Abstract
The Lactococcus lactis laboratory strain MG1363 has been described to be unable to utilize lactose. However, in a rich medium supplemented with lactose as the sole carbon source, it starts to grow after prolonged incubation periods. Transcriptome analyses showed that L. lactis MG1363 Lac+ cells expressed celB, encoding a putative cellobiose-specific phosphotransferase system (PTS) IIC component, which is normally silent in MG1363 Lac− cells. Nucleotide sequence analysis of the cel cluster of a Lac+ isolate revealed a change from one of the guanines to adenine in the promoter region. We showed here that one particular mutation, taking place at increased frequency, accounts for the lactose-utilizing phenotype occurring in MG1363 cultures. The G-to-A transition creates a −10 element at an optimal distance from the −35 element. Thus, a fully active promoter is created, allowing transcription of the otherwise cryptic cluster. Nuclear magnetic resonance (NMR) spectroscopy results show that MG1363 Lac+ uses a novel pathway of lactose utilization.
INTRODUCTION
Lactococcus lactis is an industrially important lactic acid bacterium (LAB). It is the main constituent of cheese starter cultures and is used for its ability to rapidly convert the milk sugar lactose into lactic acid. Because of the economic importance of lactose fermentation, the metabolism of this sugar is being studied extensively.
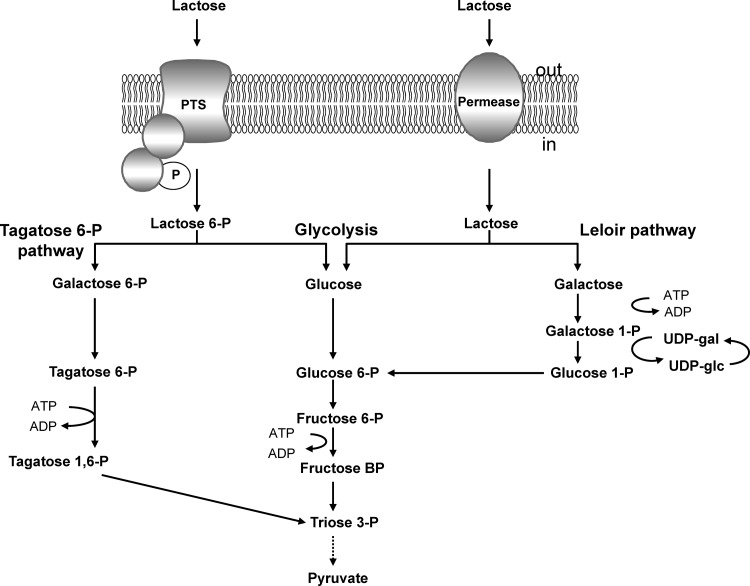
Two main systems of lactose uptake and metabolism have been described for LAB. The bioenergetically most efficient system in most strains is encoded by a plasmid and consists of the phosphoenolpyruvate:lactose phosphotransferase system (PEP-PTSLac; encoded by lacEF), phospho-β-galactosidase (lacG), and the tagatose 6-phosphate (tagatose 6-P) pathway enzymes (lacABCD) (10). During uptake, lactose is phosphorylated at the galactose moiety and then hydrolyzed. The glucose moiety enters glycolysis, while galactose 6-phosphate is degraded via the tagatose 6-P pathway, consisting of galactose 6-phosphate isomerase (lacAB), tagatose 6-phosphate kinase (lacC), and 1,6-diphosphate aldolase (lacD). The generated triosephosphates are then directed to glycolysis. All glycolytic enzymes are encoded on the lactococcal chromosome. Another way to internalize lactose is provided by the chromosomally encoded lactose-specific permease (lacY)–β-galactosidase (lacZ) system (21, 38). Internalized unphosphorylated lactose is cleaved by β-galactosidase (lacZ), and the resulting galactose molecule enters the Leloir pathway (54), while the glucose moiety is further metabolized by glycolytic enzymes. Genes coding for the galactose permease (GalP) and the Leloir pathway enzymes are clustered together in the gal operon. This pathway consists of reactions that are catalyzed by galactose mutarotase (GalM), galactokinase (GalK), galactose 1-phosphate uridylyltransferase (GalT), and UDP-galactose-4-epimerase (GalE) (14). The resulting glucose 1-phosphate is then converted to glucose 6-phosphate by α-phosphoglucomutase (PgmH) and directed to glycolysis (28). One of the main differences between these two systems is the phosphorylation state of lactose upon uptake. Therefore, the particular lactose transport system is usually coupled to a specific subsequent utilization pathway (Fig. 1).
Fig 1.
Schematic overview of the two alternative pathways for lactose transport and metabolism in L. lactis. When internalized via the PTSLac (LacEF), lactose is phosphorylated and hydrolyzed by phospho-β-galactosidase (LacG); the glucose moiety enters glycolysis, while galactose 6-phosphate is degraded via the tagatose 6-P pathway (LacABCD). When lactose is internalized via the lactose-specific permease (LacY), unphosphorylated lactose is cleaved by β-galactosidase (LacZ), and the resulting galactose molecule enters the Leloir pathway (GalMKTE), while the glucose moiety is directed to glycolysis.
The plasmid-free L. lactis laboratory strain MG1363 has been described to be unable to utilize lactose due to the loss of the lactose/proteinase plasmid pLP712 (13). This strain does not possess any of the classical systems required for transport and cleavage of lactose. However, in a rich medium supplemented with lactose as the sole carbon source, this strain starts to grow after prolonged incubation periods (3, 9, 34, 48). We noticed that the growth in lactose-containing medium was reproducible between individual cultures, and using standard mutation rates, we could not explain the frequent occurrence of the lactose utilization (Lac+) phenotype.
Here we identify the mechanism that is responsible for the appearance of the Lac+ phenotype and characterize the lactose uptake and metabolic utilization pathway in L. lactis MG1363. We show that a point mutation occurring at increased frequency in the promoter region of the cellobiose/lactose-specific PTS IIC component CelB accounts for the occurrence of the lactose-positive phenotype. Moreover, we present a novel pathway of lactose utilization, involving PTSLac-Cel, the phospho-β-glucosidases BglS and AscB, lactose 6-phosphate dephosphorylase, and the Leloir pathway enzymes.
MATERIALS AND METHODS
Microbial strains and growth conditions.
Strains and plasmids used in this study are listed in Table S1 in the supplemental material. L. lactis strains were grown as standing cultures at 30°C in M17 broth (Difco, Sparks, MD) or in chemically defined medium (CDM PC) (13a; F. Santos et al., unpublished data) or NMR-CDM (30), both supplemented with 0.5% or 1% (wt/vol) glucose, cellobiose, lactose, glucose, or galactose. When appropriate, erythromycin or chloramphenicol (Sigma-Aldrich, St. Louis, MO) was used at 3 or 5 μg ml−1, respectively.
Escherichia coli DH5α was used as a cloning host and was grown in tryptone-yeast extract medium (Difco) at 37°C or on tryptone-yeast extract medium solidified with 1.5% (wt/vol) agar. For plasmid selection, 150 μg ml−1 erythromycin (Sigma) was added.
General DNA techniques.
DNA manipulations were done essentially as described previously (42). Plasmid DNA and PCR products were isolated and purified using a High Pure plasmid isolation kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions.
Restriction enzymes, T4 DNA ligase, and Taq DNA polymerase were obtained from Fermentas (Vilnius, Lithuania) and used according to the supplier's instructions. Phusion DNA polymerase was purchased from Finnzymes Oy (Vantaa, Finland). PCR was performed in an Eppendorf thermal cycler (Hamburg, Germany) with L. lactis MG1363 chromosomal DNA as the template, unless described otherwise, using appropriate conditions. Primers used in this study are listed in Table S2 in the supplemental material.
Construction of L. lactis deletion strains.
The PCR products obtained with primer pairs KocelB1F-KocelB2R and KocelB3F-KocelB4R were cloned together as XbaI-BamHI and BamHI-XhoI restriction fragments into XbaI-XhoI-restricted integration vector pCS1966 (7, 49), resulting in pCS1966-celB′. PCR products obtained with primer pairs KoPtcBA1F-KoPtcBA2R and KoPtcBA3F-KoPtcBA4R were cloned as XbaI-BamHI and BamHI-XhoI restriction fragments into XbaI-XhoI-restricted pCS1966, resulting in pCS1966-ptcBA′. KobglS1F-KobglS2Rev and KobglS3F-KobglS4Rev PCR products were cloned as XbaI-BamHI and BamHI-XhoI restriction fragments into XbaI-XhoI-restricted pCS1966, resulting in pCS1966-bglS′. The PCR products obtained with primer pairs KoCcpA1F-KoCcpA2R and KoCcpA3F-KoCcpA4R were cloned as XbaI-BamHI and BamHI-XhoI restriction fragments into XbaI-XhoI-restricted pCS1966, resulting in pCS1966-ccpA′. All pCS1966 derivatives were obtained and maintained in E. coli DH5α (Invitrogen, Carlsbad, CA).
The pCS1966-celB′, pCS1966-ptcBA′, and pCS1966-bglS′ vectors were introduced into L. lactis MG1363 Lac+ (MGLac+) via electroporation (17); vector pCS1966-ccpA′ was introduced into L. lactis MGLac+ Pcel*-gfp, and a two-step homologous recombination event was induced by growing cells on selective SA medium plates (20) supplemented with 20 μg ml−1 5-fluoroorotic acid hydrate (7, 49). The obtained strains were labeled MGLac+ ΔcelB, MGLac+ ΔptcBA, MGLac+ ΔbglS, and MGLac+ ΔccpA Pcel*-gfp. The chromosomal structure of all deletion strains was confirmed by PCR analysis and sequencing of the modified regions.
Construction and induction of L. lactis celB overexpression strain.
One copy of the nisin-inducible two-component system encoded by nisRK was integrated into the chromosome of MGLac+ by using the vector pCS1966::pseudo10::nisRK (35). After a two-step integration event (as described above), strain MGLac+ nisRK was obtained.
For the overexpression of CelB, a fragment was amplified from the L. lactis MGLac+ chromosome by use of primers CelB_F and CelB_Rev. The resulting PCR product was digested with NcoI and XbaI and cloned into the NcoI and XbaI sites of pNZ8048 (8), downstream of the nisin-inducible promoter PnisA, yielding plasmid pNZcelB.
Expression of genes driven by PnisA was induced by addition of a supernatant (0.01% [vol/vol]) of an overnight culture of the nisin producer L. lactis NZ9700 (22, 24). Growth experiments with the celB complementation strain were performed in CDM PC supplemented with 1% (wt/vol) lactose and cellobiose.
In vivo NMR spectroscopy.
Carbon-13 spectra were acquired at 125.77 MHz on a Bruker Avance II 500-MHz spectrometer (Bruker BioSpin GmbH, Karlsruhe, Germany). All in vivo experiments were run using a quadruple-nucleus probe head at 30°C, as described before (30). HB21 cells were precultured in NMR-CDM on either cellobiose or lactose (1% [wt/vol]), harvested in the mid-logarithmic growth phase, washed twice with 5 mM KPi buffer (pH 6.6), and suspended in 100 mM KPi (pH 6.6) with 6% (vol/vol) 2H2O, to a protein concentration of approximately 15 mg protein ml−1. In vivo nuclear magnetic resonance (NMR) experiments were performed using a 10-mm NMR tube containing 3 ml of cell suspension. To avoid settling down of the cells and ensure an adequate supply of gases to the cell suspension, an air-lift system was used inside the NMR tube (43). To make the system anaerobic, argon was bubbled through the air-lift system 10 min before and continuously after acquisition was started. Lactose (15 mM) specifically labeled with 13C on carbon-1 of the galactose moiety was added to the cell suspension at time point zero, and spectra were acquired sequentially (30 s) after its addition. The time course of lactose consumption, product formation, and changes in the pools of intracellular metabolites were monitored in vivo. When the substrate was exhausted and no changes in the resonances of intracellular metabolites were observed, a perchloric acid cell extract was prepared as described previously (30). The cell extract was neutralized with potassium hydroxide to pH 6.5, and the potassium perchlorate precipitate as well as cell debris and denatured macromolecules was removed by centrifugation. The resulting supernatant was used for quantification of end products and other labeled metabolites by 1H- and 13C-NMR (30). Due to the fast pulsing conditions used for acquiring in vivo 13C spectra, correction factors were applied to convert peak intensities to concentrations (28, 30). The quantitative kinetic data for intracellular metabolites were calculated as described elsewhere (31, 32). Intracellular metabolite concentrations were calculated using a value of 2.9 μl/mg of protein for the intracellular volume of L. lactis (39). Although individual experiments are illustrated in each figure, each type of in vivo NMR experiment was repeated twice, and the results were highly reproducible.
Transcriptome analyses.
Transcriptome analysis was performed using full-genome L. lactis MG1363 DNA microarrays as described previously (23). MG1363 wild-type (MGwt) cells growing in M17 medium supplemented with 0.5% lactose were harvested at two time points. The total RNA from cells growing on residual carbon sources of rich M17 medium (OD600 of 0.25 after 2 h of growth) was compared to that from the same culture growing on lactose (OD600 of 1.85 after 120 h of growth).
In order to identify the genes involved in lactose metabolism in L. lactis MGLac+, cells were grown in CDM supplemented with 1% (wt/vol) lactose or cellobiose and harvested during the exponential phase of growth (OD600 of 0.35 for cells grown on cellobiose and 0.6 for cells grown on lactose). DNA microarray slides were scanned with a Genepix 4200 laser scanner at a 10-μm resolution. Slide images were analyzed using ArrayPro 4.5 (Media Cybernetics Inc., Silver Spring, MD). Processing and normalization (LOWESS spot pin based) of slides were done with MicroPrep software as described previously (53). Differential expression tests were performed on expression ratios with a local copy of the Cyber-T implementation of a variant of the t test. A gene was considered differentially expressed when the Bayesian P value was <0.001, and a change cutoff of 2.5-fold was applied.
Construction of lacZ expression strains.
The wild-type llmg_0186 promoter (Pcel) and a mutated version (Pcel*) carrying a G-to-A transition at position −11 were amplified by PCR with primers Pllmg_0186F and Pllmg_0186ZR, using chromosomal DNAs of L. lactis MGwt and MGLac+, respectively, as templates. The fragments were cloned as SphI-XbaI fragments into the low-copy-number expression vector pILORI4 (25). The vectors pILORI4::Pcel and pILORI4::Pcel* were introduced into L. lactis MGLac+. This yielded strains MGLac+ Pcel-lacZ and MGLac+ Pcel*-lacZ.
Construction of gfp expression vector and strains.
The green fluorescent protein (GFP) gene gfp* was amplified by PCR with the primer pair gfp_sf_F-gfp_sf_R, using pJWV102_gfp (J. W. Veening, unpublished data) as the template, and inserted into the L. lactis integration vector pSEUDO-GFP (35) as an XhoI-BamHI restriction fragment replacing the gfp-sf gene of the original vector. This yielded the pSEUDO-gfp* vector.
Pcel and Pcel* PCR fragments were amplified using primers Pllmg_0186F and Pllmg_0186R and cloned into pSEUDO-gfp* as XhoI-SphI restriction fragments. Plasmids pSEUDO::Pcel-gfp and pSEUDO::Pcel*-gfp were obtained and maintained in E. coli DH5α. One copy of each vector was integrated into the chromosome of L. lactis MGLac+, yielding strains MGLac+ Pcel-gfp and MGLac+ Pcel*-gfp.
Primer extension.
L. lactis MGLac+ was grown in M17 medium supplemented with 1% cellobiose, and cells were harvested at mid-exponential growth phase (OD600 of 0.6). RNA was isolated using a High Pure RNA isolation kit and protocol (Roche Applied Science). To determine the transcription start site of the cel cluster, a 5′/3′ RACE kit (Roche Applied Science) was used according to the manufacturer's instructions. mRNA transcribed from Pcel* was converted into cDNA by reverse transcription, and the 5′ end of the cDNA strand was subsequently labeled with a stretch of adenines. Primers 0186_1Rev, 0186_2Rev, 0186_3Rev, and 0186_4Rev were used for nested PCR to amplify the 5′ end of the cDNA obtained from the llmg_0186 transcript. The PCR product was sequenced by Macrogen Inc. (South Korea), using primers 0186_3Rev and 0186_4Rev. The first base of the transcript was determined according to its position relative to the poly(A) label.
Fluctuation analysis.
Fluctuation analysis (27) was performed as described previously (4) by inoculating 24 individual 200-μl cultures of MGwt with about 200 cells. Growth under nonselective conditions to full cell density was carried out in CDM PC medium supplemented with 25 mM glucose (CDM-glu). A fraction of each culture was plated on CDM supplemented with 12.5 mM lactose or CDM-glu supplemented with rifampin (50 mg/ml), and after 1 day of incubation at 30°C, the number of CFU was determined. The total number of cells plated was determined by plating dilutions of the cultures on CDM-glu. Mutation frequencies were calculated as described previously (4).
Enzyme assay and fluorescence intensity measurements.
L. lactis strains were grown in M17 medium supplemented with either 1% (wt/vol) glucose, lactose, cellobiose, or galactose; cells were harvested at the exponential phase of growth. β-Galactosidase activity was determined on cell suspensions that were permeabilized by chloroform as described previously (19).
To follow fluorescence intensity changes during growth, cultures of 200 μl were grown in CDM PC in 96-well microtiter plates at 30°C and monitored with an Infinite 200 Pro microplate spectrophotometer (Tecan Group Ltd., Mannedorf, Switzerland). Growth was monitored by measuring the OD600. The fluorescence intensity of GFP was monitored using an excitation wavelength of 485 nm and an emission wavelength of 535 nm. Fluorescence intensity was corrected by subtracting the autofluorescence of non-GFP-containing cells and the growth medium.
RESULTS
Transcriptome analysis of lactose-utilizing L. lactis MG1363.
When L. lactis MG1363 (MGwt) was inoculated into M17 medium supplemented with lactose (LM17), it initially grew to an optical density at 600 nm of approximately 0.3, after which a prolonged lag phase of about 90 h followed (Fig. 2A). Because the initial growth also occurred in M17 broth that was not supplemented with a carbon source, this growth is attributable to undefined carbon sources in the rich M17 medium. L. lactis MGwt cultures reproducibly started to grow after the prolonged lag phase, which was always followed by outgrowth on lactose (see Fig. S3 in the supplemental material). To identify genes involved in the appearance of the lactose-positive phenotype, DNA microarray analyses were performed in which the transcriptome of MGwt growing on undefined carbon sources in M17 medium (Fig. 2A, area I) was compared to that of later-appearing lactose-positive MG1363 (MGLac+) cells (Fig. 2A, area II). The results revealed two clusters with highly altered gene expression levels: cel and gal (see Fig. S4).
Fig 2.
(A) Growth profile of MG1363 in M17 medium supplemented with 0.5% lactose and sampling for transcriptome analysis. Growth profiles of two biological replicates are shown. Two samples of each culture were taken at time points I and II for transcriptome analysis. (B) Schematic representation of the cel cluster and significant expression changes of the genes during growth of MG1363 Lac+ on lactose compared to growth on residual carbon sources in the rich M17 medium. Arrow, putative promoter; lollipop, putative terminator structure. (C) Intragenic region containing the promoter sequence of the cel cluster. The −35 and −10 elements, the transcription start site (+1), and the ATG start codon are shown in gray. The G at position 174861 is underlined.
The galPMKTE operon, encoding enzymes for the Leloir pathway of galactose utilization, was upregulated approximately 21-fold during growth on lactose. Upregulation of the gal operon suggests that the galactose moiety of lactose is metabolized via the Leloir pathway in the tagatose 6-phosphate pathway-negative strain MG1363. Furthermore, a gene cluster coding for a putative cellobiose-specific PTS IIC component, i.e., a CelB-encoding gene (llmg_0187), was upregulated around 62-fold (Fig. 2B). Downstream of this gene cluster lays a putative phospho-β-glucosidase (BglS)-encoding gene. The higher expression level of bglS in the lactose-grown cells suggests that the enzyme is responsible for cleavage of lactose after its uptake. Phospho-β-glycohydrolases of family I (16) have a broad specificity. Structural similarities of phospho-β-glucosidase and phospho-β-galactosidase enable these enzymes to substitute for each other in catalytic reactions, as has been shown for a lacG-deficient L. lactis MG5267 derivative, which is still able to ferment lactose due to a cryptic phospho-β-glucosidase activity induced by lactose and cellobiose (48). Recently, it was demonstrated that although BglS shows phospho-β-glucosidase activity, it is also the main enzyme for lactose utilization in L. lactis IL1403 (1). In this strain, lactose is transported via a cellobiose PTS, consisting of CelB, PtcA, and PtcB. The high expression levels of the celB and bglS genes seen here suggest that an analogous system can become active in L. lactis MG1363.
A specific point mutation in the celB promoter region leads to a Lac+ phenotype.
To examine whether genetic changes had occurred, the promoter regions of various operons coding for enzymes and transporters involved in sugar metabolism were sequenced from L. lactis MGLac+. No changes in nucleotide sequence were found in the promoter regions of ptcC, ptcAB, galPMKTE, and ptnABCD. However, a single nucleotide change, from guanine to adenine, was observed at position 174861 relative to the origin of replication (ori) in the published genome sequence of L. lactis MG1363 (56) (GenBank accession no. NC_009004). According to the genome sequence, this nucleotide is in the upstream region of the celB-containing gene cluster.
Since the occurrence of the Lac+ phenotype was reproducible in batch cultures, we suspected a high mutation frequency and carried out a fluctuation analysis (27). For this analysis, 24 individual cultures were grown under nonselective conditions on CDM-glucose to full cell density and subsequently plated on CDM-lac agar plates containing lactose as the carbon source. Each of the 24 cultures was started with approximately 200 cells, making it highly unlikely that the Lac+ phenotype originated from the preculture. Single colonies picked from 10 individual cultures were sequenced, and all revealed the identical point mutation G → A at position 174861 (Fig. 2C). The fluctuation analysis revealed that the number of lactose-positive colonies was much higher than the number of rifampin-resistant colonies (see Fig. S5 in the supplemental material). Mutation rates based on rifampin resistance are relatively high in general because rifampin resistance can be achieved through many different mutations in the RNA polymerase β subunit (RpoB). Binding of rifampin to the RpoB molecule involves 12 amino acid residues. Mutagenesis of each of these residues, except for one, generates a resistant phenotype (52). The fact that we found Lac+ mutants repeatedly with the same nucleotide change at a much higher frequency than that of rifampin resistance indicates that the mutation frequency at this particular locus is increased compared to that for other loci in the genome, such as the rpoB gene.
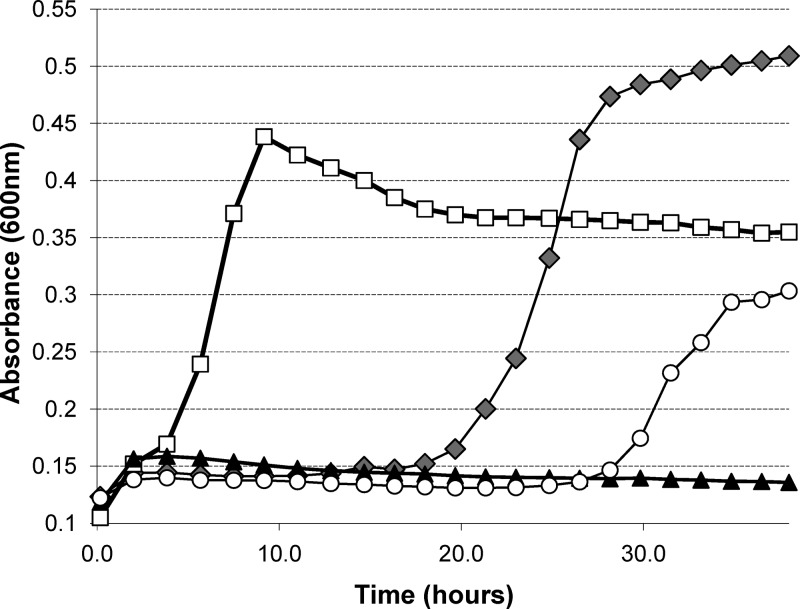
Since CelB is a putative cellobiose transporter, we tested if cultivation of L. lactis MGwt on this sugar would also select for the mutation described above. MGwt is able to transport and utilize cellobiose, but only after a prolonged lag phase (around 20 h), while MG Lac+ starts growing in CDM supplemented with cellobiose (CDM-cel) almost immediately after inoculation (Fig. 3). On M17 plates supplemented with cellobiose, L. lactis MGwt formed two types of colonies overnight: small transparent colonies resulted from the limited growth on undefined carbon sources available in rich M17 medium, while bigger and thicker colonies utilized cellobiose. Five individual large colonies were subjected to sequence analysis, and the same G → A point mutation was observed at position 174861. Thus, the availability of either cellobiose or lactose as the only carbon source in M17 medium repeatedly led to the selection of an identical mutation in the promoter region of the cel cluster. This suggests that the identified change constitutes a promoter up-mutation activating expression of celB, which allows utilization of lactose and more effective metabolism of cellobiose.
Fig 3.
Growth profiles of lactococcal strains in CDM-cel. Open squares, MGLac+; closed diamonds, MGwt; open circles, MGLac+ ΔcelB; closed triangles, MGLac+ ΔptcBA.
Mutation in the cel promoter region activates transcription of the cryptic cel cluster.
To assess the effect of the point mutation at position 174861 on the activity of the cel promoter, promoter-lacZ and promoter-gfp fusions were made. The wild-type promoter (Pcel) or a promoter with the G-to-A transition at position 174861 (Pcel*) was fused to the E. coli lacZ gene in the low-copy-number vector pILORI4 (25). These constructs were then introduced into an L. lactis MGLac+ derivative (harboring a G-to-A transition at nucleotide [nt] 174861). LacZ activity was measured after growth of strains MGLac+ Pcel-lacZ and MGLac+ Pcel*-lacZ in the presence of different sugars (Fig. 4A). The results demonstrate that the G-to-A transition is sufficient for promoter activation and represents a cel promoter up-mutation: while Pcel does not drive lacZ transcription at a detectable level, Pcel* is most active in the presence of lactose or cellobiose. In glucose- or galactose-containing medium, only a basal level of LacZ activity was detected. To verify these results without using a system involving LacZ, a β-galactosidase that might influence the metabolism of lactose in the tested strains, the green fluorescent protein-encoding gene was cloned downstream of Pcel or Pcel*. One copy of each construct was integrated into the chromosomes of L. lactis MGLac+. The obtained results were similar to those of the promoter-lacZ fusion experiments: while fluorescence of Pcel-gfp was undetectable, gfp expression driven by Pcel* was induced by cellobiose or lactose (Fig. 4B). Glucose seems to have a suppressive effect on Pcel* activity, while lactose has the strongest inducing effect on this promoter.
Fig 4.
(A) Specific β-galactosidase activity (Miller units) of HB21 containing the Pcel-lacZ or Pcel*-lacZ transcriptional fusion, grown in M17 medium supplemented with 1% glucose, galactose, cellobiose, or lactose. (B) Growth and fluorescence profiles of L. lactis HBP cel*-gfp in CDM supplemented with 1% lactose (squares), 1% cellobiose (triangles), or 1% glucose (circles). Open symbols, growth profile (OD600); closed symbols, fluorescence intensity (arbitrary units [a.u.]).
The mutation in Pcel creates a new −10 element.
In L. lactis MGwt, only −35-like elements, i.e., TTGCAA and TTGAAA, are present in the celB promoter region, while a sequence homologous to the consensus −10 element at a proper distance from a −35 element (16 to 18 nt) is not found. Sequence analysis indicated that the identified mutation might create a −10 element in the celB promoter region (Pcel*). To substantiate this notion, primer extension was used to determine the transcription start of the activated cel cluster. The results show that the transcription start site in Pcel* is at an A residue, 11 bp downstream of the transition, which apparently creates a functional −10 sequence: TGTCAT becomes TATCAT (Fig. 2C). Transition of G to A is the only mutation which is able to create a −10 element in a single step and generates a strong promoter together with the upstream −35 TTGCAA.
L. lactis MGLac+ cel cluster is under carbon catabolite repression (CCR).
The Web-based regulon prediction tool PePPeR (A. de Jong, H. Pietersma, M. Cordes, O. P. Kuipers, and J. Kok, submitted for publication) was used to analyze the upstream region of the cel cluster. Two putative CcpA-binding catabolite-responsive elements (cre sites) were found in Pcel (Fig. 5).
Fig 5.
Upstream regions of the cel cluster in L. lactis strains. The −35 and −10 elements are indicated in boxes, and the transcriptional start site is indicated as +1. Putative cre sites are underlined. Stars indicate single nucleotide deviations from the MG1363 sequence at the cre and −10 sites. MG5267 Cel+ is a spontaneous cellobiose-utilizing isolate of MG5267.
To test whether the cel cluster is under the control of CcpA-mediated CCR, the MGLac+ Pcel*-gfp ccpA deletion strain was constructed. The growth of this strain was slower than that of the wild type, as has been reported previously for MG1363 ΔccpA (57). In contrast to MGLac+ Pcel*-gfp, MGLac+ Pcel*-gfp ΔccpA expressed GFP constitutively, demonstrating that CcpA is involved in the regulation of transcription from Pcel*. However, the green fluorescence driven from Pcel* in the ccpA deletion strain was much higher in the presence of cellobiose or lactose than in the presence of glucose, supporting our previous observations that cellobiose and lactose act as inducers of Pcel* (data not shown).
L. lactis MGLac+ imports lactose via a PTS consisting of CelB-PtcA-PtcB.
To verify the role of CelB in lactose transport, a celB deletion was introduced into L. lactis MGLac+. The resulting strain, MGLac+ ΔcelB, was unable to grow on lactose (data not shown). Since CelB is annotated as a cellobiose transporter, we tested the ability of MGLac+ to grow on this sugar. While wild-type L. lactis MG1363 is characterized by a long lag phase of more than 20 h on CDM supplemented with cellobiose (CDM-cel), MGLac+ reaches stationary phase in 7 to 8 h. L. lactis MGLac+ ΔcelB showed a prolonged lag phase on CDM-cel (30 to 32 h), similar to that of L. lactis MGwt (Fig. 3). Presumably, it starts growth after induction of another chromosomally encoded cellobiose-specific PTS, namely, PtcABC (36). The reason for the long lag phase of MGwt or MGLac+ ΔcelB in CDM-cel before ptcBAC becomes active and starts transporting cellobiose is unclear. Possibly, the long activation time of ptcBAC is caused by tight regulation of the system (presumably by CCR). It has been shown that mutations in the ptcC cre site drastically shorten the lag phase of strain NZ9000 in CDM supplemented with cellobiose (26).
To complement the deletion of celB, we placed the gene under the nisin-inducible PnisA promoter in pNZ8048 (8) and introduced it in L. lactis MGLac+ nisRK ΔcelB. This strain carries the chromosome-integrated nisRK two-component system, required for nisin-inducible transcription from PnisA. The resulting strain was able to grow on lactose as the sole carbon source only in the presence of nisin, confirming the role of celB in lactose transport. Additionally, this celB complementation strain did not show a long lag phase during growth on cellobiose (data not shown).
PEP-PTS transporters are usually composed of at least three proteins or domains: a membrane-integrated sugar-specific IIC element and cytoplasmic IIA and IIB enzymes, which are responsible for transfer of a phosphoryl group to the sugar molecule (39a). It has previously been shown that the PtcAB proteins form a cellobiose-specific PTS with CelB in L. lactis IL1403 (1). To test whether this is also the case for L. lactis MG1363, we made a ptcAB knockout in the Pcel*-carrying strain. Growth experiments showed that deletion of ptcAB completely abolishes growth on lactose or cellobiose in this genetic background (Fig. 3). Therefore, we concluded that PtcAB and CelB form a complete PTS specific for lactose and cellobiose in L. lactis MGLac+.
MGLac+ uses a novel pathway of lactose utilization.
The plasmid-free L. lactis strain MG1363 lacks the tagatose 6-phosphate pathway for lactose utilization, which is normally present on plasmid pLP712 in the parental strain, NCDO712 (13). To investigate the metabolic route of lactose degradation after its internalization by PtcAB-CelB, the fate of 13C-labeled lactose was followed by NMR spectroscopy in L. lactis MGLac+ cells precultured in either CDM-cel or CDM-lac. Additionally, DNA microarrays were employed to compare the transcriptome of L. lactis MGLac+ cells growing in CDM-lac with the transcriptome of cells growing in CDM-cel. We expected that in both cultures, the lactose/cellobiose uptake system would be highly expressed, while only in lactose-grown cells would the genes involved in further metabolism of lactose be upregulated.
When MGLac+ cells were precultured on cellobiose and then exposed to lactose, they did not fully metabolize lactose (Fig. 6). Only 30% of the carbohydrate was used, and accumulation of lactose 6-phosphate and galactose 6-phosphate was observed. The fermentation end product lactate accounted for only 7.5% of the consumed lactose. Upon lactose entry into the cell, it was phosphorylated on the galactose moiety and then cleaved to glucose and galactose 6-phosphate. The latter was dephosphorylated to galactose, which accumulated in the medium and was not fermented further. For uptake and complete utilization of galactose, the Leloir pathway should be active, but in cellobiose-grown cells the gal operon is not expressed. Lactose catabolism is heavily impaired in these cells and stops, presumably because of inhibition by one of the accumulated phosphorylated products (2, 11, 40).
Fig 6.
Metabolism of lactose in suspensions of nongrowing cells of L. lactis HB21. The graphs show the kinetics of [1-13C]Lac (15 mM) consumption, pools of intracellular metabolites, and lactate formation in resting cells of L. lactis HB21 precultured in CDM-cel (A) or CDM-lac (B) under anaerobic conditions at 30°C without pH control, as monitored by in vivo 13C-NMR. Gray triangles, lactose; black circles, lactose 6-phosphate; gray diamonds, β-galactose 6-phosphate; gray squares, α-galactose 6-phosphate; gray circles, lactate.
In contrast, cells precultured on lactose can effectively metabolize this sugar and convert both moieties to fermentation end products, showing a mixed-acid fermentation pattern (a mixture of lactate, acetate, 2,3-butanediol, and ethanol is produced). The Leloir pathway is activated in these cells during preculture on lactose.
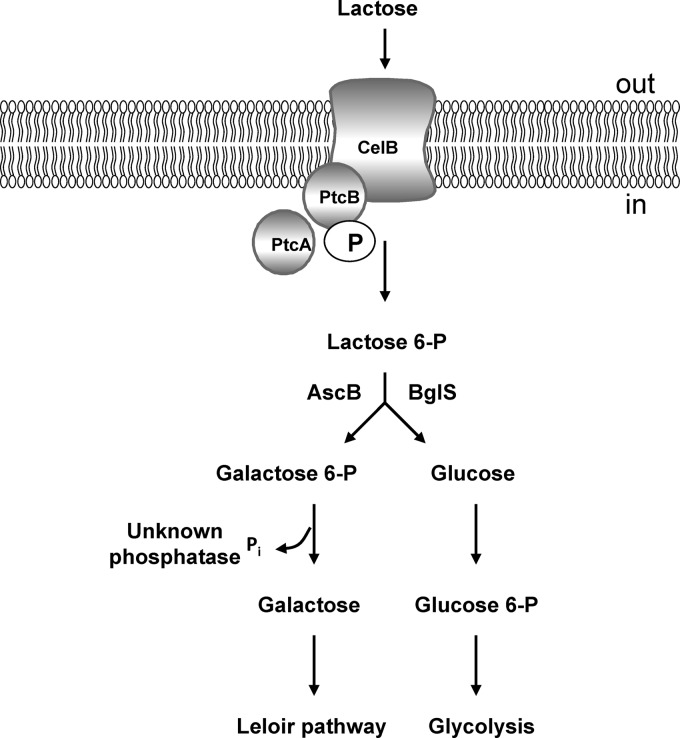
Accumulation of unphosphorylated galactose suggests the existence of an unusual dephosphorylation step prior to the Leloir pathway reactions, which has been shown to occur in NZ9000 during galactose metabolism (29). However, such a route of lactose utilization, in which the uptake by PTS is coupled to the Leloir pathway reactions via the galactose moiety dephosphorylation step, has not been described before (Fig. 7). The existence of a lactococcal cytosolic enzyme with galactose 6-phosphatase activity has been suggested earlier (50, 50a, 51). Our unsuccessful attempt to identify the phosphatase involved by transcriptome analyses suggests that the enzyme is expressed constitutively, which would be consistent with the work of Thompson and Chassy (51). It is known that sugar phosphatases often have broad specificity and might perform multiple functions in the cell. It has been suggested earlier that they might play an important role in the detoxification of potentially lethal sugar phosphates and maintaining the concentration of intracellular hexose phosphates within physiologically acceptable limits (50a). Therefore, we expect that this enzyme is present in other L. lactis strains, in addition to those possessing the Tag 6-P pathway.
Fig 7.
Proposed route for lactose uptake and further catabolism in L. lactis MG1363 Lac+. After internalization via the PTSLac-Cel (PtcBA-CelB), lactose is phosphorylated on its galactose moiety and hydrolyzed by phospho-β-glucosidases (BglS and AscB) into glucose and galactose 6-phosphate. After dephosphorylation by an unknown phosphatase, galactose is further metabolized by the Leloir pathway enzymes, and the glucose moiety enters glycolysis.
Lactose induces expression of additional enzymes in L. lactis MGLac+.
NMR spectroscopy revealed that L. lactis MGLac+ precultured in either CDM-cel or CDM-lac utilizes lactose at the same maximal rate, implying that the transporter is expressed in both types of cells and that the import of lactose is not the metabolic bottleneck. We noted the lower level of accumulation of lactose 6-phosphate in cells grown on lactose, suggesting that β-phospho-galactosidase and/or phosphatase is also upregulated under these conditions. Additionally, we were not able to detect accumulation of galactose in lactose-precultured cells. This implies that the Leloir pathway enzymes are present and rapidly metabolize galactose resulting from the dephosphorylation step. We cannot exclude the possibility of accumulation of galactose in lactose-grown cells, but its intracellular concentration is below the detection limit for intracellular metabolite resonances detected in the anomeric region of the 13C spectrum, which is 1 mM.
Consistent with our metabolic measurements, gene expression analyses showed that the gal operon is upregulated in cells grown on lactose. Curiously, one of the most upregulated genes in lactose-grown cells was arb (ascB), encoding a putative 6-phospho-β-glucosidase that belongs to glycosyl hydrolase family I. Activation of a cryptic arbutin, salicin, and cellobiose transport and metabolism operon, asc, in a cel (which consists of celABC genes encoding a PTS for β-glucosides)- and bgl (bglGFB, encoding enzymes for β-glucoside utilization)-deficient mutant by insertion of IS186 has been described for E. coli K-12 (15). Surprisingly, cellobiose does not activate the ascB promoter as strongly as lactose does in L. lactis MGLac+. Growth experiments with an L. lactis MGLac+ bglS knockout strain confirmed the presence of another enzyme(s) with β-phospho-galactosidase specificity: MGLac+ ΔbglS was still able to grow in medium containing lactose as the sole carbon source (data not shown).
DISCUSSION
L. lactis is used extensively in the dairy industry, but it has been suggested that this bacterium originates from the plant environment. Comparative genomics revealed that the transition from the plant environment to the dairy environment is characterized by the silencing of genes that are dispensable in the latter niche (46). In particular, genes involved in the utilization of typical plant carbohydrates, such as cellobiose, raffinose, and stachyose, are affected by this genome decay. The propagation of an L. lactis plant isolate for 1,000 generations in milk resulted in the downregulation of several genes involved in the utilization of plant polymers (4).
In this study, we describe how one nucleotide change in the promoter region of the cellobiose and lactose utilization cluster cel of L. lactis MG1363 creates a −10 element at an optimal distance from an existing −35 element, by which a fully active promoter is assembled, allowing transcription of the otherwise cryptic cluster. We repeatedly and independently found a G-to-A transition upstream of llmg_0186, which is the only mutation leading in a single step to a functional −10 element.
The first gene of the cel cluster, llmg_0186, is homologous to a putative outer surface protein-encoding gene of some bacilli, streptococci, enterococci, and lactobacilli. Additionally, the N terminus of the putative product of llmg_0186 contains a glycosylhydrolase motif; therefore, it is also annotated as a putative cellobiase or cellulase (KEGG database). Using Phyre 2 (Protein Homology/analogY Recognition Engine V 2.0) server, we modeled the three-dimensional structure of the putative llmg_0186 gene product. Structure alignment revealed that with a confidence of around 97%, the N-terminal fold of the protein is similar to those of glycohydrolases, especially the catalytic region of the α-amylase family. Deletion of its homologue, ybhE, in L. lactis IL1403 did not show any significant delay in growth in media containing cellobiose, lactose, a mixture of lactose and cellobiose, salicin, or glucose (1). It is plausible that the product of llmg_0186 is not functional under laboratory conditions. It has previously been noted that plant-derived lactococci and lactobacilli often possess conserved genes encoding putative cell surface proteins for carbohydrate utilization in their genomes (47), which in this case might have decayed.
Transcription from the activated Pcel promoter leads to expression of the PTSLac-Cel and to exhibition of the lactose-utilizing phenotype of the non-lactose-utilizing L. lactis strain MG1363, as well as to a shorter lag phase in medium supplemented with cellobiose as the sole carbon source. It has been described before that preculture of L. lactis MG1363 on cellobiose results in the disappearance of the long lag phase which is usually observed in CDM-cel (26). This might be explained by the occurrence and selection of Pcel* mutants that are described here as utilizing cellobiose more rapidly.
The up-mutated promoter is not transcribed constitutively in L. lactis MGLac+. Expression of the cel cluster is under carbon catabolite repression and is induced by cellobiose or lactose present in the growth medium. Recently, it was described that the cel cluster promoter in L. lactis IL1403 cannot be activated by lactose alone (1). To induce transcription from this promoter in L. lactis IL1403, addition of a trace amount of cellobiose was required. Although both L. lactis laboratory strains show a lot of similarities, they do differ in the organization of sugar metabolism genes and the utilization of sugars (56). In Streptococcus thermophilus, the lac lactose utilization genes are activated by galactose (37). Our results show that galactose has no effect on transcription of the L. lactis MGLac+ cel cluster.
Silent genes can be activated in different ways in LAB. Integration of IS981 was reported to be responsible for transcriptional activation of the silent ldhB gene in a lactate dehydrogenase-deficient derivative of L. lactis NZ9000 (6). Insertion of IS981 into the upstream region of ldhB generated a −35 element at the correct spacing from a native −10 sequence, thus creating a functional promoter. Although IS981-mediated activation was the major mechanism of restoration of ldhB expression, more lactate dehydrogenase-positive mutants with mutations of variable nature and position were isolated under selective conditions (6). Activation of silent gal operon genes by three classes of point mutations in the S. thermophilus galK-galR promoter region has been described previously (55). A mutation of G to A in one of the classes results in a −10 box with greater sequence similarity to the −10 consensus sequence (TACGAT changes into TACAAT). However, to the best of our knowledge, the formation of a −10 element, and hence a functional promoter, by one particular mutation has not been reported before. We noted a higher mutation frequency at position −11 in the cel promoter region. The mechanism by which the G-to-A substitution always occurs at this locus remains unclear. It is known that transitions are the most frequent type of point mutations because of the molecular mechanism by which they occur. Transitions are induced by spontaneous tautomeric shifts, i.e., transient changes to an alternative form of a nucleobase molecule which lead to base mispairing during replication. The structural DNA properties and/or the sequence context might play a role in increased mutability of the region (18, 41). However, sequence analysis of the Pcel region did not reveal long inverted repeats or AT-rich regions that could elevate the error rate of DNA polymerase during replication or prevent the mismatch repair system from fixing the incorporation of the mispaired base. Despite numerous attempts, we were not able to create conditions that would allow us to determine the frequency of reversion of an active Pcel* promoter to a silenced promoter in L. lactis MGLac+. Unfortunately, our data are not sufficient to unravel the mechanism of mutation formation.
To date, genome sequences of seven L. lactis strains have been published. All of them contain the cel cluster described here, and all of them carry a gene coding for a protein with unknown function that is homologous to Llmg_0186 of L. lactis MG1363, as well as celB, encoding an annotated cellobiose-specific EIIC PTS component. All of the seven upstream regions are very similar (81 to 100% nt sequence identity), and four of the seven regions contain an A instead of G at position −11 (Fig. 5). Expression of cellobiose catabolic genes is most likely advantageous in a plant environment, where cellobiose may be abundant, but could be a burden during growth on a different carbon source. L. lactis subsp. lactis KF147 was isolated from mung bean sprouts (45). The ability to grow on plant cell wall degradation products such as cellobiose is important for this organism, and thus it seems not surprising that it possesses a cel operon with an active promoter. IL1403, another laboratory strain that is also an L. lactis subsp. lactis strain, was originally isolated from a dairy niche, yet this strain is cellobiose positive and its cel promoter sequence is almost identical to that of KF147 (5). Lactococcus lactis subsp. lactis CV56 is a human isolate from the vaginas of healthy women (12). It belongs to L. lactis subsp. lactis, and its cel promoter sequence is highly homologous to that of IL1403, but at position −11 the promoter harbors a G instead of an A.
L. lactis NZ9000, an L. lactis MG1363 derivative, possesses a G at position −11 of Pcel; nevertheless, NZ9000 is a cellobiose-positive strain. This phenotype is caused by yet another mechanism: the cre site in the ptcC promoter, driving expression of the cellobiose/glucose transporter IIC component PtcC, is disrupted by two mutations. CcpA-mediated catabolite repression is thus relieved, and cells are able to import cellobiose via PtcABC and to further utilize this sugar (26). Curiously, the G-to-A transition found in MGLac+ occurs in one of the predicted cre sites in the Pcel region. Although the change occurs in a position of cre that is not highly conserved, it might additionally influence the activity and regulation of the promoter.
Lactose can be taken up and metabolized using two main systems in L. lactis. Most dairy strains possess a plasmid-encoded pathway, consisting of lactose transport via PTSLac, cleavage of lactose 6-P by phospho-β-galactosidase, and subsequent galactose moiety utilization via the tagatose 6-phosphate pathway (10). In the second case, lactose is taken up by a lactose-specific permease (21). Internalized unphosphorylated lactose is then cleaved by β-galactosidase, and the resulting galactose molecule enters the Leloir pathway (54). The plasmid-free L. lactis laboratory strain MG1363 possesses yet a different pathway. A mutation occurring in the silent promoter of the cel cluster allows transcription and translation of the cellobiose-specific PTS IIC component CelB, which together with the other two cellobiose PTS components, PtcB and PtcA, forms a PTSLac-Cel. Internalized lactose is phosphorylated on its galactose moiety, as is typical for PTSLac, and cleaved by unspecific P-β-glucosidases, and then the galactose moiety is dephosphorylated again and directed to the Leloir pathway. The way that L. lactis MG1363 couples transport via the PTS to the subsequent metabolism of galactose via the Leloir pathway has not been described before.
L. lactis MG5267 is a derivative of MG1363 with chromosomally integrated lac genes. Plating on a solid medium supplemented with cellobiose allowed us to isolate Pcel* up-mutated colonies. The growth of these MG5267 Cel+ isolates and the MG5267 Cel− (wt) strain was compared in two types of media supplemented with lactose: CDM-lac and LM17. We were not able to observe any significant differences in growth under the conditions tested; thus, the presence of two lactose transporters, namely, CelB-PtcBA and PTSLac-Cel, does not increase the rate of lactose utilization, presumably because of the limited capacity of the enzymes that catalyze the downstream reactions. It is very likely that all of the transported lactose is cleaved to glucose and galactose 6-P, which is rapidly directed to the Tag 6-P pathway. If the first reaction of the tagatose 6-P pathway is more efficient than the dephosphorylation reaction (higher affinity of galactose 6-P isomerase for the common substrate galactose 6-P), then galactose 6-P is utilized largely via the tagatose 6-P pathway reactions. It is worth mentioning that the latter pathway is more energetically efficient than our described combination of uptake by the PTSLac-Cel, dephosphorylation, and Leloir pathway reactions.
We have demonstrated a strong selection pressure on a single point mutation in the promoter region of the cel cluster in L. lactis. This mutation gives a strong selective advantage for either MG1363 (Lac−) cultured on lactose or MG5267 (Lac+) grown in cellobiose. However, experiments with MG5267 suggest that as long as one of the classical systems for lactose transport and metabolism is present in a cell, the cel cluster, providing an unusual lactose transport and utilization pathway, remains silent.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ákos T. Kovács for helpful discussions. A. R. Neves thanks P. Gaspar (ITQB-UNL) for technical support.
This work was supported by a grant from the NWO-STW Technology Foundation (project 10619).
The NMR spectrometers are part of The Portuguese National NMR Network (program REDE/1517/RMN/2005), supported by the Programa Operacional Ciência e Inovação (POCTI) 2010 and the Fundação para a Ciência e a Tecnologia (FCT). The NMR work was supported in part by FCT grant PTDC/EBB-EBI/113727/2009 and through grant PEst-OE/EQB/LA0004/2011.
Footnotes
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Aleksandrzak-Piekarczyk T, Polak J, Jezierska B, Renault P, Bardowski J. 2011. Genetic characterization of the CcpA-dependent, cellobiose-specific PTS system comprising CelB, PtcB and PtcA that transports lactose in Lactococcus lactis IL1403. Int. J. Food Microbiol. 145:186–194 [DOI] [PubMed] [Google Scholar]
- 2. Andersen HW, Solem C, Hammer K, Jensen PR. 2001. Twofold reduction of phosphofructokinase activity in Lactococcus lactis results in strong decreases in growth rate and in glycolytic flux. J. Bacteriol. 183:3458–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson DG, McKay LL. 1977. Plasmids, loss of lactose metabolism, and appearance of partial and full lactose-fermenting revertants in Streptococcus cremoris B1. J. Bacteriol. 129:367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachmann H, et al. 2012. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 22:115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bolotin A, et al. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bongers RS, et al. 2003. IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate dehydrogenase-deficient strain of Lactococcus lactis. J. Bacteriol. 185:4499–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Defoor E, Kryger MB, Martinussen J. 2007. The orotate transporter encoded by oroP from Lactococcus lactis is required for orotate utilization and has utility as a food-grade selectable marker. Microbiology 153:3645–3659 [DOI] [PubMed] [Google Scholar]
- 8. de Ruyter PG, Kuipers OP, de Vos WM. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Vos WM, Simons G. 1988. Molecular cloning of lactose genes in dairy lactic streptococci: the phospho-beta-galactosidase and beta-galactosidase genes and their expression products. Biochimie 70:461–473 [DOI] [PubMed] [Google Scholar]
- 10. de Vos WM, Vaughan EE. 1994. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol. Rev. 15:217–237 [DOI] [PubMed] [Google Scholar]
- 11. Fraenkel DG. 1968. The accumulation of glucose 6-phosphate from glucose and its effect in an Escherichia coli mutant lacking phosphoglucose isomerase and glucose 6-phosphate dehydrogenase. J. Biol. Chem. 243:6451–6457 [PubMed] [Google Scholar]
- 12. Gao Y, et al. 2011. Complete genome sequence of Lactococcus lactis subsp. lactis CV56, a probiotic strain isolated from the vaginas of healthy women. J. Bacteriol. 193:2886–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gasson MJ. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a. Goel A, Santos F, Vos WM, Teusink B, Molenaar D. 2012. Standardized assay medium to measure Lactococcus lactis enzyme activities while mimicking intracellular conditions. Appl. Environ. Microbiol. 78:134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grossiord BP, Luesink EJ, Vaughan EE, Arnaud A, de Vos WM. 2003. Characterization, expression, and mutation of the Lactococcus lactis galPMKTE genes, involved in galactose utilization via the Leloir pathway. J. Bacteriol. 185:870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hall BG, Xu L. 1992. Nucleotide sequence, function, activation, and evolution of the cryptic asc operon of Escherichia coli K12. Mol. Biol. Evol. 9:688–706 [DOI] [PubMed] [Google Scholar]
- 16. Henrissat B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holo H, Nes IF. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195–199 [DOI] [PubMed] [Google Scholar]
- 18. Hudson RE, Bergthorsson U, Roth JR, Ochman H. 2002. Effect of chromosome location on bacterial mutation rates. Mol. Biol. Evol. 19:85–92 [DOI] [PubMed] [Google Scholar]
- 19. Israelsen H, Madsen SM, Vrang A, Hansen EB, Johansen E. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jensen PR, Hammer K. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59:4363–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kashket ER, Wilson TH. 1973. Proton-coupled accumulation of galactoside in Streptococcus lactis 7962. Proc. Natl. Acad. Sci. U. S. A. 70:2866–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281–291 [DOI] [PubMed] [Google Scholar]
- 23. Kuipers OP, et al. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie Van Leeuwenhoek 82:113–122 [PubMed] [Google Scholar]
- 24. Kuipers OP, De Ruyter PGGA, Kleerebezem de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15–21 [Google Scholar]
- 25. Larsen R, Buist G, Kuipers OP, Kok J. 2004. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. J. Bacteriol. 186:1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Linares DM, Kok J, Poolman B. 2010. Genome sequences of Lactococcus lactis MG1363 (revised) and NZ9000 and comparative physiological studies. J. Bacteriol. 192:5806–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luria SE, Delbruck M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neves AR, et al. 2006. The alpha-phosphoglucomutase of Lactococcus lactis is unrelated to the alpha-d-phosphohexomutase superfamily and is encoded by the essential gene pgmH. J. Biol. Chem. 281:36864–36873 [DOI] [PubMed] [Google Scholar]
- 29. Neves AR, et al. 2010. Towards enhanced galactose utilization by Lactococcus lactis. Appl. Environ. Microbiol. 76:7048–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neves AR, et al. 1999. In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 64:200–212 [DOI] [PubMed] [Google Scholar]
- 31. Neves AR, et al. 2000. Metabolic characterization of Lactococcus lactis deficient in lactate dehydrogenase using in vivo 13C-NMR. Eur. J. Biochem. 267:3859–3868 [DOI] [PubMed] [Google Scholar]
- 32. Neves AR, et al. 2002. Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? Kinetics of NAD(+) and NADH pools determined in vivo by 13C NMR. J. Biol. Chem. 277:28088–28098 [DOI] [PubMed] [Google Scholar]
- 33. Reference deleted.
- 34. Payne J, MacCormick CA, Griffin HG, Gasson MJ. 1996. Exploitation of a chromosomally integrated lactose operon for controlled gene expression in Lactococcus lactis. FEMS Microbiol. Lett. 136:19–24 [DOI] [PubMed] [Google Scholar]
- 35. Pinto JP, et al. 2011. pSEUDO, a genetic integration standard for Lactococcus lactis. Appl. Environ. Microbiol. 77:6687–6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pool WA, Neves AR, Kok J, Santos H, Kuipers OP. 2006. Natural sweetening of food products by engineering Lactococcus lactis for glucose production. Metab. Eng. 8:456–464 [DOI] [PubMed] [Google Scholar]
- 37. Poolman B. 1993. Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 12:125–147 [DOI] [PubMed] [Google Scholar]
- 38. Poolman B, Royer TJ, Mainzer SE, Schmidt BF. 1989. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J. Bacteriol. 171:244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poolman B, Smid EJ, Veldkamp H, Konings WN. 1987. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J. Bacteriol. 169:1460–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a. Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prasad C, Freese E. 1974. Cell lysis of Bacillus subtilis caused by intracellular accumulation of glucose-1-phosphate. J. Bacteriol. 118:1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rogozin IB, Pavlov YI. 2003. Theoretical analysis of mutation hotspots and their DNA sequence context specificity. Mutat. Res. 544:65–85 [DOI] [PubMed] [Google Scholar]
- 42. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43. Santos H, Turner DL. 1986. 13C and proton NMR studies of horse cytochrome c. Assignment and temperature dependence of methyl resonances. FEBS Lett. 194:73–77 [DOI] [PubMed] [Google Scholar]
- 44. Reference deleted.
- 45. Siezen RJ, et al. 2010. Complete genome sequence of Lactococcus lactis subsp. lactis KF147, a plant-associated lactic acid bacterium. J. Bacteriol. 192:2649–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Siezen RJ, et al. 2008. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 74:424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Siezen R, et al. 2006. Lactobacillus plantarum gene clusters encoding putative cell-surface protein complexes for carbohydrate utilization are conserved in specific gram-positive bacteria. BMC Genomics 7:126 doi:10.1186/1471-2164-7-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simons G, Nijhuis M, de Vos WM. 1993. Integration and gene replacement in the Lactococcus lactis lac operon: induction of a cryptic phospho-beta-glucosidase in LacG-deficient strains. J. Bacteriol. 175:5168–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Solem C, Defoor E, Jensen PR, Martinussen J. 2008. Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis. Appl. Environ. Microbiol. 74:4772–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thompson J. 1980. Galactose transport systems in Streptococcus lactis. J. Bacteriol. 144:683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a. Thompson J, Chassy BM. 1982. Novel phosphoenolpyruvate-dependent futile cycle in Streptococcus lactis: 2-deoxy-D-glucose uncouples energy production from growth. J. Bacteriol. 151:1454–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thompson J, Chassy BM. 1983. Intracellular hexose-6-phosphate:phosphohydrolase from Streptococcus lactis: purification, properties, and function. J. Bacteriol. 156:70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tupin A, et al. 2010. Resistance to rifampicin: at the crossroads between ecological, genomic and medical concerns. Int. J. Antimicrob. Agents 35:519–523 [DOI] [PubMed] [Google Scholar]
- 53. van Hijum SA, Garcia de la Nava J, Trelles O, Kok J, Kuipers OP. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2:241–244 [PubMed] [Google Scholar]
- 54. Vaughan EE, Pridmore RD, Mollet B. 1998. Transcriptional regulation and evolution of lactose genes in the galactose-lactose operon of Lactococcus lactis NCDO2054. J. Bacteriol. 180:4893–4902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vaughan EE, van den Bogaard PT, Catzeddu P, Kuipers OP, de Vos WM. 2001. Activation of silent gal genes in the lac-gal regulon of Streptococcus thermophilus. J. Bacteriol. 183:1184–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wegmann U, et al. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:1366–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.