Abstract
Although Japanese morning glory (Ipomoea nil (L.) Roth.) has been used intensively for genetic studies, DNA markers have not been developed in Ipomoea nil sufficient to cover all chromosomes. Therefore, we conducted microsatellite (simple sequence repeats, SSR) marker development in I. nil for future genetic studies. From 92,662 expressed sequence tag (EST) sequences, 514 unique microsatellite-containing ESTs were identified. Primer pairs were designed automatically in 326 SSRs. Of 150 SSRs examined, 75 showed polymorphisms among strains. A phenogram based on the SSR genotypes revealed the genetic relation among seven Japanese morning glories from five different regions of the world and an ivyleaf morning glory (I. hederacea Jacq.). The developed SSR markers might be applicable for genetic studies of morning glories and their relatives.
Keywords: EST, SSR, morning glory, genetic diversity, DNA marker
Introduction
Japanese morning glory (Ipomoea nil) is a popular ornamental plant in Japan; classical genetic studies of this species have been conducted intensively (Hagiwara 1956, Imai 1927, 1929, 1931, 1938). Nitasaka (http://mg.biology.kyushu-u.ac.jp) made a linkage map derived from the cross between I. nil and I. hederacea based on amplified fragment length polymorphism (AFLP). However, the AFLP marker is a dominant marker and necessitates the use of expensive facilities for fragments detection and analysis. In contrast, SSR markers are co-dominant markers; they are easy to implement in most laboratories and are highly polymorphic. Consequently, SSRs are applied widely in plant genome mapping and genetic analysis (Akkaya et al. 1992, 1995, Jarret and Bowen 1994, Roder et al. 1995, Rongwen et al. 1995). In recent years, as a by-product of EST projects in many organisms in which a vast amount of sequence data were generated, microsatellite mining from SSR-containing ESTs is inexpensive and time-saving. It is an effective approach to develop microsatellites for genetic mapping and population genetics in plants (Chen et al. 2006, Cordeiro et al. 2001, Kantety et al. 2002, Temnykh et al. 2001). In this study, EST-SSR markers were developed from ESTs of Japanese morning glory. Then their polymorphisms were estimated and used for analyzing genetic relations among seven accessions that originated from five regions of the world and an accession of ivyleaf morning glory, I. hederacea, which is endemic to the southern part of north America and the closest relative species of I. nil.
Materials and Methods
Data mining for SSR markers and primer design
We used 92,662 ESTs that were assembled into 24,546 unigenes, which included 18,530 contigs and 6,016 singletons, from a strain of Ipomoea nil: Tokyo Kokei Standard (Hoshino et al. unpublished, Japanese morning glory cDNA database http://ipomoeanil.nibb.ac.jp/). Sequence and annotation of unigenes are available in the Japanese morning glory cDNA database by searching “a previous clone, pair, or contig”, and details of EST will be published elsewhere. SSRs that contained more than 10 repeat units or SSRs that contained multiple repeat units and the sum of repeat is more than 20 were screened from the unigenes using srchssr2.pl, a module script that constitutes an SSR marker-designing pipeline, read2Marker (Fukuoka et al. 2005). Polymerase chain reaction (PCR) primer pairs for the SSR markers were also designed using read2Marker automatically (Table 1 and Supplement Table 1).
Table 1.
Microsatellite markers derived from ESTs of Japanese morning glory
| Unigene IDa | Core motifs | Primer sequencesb | PIC value | Product size (bp)c | Polymorphism among eight accessions | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| A2 | A3 | A4-1 | A4-2 | Q63 | Q65 | Q931 | Q1176 | |||||
| Contig00659 | (ag)15 | ccttcgtgagtgctcaatacagaca tcggcaatccagtgaacacatagt |
0.47 | 130 | 2d | 2 | 2 | 2 | 3 | 1 | 2 | 3 |
| Contig01174 | (tc)12 | tcaaactccctttcctctcttgct tagacgaccgcctcttcttgaaac |
0.41 | 201 | –e | 2 | 2 | 1 | 2 | 3 | 2 | 2 |
| Contig02174 | (ag)13 | agaacccaccaggatattttccgt ccatttcattggcttcttctttgg |
0.30 | 225 | – | – | 2 | 2 | 1 | – | – | 2 |
| Contig02293 | (tg)14 | tggaaatgaagctgctggtattga tacaagggcagtacagcacaaacc |
0.41 | 210 | 3 | – | 3 | 2 | 3 | 1 | 3 | 3 |
| Contig02441 | (ta)18 | ccaacaccaatatccacaaccaga cgaattaaaatccacacacgaca |
0.19 | 284 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| Contig02586 | (ta)19 | atgatgtcacgatgagattgagcc tatcaccaccttagctccatgcc |
0.30 | 220 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 |
| Contig02903 | (ag)14 | cacgagggaaagagataggagcaa gctttagcagcagcaaaactccat |
0.37 | 154 | 2 | 2 | 2 | 2 | 3 | 1 | 2 | 2 |
| Contig03049 | (tc)12 | ttcagaaagcaccagcaaagatgt gtgactcccaactgaggaggaaag |
0.41 | 154 | – | 3 | 3 | 3 | 2 | 1 | 3 | 3 |
| Contig04019 | (tat)15 | cttgttgattgccaacaaaggcta ttaaggctacagcgtgcaaaatga |
0.62 | 222 | – | 3 | 3 | 3 | 4 | 2 | – | 1 |
| Contig04087 | (tc)12 | tcttctctatctggagctctcggtc gagaagcaaatccaagaaaacgga |
0.21 | 214 | – | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| Contig04156 | (ag)12 | ccccatcccccaatatacacttct caaccagaacaacaccaaacaagc |
0.21 | 210 | – | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| Contig04592 | (tc)12 | tgtatattaatcagcccaccaccc acttgggacttgggagtggtgata |
0.60 | 120 | 3 | 2 | 3 | 2 | 1 | 4 | 3 | 3 |
| Contig04657 | (ag)17 | aatgtgctaaactctccttcgcca agagcggcaaataccacagaactc |
0.41 | 203 | 3 | 3 | 3 | 3 | 2 | 1 | – | 3 |
| Contig05021 | (tc)14 | tctcatcaaccaattcaacaccct gtatggatcatccctcgacagctt |
0.47 | 127 | 2 | 2 | 2 | 2 | 1 | 3 | 2 | 1 |
| Contig05098 | (ag)17 | tcggttcggtttggtatttacagc agattgaggctttgttgatcgc |
0.21 | 173 | 2 | 2 | – | 2 | 2 | 1 | 2 | 2 |
| Contig05104 | (ag)14 | gggcagaaaagaaaaagaagaaagc gccaacaatgctagggaatacagc |
0.30 | 249 | – | – | – | – | 1 | 1 | 2 | 1 |
| Contig05275 | (tc)12 | gccacaatcctcgaagaagaagaa gccatatccaaaggttcccttagc |
0.41 | 250 | – | 3 | 3 | 3 | 2 | 1 | 3 | 3 |
| Contig05282 | (tc)13 | acggcctgacagccagataaataa aacaacgcagagtcggagagagat |
0.51 | 129 | 3 | 3 | 2 | 2 | 3 | 1 | 3 | 2 |
| Contig05407 | (ag)14 | tccttcaccagacacgagtcgtta cacctcccttcactctctccactc |
0.19 | 128 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| Contig05421 | (ag)12 | cgcctatacatacacgctgctctg tttgtttctcgctctacctcaggg |
0.36 | 167 | – | 2 | – | 2 | 1 | 1 | – | 2 |
| Contig05448 | (tc)13 | ctcacaatcacaatcctcttcccc ttgaatttgccgtaccagatgaga |
0.21 | 279 | – | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| Contig05536 | (ag)12 | ttgaatttcccagtttggaccact accgaactgcccactcatgttatt |
0.57 | 270 | – | 2 | 2 | 2 | 4 | 1 | 2 | 3 |
| Contig05575 | (ag)17 | caagagagagcgaatctttctttaggg aggtcatcgttgctttcttcttcg |
0.60 | 180 | 3 | 3 | 2 | 2 | 3 | 1 | 4 | 3 |
| Contig05644 | (tc)13 | cgctgcttctttcttcctctgttc caaagagtcgacgaaccttagcgt |
0.19 | 268 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| Contig05652 | (ag)13 | gagcgtcctcgatcaacccta aagcatcggtacctgtgagagacc |
0.47 | 235 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 1 |
| Contig05911 | (ag)17 | cgtcctcgtaaacggaaagtaacg tacctcttacggagcaaagcctca |
0.41 | 242 | 1 | 1 | – | 1 | 2 | 3 | 1 | 1 |
| Contig06063 | (tc)13 | ttaacaaaagggagtacggagcga tggagaaggtagtaggcaacggag |
0.21 | 300 | – | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| Contig06087 | (tc)13 | tgatgattcttgaaggggagcact agccttagctccttctctcccttg |
0.51 | 193 | 3 | 3 | 3 | 2 | 2 | 1 | 3 | 2 |
| Contig06422 | (ag)3aa(ag)14 | tccgccacttcagagcatacataa tagcaatgagcttttgcttctccc |
0.30 | 192 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 |
| Contig06449 | (ta)13 | ctttgcctttgttttgggttacga gagacgcaaccagaaaaggaacat |
0.55 | 213 | – | – | – | 3 | 1 | 1 | – | 2 |
| Contig06459 | (ag)4at(ag)12 | ccagtgtcttccctgcaaagtgta ttctcttctagctcccccaaaacc |
0.41 | 297 | 2 | 2 | – | 2 | 3 | 2 | 2 | 1 |
| Contig06566 | (ag)17 | tcatagctattacagggaaacactgga tgctgtagatcatcaggaagcacc |
0.58 | 157 | 2 | 3 | 3 | 2 | 2 | 1 | 1 | 3 |
| Contig06590 | (tc)16 | cttggggttaccctttttctccac gccgctaattcacggaatgtaaaa |
0.55 | 288 | 3 | 2 | 3 | 3 | 1 | 1 | 3 | 2 |
| Contig06640 | (ag)17 | gctgcttccgcaaagttcatattc tcaacaatcacgaagcaaccactt |
0.60 | 117 | 4 | 4 | 3 | 3 | 2 | 1 | 4 | 4 |
| Contig06657 | (tc)4cc(tc)12 | cccgcccttctattattcatccat cttattgattccgtttccgagtgc |
0.24 | 168 | 1 | 1 | – | 1 | – | 2 | 1 | 1 |
| Contig06713 | (ag)3(ag)12 | ctgccattgaaaagctcagtagcc ttgatttcgcccttccttcaacta |
0.21 | 255 | 1 | 1 | – | 1 | 1 | 1 | 2 | 1 |
| Contig06796 | (ag)13 | ggggttttgcttagctcactgaa gaaagccgcctttatagacagcaa |
0.30 | 256 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 |
| Contig07002 | (tc)13 | gactctctcgctaaacacagcttagta cgtgtagacaagaccggtcaaaga |
0.67 | 158 | 4 | – | 3 | 3 | 2 | 1 | – | 4 |
| Contig07004 | (tg)6(ag)12 | gacactttttctcctgcacaaacg cggagatgatcagcaaggaagaat |
0.27 | 196 | 2 | 2 | 2 | 2 | 1 | – | – | – |
| Contig07202 | (ag)17 | ggcctctcgtcttcgtctaaccta tcccttagcttcgtctgccataag |
0.37 | 150 | 3 | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
| Contig07219 | (tct)16 | ttgtggtgatggcatcttgattct tcagatcttcttcgaaatctccgc |
0.37 | 186 | 3 | 3 | 3 | 3 | 1 | 2 | 3 | 3 |
| Contig07368 | (tc)17 | aaagcccacaaagctctcatgtgt gctgtaaccaattgggagcaaaag |
0.51 | 255 | 1 | 1 | 2 | 2 | 3 | 2 | 1 | 2 |
| Contig07568 | (ag)16 | gactaccctgtgtattgagagtggg tctccttcaagctctccaccatct |
0.50 | 269 | 3 | 3 | – | 3 | 3 | 1 | 2 | 2 |
| Contig07663 | (ag)12 | gggagtgagggagggagaaagata tgagggacagaatcctctcaaacc |
0.37 | 217 | 3 | 3 | 3 | 3 | 2 | 1 | 3 | 3 |
| Contig07664 | (ag)16 | cctaaagttcaagaattgtggagga ggaagagagagaatggtacccacg |
0.41 | 101 | 3 | 3 | 3 | 3 | – | 1 | 3 | 2 |
| Contig07718 | (tc)16 | tcttccaaccccacatccttaact ccgatctccttccacactgaagat |
0.50 | 275 | 2 | 1 | – | 2 | 3 | 1 | 1 | 1 |
| Contig07725 | (ag)15 | cttctggtctccatacgtgagtcg ttctgattctccattttctccccc |
0.58 | 194 | 3 | 3 | 2 | 3 | 1 | 1 | 1 | 2 |
| Contig10368 | (ag)13 | gcaagcttcgccatgtacacatac aaaccaaaaaccgctccttcactt |
0.57 | 147 | – | 3 | 3 | 3 | 1 | 2 | 3 | 4 |
| Contig10405 | (ta)17 | ctgcacagcaaggatttaacaatga tggagaagaaacatggtttgaagg |
0.21 | 263 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | – |
| Contig10425 | (ag)18 | cgtccttgaccttcagttccttgt gtatccaaccacactgccgcacta |
0.41 | 268 | 1 | 1 | 1 | 1 | 2 | 3 | 1 | – |
| Contig10511 | (ag)17 | tataggatgatgcaaggcagagca tgccgaggaatgaaaagaggaata |
0.60 | 151 | 3 | 3 | 4 | 4 | 2 | 1 | 3 | 3 |
| Contig10818 | (tc)13 | gcacctcagattaacaacactttcagg tccatatctttgccttagctccca |
0.37 | 141 | 2 | 2 | 2 | 2 | 3 | 1 | 2 | 2 |
| Contig11626 | (ag)15 | gttttgaggtcggacattggaatc tgcaaccacatgctcactatcctt |
0.30 | 162 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 |
| Contig11987 | (gaa)14 | tgaagaaacttccccaaccacttc ttggggcctccaaatgtgatatag |
0.47 | 293 | 2 | 2 | 2 | 2 | 3 | 3 | 2 | 1 |
| Contig12195 | (gaa)19 | ctgattgttcaaaacccacactcg gcagtcctgtatttgcagatccct |
0.70 | 249 | 4 | 1 | – | 2 | 3 | 5 | 3 | 3 |
| Contig12319 | (ag)13 | cagcaaatgtgacaacacagatcg ttattggtcggagtgggtatggac |
0.21 | 290 | – | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| Contig13374 | (ta)33 | gccatggaggaagattgaagaaga acaagacatcaactgtggctggag |
0.49 | 255 | 1 | 1 | – | 1 | 1,2 | 1,2 | 1 | 1 |
| Contig13620 | (ag)12 | acgtttccagatcaacctgaggag gatgcagtagattttcacgtcgct |
0.21 | 210 | 2 | 2 | – | 2 | 2 | 1 | 2 | 2 |
| JMFF003B05 | (ag)16 | cacaagaaaaacaccacagtaagagag tcttgtgaggttgttgaaggagca |
0.41 | 273 | 2 | 2 | – | 2 | 1 | 3 | 2 | 2 |
| JMFF039J15 | (ag)15 | gggtgctctttgattctgcatttc ggatgagaaactgctagggggaag |
0.32 | 209 | – | 2 | 2 | 2 | 1 | 1 | 2 | 2 |
| JMFN008E02 | (ag)13 | gggggaatcgtgacagagttta ggaaagcttacaccagaaacggaa |
0.19 | 173 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 |
| JMFN029H19 | (ta)22 | tatcttcttcaccctcccggaatc ataaaaggtgacgaacagcgacga |
0.60 | 271 | 2 | 4 | 4 | 4 | 1 | 3 | 2 | 4 |
| JMFS049C04 | (ac)14 | atatcacctgaagcctcaacggaa tgcagaatcattcgaccatttcag |
0.21 | 254 | 2 | 2 | 2 | 2 | – | 1 | 2 | 2 |
| JMFS155F04 | (ta)14 | tcatatggaaactggtgtctccca attgttttcgggacagacaaggaa |
0.37 | 233 | 3 | 3 | 3 | 3 | 2 | 1 | 3 | 3 |
| JMSF002G14 | (tct)14 | gtttatggttaaaatgggcgagca ggcttggaagaaggatcaacttca |
0.30 | 265 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 |
| JMSF005H09 | (tct)13 | tttgtgtcgagaagatttgggtca ctcctgttcaaaacctccagcttc |
0.58 | 254 | – | 2 | 3 | 3 | 2 | 3 | 1 | 1 |
| JMSF015H22 | (ag)17 | agtctttcaagaatcaacgcggag aactgtttaccgacatcgctctct |
0.19 | 137 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 |
| JMSF018H02 | (ag)12 | gaggcagctgctttgctttgtatt gctttcccttctctctccatctcc |
0.52 | 211 | 3 | 3 | 1 | 2 | 4 | 3 | 3 | 3 |
| JMSF026B01 | (tc)13 | caggagcataggactataggaggc tatgggtgggaaatcagaggagaa |
0.67 | 219 | – | – | 3 | 3 | 2 | 1 | 2 | 4 |
| JMSF033C08 | (tc)20 | agatgcagaaatgtggacaacgaa tatgagaatcatcatcaggtcccg |
0.50 | 294 | 1,3 | 3 | 1 | 2 | 3 | 3 | 1,3 | – |
| JMSF033G03 | (ag)16 | aataccaaaaccccaaccagcttt cggatagagagaggaggaaggagg |
0.51 | 300 | 3 | 2 | 3 | 3 | 2 | 1 | 2 | 3 |
| JMSF046J01 | (tc)16 | acggagcatatctatacactgaagc tccggttaaactcagggaaagaaa |
0.37 | 172 | 2 | 2 | 2 | 2 | 3 | 1 | 2 | 2 |
| JMFF041I11 | (ta)12 | attctcttccgcttttggcttagg ctcttcctttccagcgtaaccctt |
0.51 | 171 | 2 | 3 | 3 | 3 | 1 | 2 | 2 | 2 |
| JMFF043B04 | (tc)8(ta)12 | agcagccccagaactaaccctaac agaaaacaaccagcgaactcaagc |
0.51 | 228 | 3 | 1 | 1 | 2 | 1 | 2 | 2 | 2 |
| JMSF044O11 | (ag)21 | ggtctgcagttctcacttcaatgc ttgtgtgataaaggcagcagcagt |
0.50 | 297 | 2 | 2 | 2 | – | 2 | 1 | 2 | 2 |
Unigene ID is based on the Japanese morning glory cDNA Database (http://ipomoeanil.nibb.ac.jp/).
Upper sequences show forward primers and lower sequences show reverse primers.
PCR product size using I. nil var ‘Tokyo Kokei Standard’ as a template.
Number 1–5 refer to fragment size. As the number is higher, the fragment size is bigger.
PCR amplification was not observed in this study.
Plant material
We used eight accessions in this study. Seven accessions of Ipomoea nil were Q931, Shirohanagenkei (A2), Peking Tendan (A3), A4-1, A4-2, Q63 and Q1176. The one accession of Ipomoea hederacea was (Q65). In fact, A2, A3, A4-1 and A4-2 were from a collection at Ibaraki University. The others––Q63, Q65, Q931 and Q1176––were from a collection at Kyushu University. Both Q931 and A2 were Japanese varieties. A3 was collected from Beijing, China by Kihara in 1938 (Yoneda and Takenaka 1981). Austin collected Q1176 from Iran in 2002. Both A4-1 and A4-2 were collected from Nepal by Nakao in 1952 (Yoneda and Takenaka 1981). Furusato collected Q63 from Guinea, Africa at latitude of 10 °N in 1956 (Yoneda and Takenaka 1981).
SSR analysis
Tender leaves from each plant were sampled and total DNA was extracted using CTAB method (Murray and Thompson 1980). Then PCR amplifications were performed with PCR System 2700 (Applied Biosystems). Each 6 μl amplification mixture contained 10 mM Tris-HCl(pH 8.3), 50 mM KCl, 1.5 mM of MgCl2, 0.2 mM of dNTP, 0.4 μM primer, 0.15 U Taq polymerase (Ampliqon, Skovlunde, Denmark) and 10 ng of template DNA. The PCR amplification condition included initial denaturation at 94°C for 3 min, and subsequently 40 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 1 min and terminal extension at 72°C for 5 min. Electrophoresis of PCR products was done in 10% acrylamide gel at constant voltage (300 V). The running buffer used was 0.15 M Tris-Glycine (pH 8.8). Amplified fragments were stained with ethidium bromide, visualized under UV illumination and photographed. The degree of DNA polymorphism at each SSR locus was evaluated, based on the polymorphic information content (PIC). The PIC value was calculated for each SSR locus according to the formula (Botstein et al. 1980);
where n is the total number of alleles detected for a locus and Pi is the frequency of the ith allele in the set of eight accessions tested.
Analysis of genetic relation
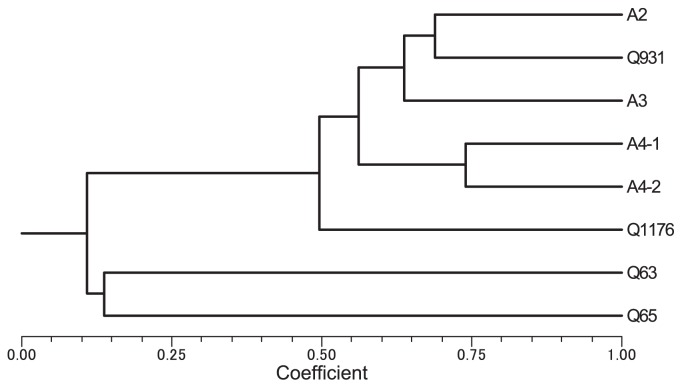
Genetic relations among eight accessions of morning glory were evaluated using 75 SSR markers. Each size of amplified band of EST-SSRs was recorded as present (1) or absent (0) and compiled into a binary matrix file that was used to calculate pairwise Jaccard’s similarity coefficients (1908). A dendrogram of eight genotypes was built using the unweighted pair-group method with arithmetric average (UPGMA) software (NTSYS-pc, ver. 2.2; Rohlf (2000)) (Fig. 1).
Fig. 1.
Phenogram for eight accessions of morning glory obtained with the similarity coefficient of Jaccard and UPGMA using 75 EST-SSR markers. Q931 and A2 (Shirohanagenkei) are from Japan. A3 (Peking-Tendan) is from China. A4-1 and A4-2 are both from Nepal. Q1176 is from Iran. Q63 is from Africa. Q65 (I. hederacea) was collected from Japan. This strain probably came to Japan from North America as an invasive species.
Results
SSR marker development
In all, 514 unique microsatellite-containing expressed sequence tags (EST) were identified from the EST of Japanese morning glory using srchssr2.pl (Table 1 and Supplement Table 1). SSRs more than seven repeat units, were counted and characterized the feature of these EST-SSRs. In total, 90% (465/514) of the SSRs consisted of a di-nucleotide repeat unit, and out of them, TC repeats were the most frequent, accounting for 36.8% (189/514), followed by AG repeats (33.3%, 171/514) and TA repeats (18.3%, 98/514). Tri-nucleotide repeat were found in 9.5% of the SSRs and GAA repeats were the most frequent (2.5%, 13/514), followed by TCT repeats (2.1%, 11/514) and TAA repeats (1.9%. 10/514).
Primer pairs were automatically designed in 326 SSRs. 150 primer pairs of which SSRs had longer repeat were synthesized and examined for amplification and polymorphism among the eight accessions. Of 150 primer pairs, amplification products were obtained in 143 primer pairs; 75 primer pairs showed polymorphism among accessions (Table 1). In these SSRs, AG repeats were the most polymorphic (86%, 38 were polymorphic of 44 examined). While TC repeats and AC repeats were less polymorphic; 21 were polymorphic of 40 examined (53%), and 9 were polymorphic of 34 examined (26%) respectively. The sum of these three motifs occupied 91% of all polymorphic SSRs. The PIC of 75 SSR was from 0.19 to 0.70. The mean PIC was 0.41 ± 0.14.
Genetic relationship of eight accessions
To find suitable cross combinations for constructing a linkage map, genetic similarities among accessions were estimated based on Jaccard’s similarity using 75 polymorphic SSRs. A wide range of genetic similarity among these genotypes was revealed with similarity of 0.11–0.74. A UPGMA phenogram was constructed for eight accessions (Fig. 1). Q63 and Q65, American and African accessions respectively were distinguished from Asian accessions. Asian accessions were divided into three groups, East Asian, Nepalese and Iranian.
Discussion
A set of functional SSR markers was developed using data mining from Japanese morning glory EST sequences. Approximately 2% of Japanese morning glory ESTs contained SSR, and this is comparable to the result of Solanaceae species from 1% to 3% (Fukuoka et al. 2010) and onion’s 2.8% (Khul et al. 2004).
The result of genetic similarities reveals a collinear relation between geographic distance and genetic relation. The two accessions from Japan and Peking-Tendan, namely A2, Q931 and A3, have mutually similar genetic backgrounds each other. On the other hand, Q63 from Africa shows quite different genetic relation with other accessions in I. nil. All these results coincide with the analysis based on AFLP-marker genotype data by Nitasaka (http://mg.biology.kyushu-u.ac.jp). These results indicate that Q63 is well diverged from Japanese accessions as is I. hederacea, Q65. Interspecific hybridizations between I. hederacea and I. nil rarely succeeds due to pre-zygotic reproductive barriers (Hagiwara 1946). On the other hand, Q63 can be easily hybridize with Japanese morning glory. Thus Q63 seems to be more suitable to construct segregating populations for linkage analysis, although the flowering time of Q63 is very late, as it does not flower before late September in Ibaraki, Japan.
Although it was predicted that I. nil was originally native to American tropics (Austin et al. 2001), I. nil already appeared in pre-Columbian documents and it is distributed in tropical region of Asia, Africa and America. So it is an interesting question: How did I. nil leave America and reach Japan? If we could compare the genotype between American accessions and Japanese accessions, we might be able to predict whether I. nil came to Japan through Africa or over the Pacific Ocean.
The average PIC (0.41) of the EST-SSRs developed in this study is comparable to 0.37 of tomato varieties (He et al. 2003) and 0.44 of cultivated beans (Hanai et al. 2007). Thus they might be useful as in tomato and cultivated beans and applicable not only for estimating the genetic relationship of morning glories, but also for linkage map construction and genetic studies of Ipomoea species. The physiological studies of Japanese morning glory have been long history, so that the synergistic effect of genetics and physiology might be expected if these studies were reexamined in genetical point of view.
Supplementary Material
Literature Cited
- Akkaya M.S., Bhagwat A.A., Cregan P.B. (1992) Length polymorphisms of simple sequence repeat DNA in soybean. Genetics 132: 1131–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkaya M.S., Shoemaker R.C., Specht J.E., Bhagwat A.A., Cregan P.B. (1995) Integration of simple sequence repeat DNA markers into a soybean linkage map. Crop Sci. 35: 1439–1445 [Google Scholar]
- Austin D.F., Kitajima K., Yoneda Y., Qian L. (2001) A putative tropical American plant, Ipomoea nil (Convolvulaceae), in pre-columbian Japanese art. Economic Botany 55: 515–527 [Google Scholar]
- Botstein D., White R.L., Skoinck M., Davis R.W. (1980) Construction of genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32: 314–331 [PMC free article] [PubMed] [Google Scholar]
- Chen C.X., Zhou P., Choi Y.A., Huang S., Gmitter F.G., Jr. (2006) Mining and characterizing microsatellites from citrus ESTs. Theor. Appl. Genet. 112: 1248–1257 [DOI] [PubMed] [Google Scholar]
- Cordeiro G.M., Casu R., McIntyre C.L., Manners J.M., Henry R.J. (2001) Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci. 160: 1115–1123 [DOI] [PubMed] [Google Scholar]
- Fukuoka H., Nunome T., Minamiyama Y., Kono I., Namiki N., Kojima A. (2005) read2Marker: a data processing tool for microsatellite marker development from a large data set. Biotechniques 39: 472–476 [DOI] [PubMed] [Google Scholar]
- Fukuoka H., Yamaguchi H., Nunome T., Negoro S., Miyatake K., Ohyama A. (2010) Accumulation, functional annotation, and comparative analysis of expressed sequence tags in eggplant (Solanum melongena L.), the third pole of the genus Solanum species after tomato and potato. Gene 450: 76–84 [DOI] [PubMed] [Google Scholar]
- Hagiwara T. (1946) Interspecific crossings in Pharbitis nil. Japanese Journal of Genetics 21: 50–52 [Google Scholar]
- Hagiwara T. (1956) Genes and chromosome maps in the Japanese morning glory. Bull. Res. Coll. Agr. Vet. Sci. Nihon Univ. 5: 34–56 [Google Scholar]
- Hanai L.R., de Campos T., Camargo L.E., Benchimol L.L., de Souza A.P., Melotto M., Carbonell S.A., Chioratto A.F., Consoli L., Formighieri E.F., et al. (2007) Development, characterization, and comparative analysis of polymorphism at common bean SSR loci isolated from genic and genomic sources. Genome 50: 266–277 [DOI] [PubMed] [Google Scholar]
- He C., Poysa V., Yu K. (2003) Development and characterization of simple sequence repeat (SSR) markers and their use in determining relationships among Lycopersicon esculentum cultivars. Theor. Appl. Genet. 106: 363–373 [DOI] [PubMed] [Google Scholar]
- Imai Y. (1927) A genetic analysis of white-margined flowers in the Japanese morning-glory. Genet. 12: 199–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y. (1929) Linkage group of the Japanese morning glory. Genetics. 14: 223–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y. (1931) Description of the genes found in Pharbitis nil. Genetica 12: 297–318 [Google Scholar]
- Imai Y. (1938) Genetic literature of the Japanese morning glory. Jpn. J. Genet. 14: 91–96 [Google Scholar]
- Jarret R.L., Bowen N. (1994) Simple sequence repeats (SSRs) for sweet potato germplasm characterization. Plant Genet. Res. Newslet. 100: 9–11 [Google Scholar]
- Jaccard P. (1908) Nouvelles recherches sur la distribution florale. Bull. Soc. Vaudoise Sci. Nat. 44: 223–270 [Google Scholar]
- Kantety R.V., Rota M.L., Matthews D.E., Sorrells M.E. (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol. Biol. 48: 501–510 [DOI] [PubMed] [Google Scholar]
- Kuhl J.C., Cheung F., Yuan Q., Martin W., Zewdie Y., McCallum J., Catanach A., Rutherford P., Sink K.C., Jenderek M., et al. (2004) A unique set of 11,008 onion expressed sequence tags reveals expressed sequence and genomic differences between the monocot orders Asparagales and Poales. Plant Cell 16: 114–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röder M.S., Plaschke J., König S.U., Börner A., Sorrels M.E., Tanksley S.D., Ganal M.W. (1995) Abundance, variability and chromosomal location of microsatellites in wheat. Mol. Gen. Genet. 246: 327–333 [DOI] [PubMed] [Google Scholar]
- Rohlf F.J. (2000) NTSYS-pc: Numerical taxonomy and multivariate analysis system, ver. 2.2. Exeter Software: Setauket, New York [Google Scholar]
- Rongwen J., Akkaya M.S., Bhagwat A.A., Lavi U., Cregan P.B. (1995) The use of microsatellite DNA markers for soybean genotype identification. Theor. Appl. Genet. 90: 43–48 [DOI] [PubMed] [Google Scholar]
- Temnykh S., DeClerck G., Lukashova A., Lipovich L., Cartinhour S., McCouch S. (2001). Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11: 1441–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda Y., Takenaka Y. (1981) Genshoku asagao kensaku zukan (Natural-Color Illustrated monograph of Japanese morning glory) Second edition Hokuryukan Publ. Co., Tokyo, Japan [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.