Abstract
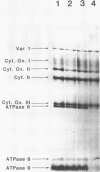
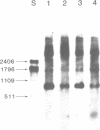
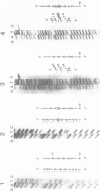
A temperature-conditional mit- mutant of Saccharomyces cerevisiae has been characterized; the mutant strain h45 cannot grow at 36 degrees C on nonfermentable substrates yet appears to be normal at 28 degrees C. The mutation in strain h45 maps genetically to the oli1 region of the mitochondrial DNA (mtDNA) genome, and prevents the synthesis at 36 degrees C of the oli1 gene product, subunit 9 of the mitochondrial ATPase complex. Since the level of oli1 mRNA in mutant h45 is close to normal at 36 degrees C, it is concluded that there is a specific block in translation of this mRNA at the non-permissive temperature. DNA sequence analysis of mtDNA from strain h45 reveals an additional T residue inserted 88 bp upstream of the oli1 coding region, in the A,T-rich sequence that is transcribed into the 5'-untranslated region of the oli1 mRNA. Sequence data on two revertants show that one returns to wild-type parental (J69-1B) mtDNA sequence, whilst the other contains an inserted A residue adjacent to the T inserted in the original h45 mutant. The results are discussed in terms of the stability of folds in RNA upstream of putative ribosome-binding sites in mitochondrial mRNA, and the potential action of nuclear-coded proteins that might be activators of the translation of specific mitochondrial mRNAs in yeast mitochondria.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Costanzo M. C., Mueller P. P., Strick C. A., Fox T. D. Primary structure of wild-type and mutant alleles of the PET494 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1986 Feb;202(2):294–301. doi: 10.1007/BF00331654. [DOI] [PubMed] [Google Scholar]
- Devenish R. J., Englisn K. J., Hall R. M., Linnase A. W., Lukins H. B. Biogenesis of mitochondria 49 identification and mapping of a new mitochondrial locus (tsr1) which maps within polar region of yeast mitochondrial genome. Mol Gen Genet. 1978 May 31;161(3):251–259. doi: 10.1007/BF00330998. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Edwards J. C., Osinga K. A., Christianson T., Hensgens L. A., Janssens P. M., Rabinowitz M., Tabak H. F. Initiation of transcription of the yeast mitochondrial gene coding for ATPase subunit 9. Nucleic Acids Res. 1983 Dec 10;11(23):8269–8282. doi: 10.1093/nar/11.23.8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gren E. J. Recognition of messenger RNA during translational initiation in Escherichia coli. Biochimie. 1984 Jan;66(1):1–29. doi: 10.1016/0300-9084(84)90188-3. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Grivell L. A., Borst P., Bos J. L. Nucleotide sequence of the mitochondrial structural gene for subunit 9 of yeast ATPase complex. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1663–1667. doi: 10.1073/pnas.76.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B., Burger G., Doxiadis I., Thomas D. Y., Bandlow W., Kaudewitz F. A simple method for the large-scale preparation of mitochondria from microorganisms. Anal Biochem. 1977 Jan;77(1):110–121. doi: 10.1016/0003-2697(77)90295-0. [DOI] [PubMed] [Google Scholar]
- Li M., Tzagoloff A., Underbrink-Lyon K., Martin N. C. Identification of the paromomycin-resistance mutation in the 15 S rRNA gene of yeast mitochondria. J Biol Chem. 1982 May 25;257(10):5921–5928. [PubMed] [Google Scholar]
- Lin C. K., Goldfarb D. S., Doi R. H., Rodriguez R. L. Mutations that affect the translation efficiency of Tn9-derived cat gene in Bacillus subtilis. Proc Natl Acad Sci U S A. 1985 Jan;82(1):173–177. doi: 10.1073/pnas.82.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane A. W., Lukins H. B., Molloy P. L., Nagley P., Rytka J., Sriprakash K. S., Trembath M. K. Biogenesis of mitochondria: molecular mapping of the mitochondrial genome of yeast. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2082–2085. doi: 10.1073/pnas.73.6.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macino G., Tzagoloff A. Assembly of the mitochondrial membrane system. The DNA sequence of a mitochondrial ATPase gene in Saccharomyces cerevisiae. J Biol Chem. 1979 Jun 10;254(11):4617–4623. [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M., Choo K. B., Macreadie I., Marzuki S., Lukins H. B., Nagley P., Linnane A. W. Biogenesis of mitochondria: a temperature sensitivity mutation affecting the mitochondrially synthesized var1 protein of Saccharomyces cerevisiae. Arch Biochem Biophys. 1980 Aug;203(1):260–270. doi: 10.1016/0003-9861(80)90176-9. [DOI] [PubMed] [Google Scholar]
- Müller P. P., Reif M. K., Zonghou S., Sengstag C., Mason T. L., Fox T. D. A nuclear mutation that post-transcriptionally blocks accumulation of a yeast mitochondrial gene product can be suppressed by a mitochondrial gene rearrangement. J Mol Biol. 1984 Jun 5;175(4):431–452. doi: 10.1016/0022-2836(84)90178-5. [DOI] [PubMed] [Google Scholar]
- Nagley P., Hall R. M., Ooi B. G. Amino acid substitutions in mitochondrial ATPase subunit 9 of Saccharomyces cerevisiae leading to oligomycin or venturicidin resistance. FEBS Lett. 1986 Jan 20;195(1-2):159–163. doi: 10.1016/0014-5793(86)80152-1. [DOI] [PubMed] [Google Scholar]
- Ooi B. G., McMullen G. L., Linnane A. W., Nagley P., Novitski C. E. Biogenesis of mitochondria: DNA sequence analysis of mit- mutations in the mitochondrial oli1 gene coding for mitochondrial ATPase subunit 9 in Saccharomyces cerevisiae. Nucleic Acids Res. 1985 Feb 25;13(4):1327–1339. doi: 10.1093/nar/13.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi B. G., Nagley P. The oli1 gene and flanking sequences in mitochondrial DNA of Saccharomyces cerevisiae: the complete nucleotide sequence of a 1.35 kilobase petite mitochondrial DNA genome covering the oli1 gene. Curr Genet. 1986;10(10):713–723. doi: 10.1007/BF00405093. [DOI] [PubMed] [Google Scholar]
- Ooi B. G., Novitski C. E., Nagley P. DNA sequence analysis of the oli1 gene reveals amino acid changes in mitochondrial ATPase subunit 9 from oligomycin-resistant mutants of Saccharomyces cerevisiae. Eur J Biochem. 1985 Nov 4;152(3):709–714. doi: 10.1111/j.1432-1033.1985.tb09251.x. [DOI] [PubMed] [Google Scholar]
- Osinga K. A., De Vries E., Van der Horst G., Tabak H. F. Processing of yeast mitochondrial messenger RNAs at a conserved dodecamer sequence. EMBO J. 1984 Apr;3(4):829–834. doi: 10.1002/j.1460-2075.1984.tb01892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putrament A., Baranowska H., Prazmo W. Induction by manganese of mitochondrial antibiotic resistance mutations in yeast. Mol Gen Genet. 1973 Nov 22;126(4):357–366. doi: 10.1007/BF00269445. [DOI] [PubMed] [Google Scholar]
- Rödel G., Körte A., Kaudewitz F. Mitochondrial suppression of a yeast nuclear mutation which affects the translation of the mitochondrial apocytochrome b transcript. Curr Genet. 1985;9(8):641–648. doi: 10.1007/BF00449816. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanssens P., Remaut E., Fiers W. Alterations upstream from the Shine-Dalgarno region and their effect on bacterial gene expression. Gene. 1985;36(3):211–223. doi: 10.1016/0378-1119(85)90176-3. [DOI] [PubMed] [Google Scholar]
- Thalenfeld B. E., Bonitz S. G., Nobrega F. G., Macino G., Tzagoloff A. oli1 Transcripts in wild type and in a cytoplasmic "petite" mutant of yeast. J Biol Chem. 1983 Dec 10;258(23):14065–14068. [PubMed] [Google Scholar]
- Tzagoloff A., Myers A. M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Willson T. A., Ooi B. G., Lukins H. B., Linnane A. W., Nagley P. Mutations in the mitochondrial oli1 gene of Saccharomyces cerevisiae affecting subunit 9 of the mitochondrial ATPase complex. Nucleic Acids Res. 1986 Oct 24;14(20):8228–8228. doi: 10.1093/nar/14.20.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]