of ΔN63 Atg4D in primary human erythroblasts. We found that erythroblasts stably expressing ΔN63 Atg4D differentiated normally, but had substantially decreased mitochondrial populations at the PCE stage. Notably, in common with the data obtained from HeLa cells, the mitochondria within these cells showed evidence of crista disorganization, supporting a role for ΔN63 Atg4D in mitochondrial organization and/or bioenergetics in physiological cell differentiation systems.
During erythropoiesis, mitochondria are targeted for mitophagy by the actions of Nix24—an adaptor for Atg8 family members.22 Intriguingly, mitochondria that persist in Nix−/− mice that are deficient in erythroid mitophagy retain high membrane potential,19,25 meanwhile the requirement for Nix during erythroid mitophagy can be bypassed by the addition of CCCP.25 This suggests that Nix might initially be required for mitochondrial depolarization during erythroid terminal differentiation. Indeed, a recent study provided evidence that Nix is needed for mitochondrial depolarization following acute CCCP treatment, and that CCCP-stimulated autophagy required ROS.43 It is therefore interesting that in human erythroid cells stably expressing ΔN63 Atg4D, ROS levels were significantly increased, perhaps linked to the increase in the proportion of mitochondria with disorganized cristae and the possible advancement of mitochondrial clearance in these cells.
Our finding that Atg4C and Atg4D can be imported into mitochondria raises questions concerning the roles of these proteins as regulators of autophagy. Data show that full-length Atg4C and Atg4D have very weak Atg8 priming activities in vitro against multiple Atg8 substrates,9,38 suggesting that they may not be functional autophagic regulators. The two available mouse Atg4 knockout models have helped us understand the roles of these family members, as well as providing some clues as to the redundancy that exists within the Atg4 family. The Atg4B knockout mouse is viable, suggesting redundancy, but has dramatically reduced autophagic response in vivo and in vitro.44 Its primary phenotype is in the development of otoconia and balance pathways,44 implying that the dampening of autophagy can have profound effects upon certain differentiation pathways. The interpretation of the phenotype of the Atg4C knockout mouse is less clear, particularly in the light of our findings that this protein has the potential to be imported into mitochondria. Atg4C−/− mice have mild autophagy phenotypes, largely restricted to the diaphragm, and are more susceptible to fibrosarcoma.37 Perhaps significantly, Atg4C−/− mice display reduced locomotor activity only under starvation conditions when compared with their wild-type littermates.37 A general decline in animal physiology due to reduced autophagy in the diagram is the suggested explanation;37 however, it is tempting to speculate that altered mitochondria and disturbed energy homeostasis due to the lack of Atg4C in stressed mice might also contribute. Clearly, more research into the relative functions of Atg4 family members during autophagy and cell stress is needed.
Materials and Methods
Antibodies and reagents
Unless stated otherwise, all reagents were from Sigma. Stock solutions of CCCP (carbonyl cyanide m-chloro phenyl hydrazone; C2759; 10 mM), antimycin A (A8674; 1 mg/ml in ethanol), anisomycin (A9789; 5 mg/ml), staurosporine (S4400; 1 mM), DAPI (4′, 6-diamidino-2-phenylindole; D9542; 1 mg/ml), proteinase K (P6556; 10 mg/ml), puromycin (P7255; 10 mg/ml) were stored at -20°C. The following primary antibodies were used: anti-myc (9E10; M4439); anti-HSP60 (H4149); anti-actin (Santa Cruz Biotechnology, sc-1616); anti-PARP (Calbiochem, AM30); anti-GFP for immunoblotting (Covance, MMS-118R); anti-GFP for immunoEM (Rockland, 600-401-215); anti-Tom20 (BD Biosciences, 612278); anti-OPA1 (BD Biosciences, 612607); anti-tubulin (Sigma, T5168). Secondary antibodies for immunoblotting (HRP-tagged) were from Jackson Immunochemicals (mouse, 715-035-150; rabbit, 711-035-152, goat: 705-035-147); for immunofluorescence were from Molecular Probes (anti-mouse Alexa 594, A-11032); for immunoEM were from Aurion (6 nm gold; 806.011).
HeLa cell culture and transient transfection
HeLa cells were maintained in DMEM supplemented with 10% fetal bovine serum, at 37°C and 5% CO2. Cells were transfected using Genejuice (Novagen, 70967) according to the manufacturer’s instructions.
Lentiviral cloning
Domains of Atg4D were PCR amplified and inserted in frame into pEGFP or pEYFP plasmids (Clontech). Full-length and caspase-truncated Atg4D were inserted into pcDNA3.1 myc/his (Invitrogen, V800-20). Lentiviruses were generated by digestion of the relevant pEGFP (-C1) constructs (wild-type and C144A ΔN63 Atg4D-GFP; 64-105 Atg4D-GFP) using the restriction enzymes Afe1 and BamH1, followed by sub-cloning into pLVX-Puro vector (Clontech, 632164). Viruses were produced in HEK293T cells according to the manufacturer’s instructions (Lenti-XTM HTX packaging system; Clontech, 631247) and these were used to infect HeLa cells. Selection of stable clones was performed by addition of puromycin (1 μg/ml).
For lentiviral transduction of erythroid cells (see below), vectors containing 64-105 Atg4D-GFP and ΔN63 Atg4D-GFP were obtained by sub-cloning into pxlg3-gfp (a modified pSEW sin vector kindly provided by Dr. G. Cory, Exeter University, UK)45 after removal of GFP from the pxlg3 vector backbone. Lentiviruses were produced by cotransfection of the pxlg3 constructs in HEK 293T cells as described previously.46
Erythroid cell differentiation and lentiviral transformation
Peripheral, human blood cells were isolated from waste buffy-coat material or from waste apheresis cones from anonymous blood and platelet donors (National Blood Services, Bristol, UK); a provision that complies with the Nuffield Council on Bioethics Guidance on Human Tissue Ethical and Legal Issues 1954, the Medical Research Council’s Operational and Ethical Guidance on Human Tissue and Biological Samples for use in Research 2005 and the Royal College of Pathologists Transitional Guidelines to facilitate changes in procedures for handling “surplus” and archival material from human biological samples.
Samples were diluted 1:1 with Hanks balanced salt solution (HBSS, H6648) and layered on Histopaque-1077 (H8889). Mononuclear cells were harvested from the gradient, washed with HBSS and red blood cells were lysed in lysis buffer [150 mM ammonium chloride, 1 mM K2EDTA.2 H2O, 10 mM potassium bicarbonate buffer (pH 7.5)]. CD34+ cells were then isolated using the Direct CD34+ Progenitor Cell Isolation kit (Miltenyi Biotech Ltd., 130-046-072) according to manufacturer’s instructions. Isolated CD34+ cells were cultured using a modified two-stage system.28,29 They were first cultured for up to 10 d in serum-free Stemspan media (Stem Cell Technologies, 09650) supplemented with Stem Cell Factor (SCF; 10 ng/ml; R&D Systems, 255-SC), IL-3 (1ng/ml; R&D Systems, 203-IL), EPO (3 UI/ml; Roche, NeoRecormon), bovine lipoprotein (1 μl/μl; L4646) and Prograf (0.1 ng/ml; Fujisawa). Cells were transduced by lentivirus overnight on day 3 of culture in the presence of polybrene (8 μg/ml; H9268). The media was supplemented with dexamethasone (1 μM; D4902) in the absence of IL-3 from day 4, and GFP expressing cells were sorted by flow cytometry (BD Influx Cell Sorter) at day 7, then cultured for a further 3 d. Cells were then transferred to a Stemspan-based differentiation media supplemented with 1 mg/ml holotransferrin (T0665), 3% AB human serum (H4522), 10 UI/ml EPO, 10 ng/ml insulin (I9278) and 1 μM 3,5,3′-triiodo-L-thyronine sodium salt (T6397).
Subcellular fractionation and proteinase K treatment
HeLa cells transiently expressing myc-tagged Atg4D constructs were trypsinized, washed in PBS and resuspended in mitochondrial import buffer (50 mM hepes, pH 7.1; 0.6 M sorbitol; 2 mM KH2PO4; 50 mM KCl; 10 mM MgCl2; protease inhibitors).47 Cells were lysed on ice using a cell cracker (eight passes at 8 μm clearance) then centrifuged at 800 g for 5 min to remove the heavy nuclear pellet. The postnuclear supernatant was then centrifuged for 10 min at 10000 g, and the resulting mitochondria-containing pellet was washed with 500 μl import buffer and incubated in the absence or presence of 0.1 mg/ml proteinase K for 15 min on ice. The reaction was stopped by the addition of PMSF to 1 mM.
Epifluorescence microscopy
Wild-field epifluorescence images were obtained using an Olympus IX-71 inverted microscope (60 × Uplan Fluorite objective 0.65–1.25 NA, at maximum aperture) fitted with a CoolSNAP HQ CCD camera (Photometrics) driven by MetaMorph software (Molecular Devices). Confocal images were obtained using a Leica AOBS SP2 microscope (63 × PLAPO objective 1.4 NA) at 0.2 μm z-steps. For immunofluorescence, cells were fixed in 2% formaldehyde followed by permeabilization with 0.1% Triton X-100, or in -20°C methanol. Cells were routinely stained with DAPI and mounted in Mowiol containing 25 mg/ml DABCO (D2522) anti-fade. Images were processed using Adobe Photoshop CS3 for MacIntosh.
Cell death assays
To analyze the level of cell death in HeLa cell-lines treated with mitochondrial poisons (CCCP: 10 µM; antimycin A: 1 µg/ml), cell supernatants were analyzed for LDH concentration—an indirect reporter of cell lysis—using a CytoTox 96 LDH release assay kit (Promega, G1780, according to the manufacturer’s instructions). Data were obtained using an L Max II luminometer, and cell death was determined as relative LDH release normalized to total sample LDH content. For the analysis of cell death in HeLa cells treated with anisomycin (1 µg/ml) or staurosporine (1 µM), cells were fixed in formaldehyde, stained with DAPI, and the proportion of cells with pyknotic/fragmented chromatin in 20 random microscope fields (60 × objective) was recorded.
High-pressure freezing and immuno EM
For high-pressure freezing of erythroid cells, samples of up to 5 × 106 cells were resuspended in a small volume of media and fixed using a Leica EMPACT2+ RTS high-pressure freezer. Freeze-substitution was then performed using 1.1% osmium/0.1% uranyl acetone using a Leica AFS freeze substitution and low temperature embedding system. Samples were dehydrated with acetone and infiltrated with increasing amounts of resin (25%, 50%, 75% Epon-acetone for 1 h each followed by 2 h in 100% Epon resin). They were finally embedded in fresh Epon and left to harden for 48 h at 60°C. Thin sections of 70 nm were obtained using a diamond knife, counter stained then observed using a Tecnai 12 -FEI 120 kV BioTwin Spirit Transmission Electron Microscope. Images of randomly selected cells (minimum 22 cells per sample) were recorded with a FEI Eagle 4 k × 4 k CCD camera. For analysis, the number of mitochondria was measured relative to cytoplasmic area using MetaMorph software.
For immunoEM detection of mitochondrial Atg4D, frozen samples were embedded in lowicryl media using a Lieca AFS2 freeze substitution apparatus. These were sectioned, then stained with anti-GFP antibodies and anti-goat 6nm gold secondary antibodies. Sections were counterstained in uranyl acetate (3%) before being imaged as above.
Aldehyde fixation and Epon embedding for transmission EM
HeLa cells stably expressing 64-105 Atg4D-GFP, wild-type ΔN63 Atg4D-GFP of C144A ΔN63 Atg4D-GFP were fixed in gluteraldehyde and processed for transmission EM by embedding in Epon as described previously.48 To analyze mitochondrial cristae density, imaged obtained as described above were imported into Metamorph software, and integrated crista length was caculated as a function of mitochondrial cross-sectional area.
Supplementary Material
Acknowledgments
We acknowledge the support of the Medical Research Council in providing an Infrastructure Award to establish the School of Medical Sciences Cell Imaging Facility at Bristol University, and the Wolfson Foundation for recent funds to modernize and expand the facility. The also authors wish to acknowledge the assistance of Dr. Andrew Herman for cell sorting, and the University of Bristol Faculty of Medical and Veterinary Sciences Flow Cytometry Facility. This work is funded by a project grant from the NHS Blood and Transplant (Ref: PG07/1). J.D.L. is a RCUK Academic Fellow. T.D.B.M. is funded by a Ph.D. studentship from the Wellcome Trust.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/19227
References
- 1.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim J, Huang WP, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–8. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariño G, Uri´a JA, Puente XS, Quesada V, Bordallo J, Lo´pez-Oti´n C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–8. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 5.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weidberg H, Shpilka T, Shvets E, Elazar Z. Mammalian Atg8s: one is simply not enough. Autophagy. 2010;6:808–9. doi: 10.4161/auto.6.6.12579. [DOI] [PubMed] [Google Scholar]
- 7.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–76. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 8.Scherz-Shouval R, Sagiv Y, Shorer H, Elazar Z. The COOH terminus of GATE-16, an intra-Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J Biol Chem. 2003;278:14053–8. doi: 10.1074/jbc.M212108200. [DOI] [PubMed] [Google Scholar]
- 9.Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122:2554–66. doi: 10.1242/jcs.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–96. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–60. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherz-Shouval R, Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci. 2011;36:30–8. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norman JM, Cohen GM, Bampton ET. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy. 2010;6:1042–56. doi: 10.4161/auto.6.8.13337. [DOI] [PubMed] [Google Scholar]
- 15.Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341–50. doi: 10.1038/emboj.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betin VM, Lane JD. Atg4D at the interface between autophagy and apoptosis. Autophagy. 2009;5:1057–9. doi: 10.4161/auto.5.7.9684. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol. 2010;11:655–67. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 18.Mortensen M, Ferguson DJ, Simon AK. Mitochondrial clearance by autophagy in developing erythrocytes: clearly important, but just how much so? Cell Cycle. 2010;9:1901–6. doi: 10.4161/cc.9.10.11603. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–64. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U S A. 2010;107:832–7. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–67. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M, Sandoval H, Wang J. Selective mitochondrial autophagy during erythroid maturation. Autophagy. 2008;4:926–8. doi: 10.4161/auto.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–5. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–5. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zermati Y, Garrido C, Amsellem S, Fishelson S, Bouscary D, Valensi F, et al. Caspase activation is required for terminal erythroid differentiation. J Exp Med. 2001;193:247–54. doi: 10.1084/jem.193.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeil JA, Zermati Y, Vandekerckhove J, Cathelin S, Kersual J, Dussiot M, et al. Hsp70 regulates erythropoiesis by preventing caspase-3-mediated cleavage of GATA-1. Nature. 2007;445:102–5. doi: 10.1038/nature05378. [DOI] [PubMed] [Google Scholar]
- 28.Leberbauer C, Boulme´ F, Unfried G, Huber J, Beug H, Müllner EW. Different steroids co-regulate long-term expansion versus terminal differentiation in primary human erythroid progenitors. Blood. 2005;105:85–94. doi: 10.1182/blood-2004-03-1002. [DOI] [PubMed] [Google Scholar]
- 29.Southcott MJ, Tanner MJ, Anstee DJ. The expression of human blood group antigens during erythropoiesis in a cell culture system. Blood. 1999;93:4425–35. [PubMed] [Google Scholar]
- 30.Gronowicz G, Swift H, Steck TL. Maturation of the reticulocyte in vitro. J Cell Sci. 1984;71:177–97. doi: 10.1242/jcs.71.1.177. [DOI] [PubMed] [Google Scholar]
- 31.Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT, et al. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22:476–88. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–60. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–98. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulivieri C. Cell death: insights into the ultrastructure of mitochondria. Tissue Cell. 2010;42:339–47. doi: 10.1016/j.tice.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Griparic L, van der Wel NN, Orozco IJ, Peters PJ, van der Bliek AM. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J Biol Chem. 2004;279:18792–8. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- 36.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lo´pez-Oti´n C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–83. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 38.Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011;286:7327–38. doi: 10.1074/jbc.M110.199059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabrera S, Mariño G, Fern´ndez AF, Lo´pez-Oti´n C. Autophagy, proteases and the sense of balance. Autophagy. 2010;6:961–3. doi: 10.4161/auto.6.7.13065. [DOI] [PubMed] [Google Scholar]
- 40.Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol. 2009;187:959–66. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–42. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–90. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mariño G, Fern´ndez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodri´guez F, et al. Autophagy is essential for mouse sense of balance. J Clin Invest. 2010;120:2331–44. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danson CM, Pocha SM, Bloomberg GB, Cory GO. Phosphorylation of WAVE2 by MAP kinases regulates persistent cell migration and polarity. J Cell Sci. 2007;120:4144–54. doi: 10.1242/jcs.013714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–13. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 47.Vergnolle MA, Sawney H, Junne T, Dolfini L, Tokatlidis K. A cryptic matrix targeting signal of the yeast ADP/ATP carrier normally inserted by the TIM22 complex is recognized by the TIM23 machinery. Biochem J. 2005;385:173–80. doi: 10.1042/BJ20040650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lane JD, Allan VJ, Woodman PG. Active relocation of chromatin and endoplasmic reticulum into blebs in late apoptotic cells. J Cell Sci. 2005;118:4059–71. doi: 10.1242/jcs.02529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.