Abstract
Spacecraft hardware and assembly cleanroom surfaces (233 m2 in total) were sampled, total genomic DNA was extracted, hypervariable regions of the 16S rRNA gene (bacteria and archaea) and ribosomal internal transcribed spacer (ITS) region (fungi) were subjected to 454 tag-encoded pyrosequencing PCR amplification, and 203,852 resulting high-quality sequences were analyzed. Bioinformatic analyses revealed correlations between operational taxonomic unit (OTU) abundance and certain sample characteristics, such as source (cleanroom floor, ground support equipment [GSE], or spacecraft hardware), cleaning regimen applied, and location about the facility or spacecraft. National Aeronautics and Space Administration (NASA) cleanroom floor and GSE surfaces gave rise to a larger number of diverse bacterial communities (619 OTU; 20 m2) than colocated spacecraft hardware (187 OTU; 162 m2). In contrast to the results of bacterial pyrosequencing, where at least some sequences were generated from each of the 31 sample sets examined, only 13 and 18 of these sample sets gave rise to archaeal and fungal sequences, respectively. As was the case for bacteria, the abundance of fungal OTU in the GSE surface samples dramatically diminished (9× less) once cleaning protocols had been applied. The presence of OTU representative of actinobacteria, deinococci, acidobacteria, firmicutes, and proteobacteria on spacecraft surfaces suggests that certain bacterial lineages persist even following rigorous quality control and cleaning practices. The majority of bacterial OTU observed as being recurrent belonged to actinobacteria and alphaproteobacteria, supporting the hypothesis that the measures of cleanliness exerted in spacecraft assembly cleanrooms (SAC) inadvertently select for the organisms which are the most fit to survive long journeys in space.
INTRODUCTION
The introduction of terrestrial microbiota to extraterrestrial settings could have profound repercussions on (i) the scientific integrity of in situ and sample return-based life detection experiments and (ii) the pristine condition of such environments. A key challenge for sample return missions lies in discriminating between authentic and contaminant biosignatures detected within a sample of extraterrestrial origin. The monitoring of biodiversity associated with outbound spacecraft is therefore important for the successful exploration of other solar-system bodies (e.g., Mars, Europa) (6). At present, the National Aeronautics and Space Administration (NASA) is supporting efforts to select and validate technologies capable of providing a comprehensive census of the microbes present on spacecraft surfaces. There is a great need for this capability to pursue solar system exploration with the future mission set.
Historically, bacterial endospores have been the logical candidates in considering terrestrial life forms potentially capable of surviving interplanetary transport (12, 26, 47). However, recent studies have shown that fungal cells are also capable of withstanding space conditions for extended periods of time (42), rendering these organisms also of concern to astrobiologists. Similarly, due to their ubiquitous nature and physiological flexibility, the archaeal cells have been proposed as being capable of tolerating the Martian environment (23, 34). In light of this, the breadth of current spacecraft-associated microbial diversity assessments must expand to include eukaryotes and archaea. Only by shifting the current focal plane and relative depth of field of such assessments and distributing them more uniformly across the three domains of life can a truly comprehensive census be achieved.
High-throughput next-generation sequencing and phylogenetic microarray techniques have dramatically increased the resolution and detectable spectrum of diverse microbial phylotypes from environmental samples. These and other innovative methodologies capable of providing an all-inclusive account of the “rare biosphere” (i.e., low-abundance taxa) (31, 38, 53) within a given sample are in high demand and are of utmost importance to a variety of research endeavors. The 454 tag-encoded pyrosequencing method employed in this study is an attractive tool for ascertaining the phylogenetic affiliations of members of the complex microbial communities found on spacecraft hardware. Further molecular microbiological studies and downstream computational population modeling can then facilitate inferences regarding the proportions of taxa present on the spacecraft surface that are sensitive versus resistant, transient versus resident, and ecologically inconsequential versus relevant to astrobiological endeavors. To date, an all-inclusive rRNA gene-based inventory of the total microbial population associated with spacecraft hardware has yet to be provided. We present here for the first time the results of pyrosequencing studies that comprehensively elucidate the microbial diversity (bacterial, archaeal, and fungal) associated with extremely low-biomass spacecraft surfaces.
MATERIALS AND METHODS
Sample collection. (i) Spacecraft hardware.
Samples were collected from spacecraft hardware of a recent Mars mission via wiping ∼1-m2 surface areas with sterile, water-moistened polyester wipes (catalog no. TX761; Texwipe, Upper Saddle River, NJ). After sampling, wipes were immediately placed into presterilized 0.5-liter Corning bottles. Negative controls, handling controls (sampling devices briefly exposed to the ambient sampling environment), and other reagents were also prepared and analyzed.
(ii) SACs and GSE.
Surface sampling of spacecraft assembly cleanroom (SAC) floors and ground support equipment (GSE) was performed using either premoistened polyester wipes or biological sampling kits (BiSKit; QuickSilver Analytics, Abingdon, MD) as previously described (4, 24). The two cleanroom facilities examined during this study were both certified as class 8 per ISO 14644-1. Spacecraft hardware and components were undergoing active assembly in the first cleanroom site, the Jet Propulsion Laboratory Spacecraft Assembly Facility (JPL-SAF) (sample GI-36; Table 1), whereas the second cleanroom site, JPL cleanroom 144 (samples GI-42 and GI-43; Table 1), was not supporting spacecraft assembly activities at the time of sampling but was maintaining its class 100K certification. Both of those cleanroom facilities were operated under conditions of positive air pressure, with temperatures in the range of 20 ± 4°C and relative humidity ranging from 30% to 50%. In both cases, the total hydrocarbon content of the facility air (gases and vapors) was below 15 ppm (calculated as methane content). The GSE consisted of nonflight hardware items associated with the project, employed prior to or during hardware receipt, assembly, integration, testing, storage, shipment, and prelaunch activities. All GSE materials used in direct contact with flight hardware or inside the cleanroom were inspected for compliance with visible cleanliness levels. To ensure that contamination was not transferred to flight hardware, GSE such as mating gas/liquid lines and electrical connectors were also precleaned according to established in-house protocols.
Table 1.
Sample characteristics of various spacecraft and associated surfacesa
| Sample (total area sampled) | Sample | Sampling device | Sample type | Area (m2) | Mission | Cleanroom type | Description |
|---|---|---|---|---|---|---|---|
| Group I: cleanroom types (33 m2) | |||||||
| GI-37 | 150 | BisKit | Floor 70A | 10 | None | Non-NASA | Non-NASA cleanroom (#70A) |
| GI-35-6 | 143 | BisKit | Entrance floor | 1 | None | Ordinary room | Ordinary room adjacent to JPL-SAF |
| GI-35-4 | 141 | BisKit | Shoe cleaner | 1 | None | Ordinary room | Ordinary room adjacent to JPL-SAF |
| GI-35-7 | 144 | BisKit | Floor 1 | 1 | None | Ordinary room | Ordinary room adjacent to JPL-SAF |
| GI-35-8 | 145 | BisKit | Floor 2 | 1 | None | Ordinary room | Ordinary room adjacent to JPL-SAF |
| GI-35-5 | 142 | BisKit | Air lock | 1 | None | Ordinary room | Ordinary room adjacent to JPL-SAF |
| GI-36-3 | 146 | BisKit | JPL-SAF GSE | 9 | Mars | Class 100K | During spacecraft assembly (JPL-SAF) |
| GI-36-4 | 148 | BisKit | JPL-SAF floor | 9 | Mars | Class 100K | During spacecraft assembly (JPL-SAF) |
| Group II: postcleaning vs precleaning (38 m2) | |||||||
| GI-42-1 | 155 | BisKit | Floor | 9 | None | Class 100K | JPL-144 cleanroom prior to cleaning |
| GI-42-2 | 157 | BisKit | GSE | 9 | None | Class 100K | JPL-144 cleanroom prior to cleaning |
| GI-43-1 | 159 | Polyester wipe | Floor | 10 | Mars | Class 100K | JPL-144 cleanroom postcleaning |
| GI-43-2 | 161 | Polyester wipe | GSE | 10 | Mars | Class 100K | JPL-144 cleanroom postcleaning |
| Group III: spacecraft surfaces (spore count-based determinations) (110 m2) | |||||||
| GI-16 | 124 | Polyester wipe | Spacecraft | 6 | Mars | Class 100K | No spore count |
| GI-17 | 125 | Polyester wipe | Spacecraft | 10 | Mars | Class 100K | No spore count |
| GI-25 | 133 | Polyester wipe | Spacecraft | 8 | Mars | Class 100K | No spore count |
| GI-26 | 134 | Polyester wipe | Spacecraft | 7 | Mars | Class 100K | No spore count |
| GI-27 | 135 | Polyester wipe | Spacecraft | 4 | Mars | Class 100K | No spore count |
| GI-28 | 136 | Polyester wipe | Spacecraft | 6 | Mars | Class 100K | No spore count |
| GI-29 | 137 | Polyester wipe | Spacecraft | 18 | Mars | Class 100K | No spore count |
| GI-18 | 126 | Polyester wipe | Spacecraft | 10 | Mars | Class 100K | 1 to 5 spores per m2 |
| GI-19 | 127 | Polyester wipe | Spacecraft | 14 | Mars | Class 100K | 1 to 5 spores per m2 |
| GI-20 | 128 | Polyester wipe | Spacecraft | 5 | Mars | Class 100K | 1 to 5 spores per m2 |
| GI-21 | 129 | Polyester wipe | Spacecraft | 4 | Mars | Class 100K | 1 to 5 spores per m2 |
| GI-22 | 130 | Polyester wipe | Spacecraft | 1 | Mars | Class 100K | 1 to 5 spores per m2 |
| GI-30 | 138 | Polyester wipe | Spacecraft | 13 | Mars | Class 100K | 1 to 5 spores per m2 |
| GI-32 | 140 | Polyester wipe | Spacecraft | 3 | Mars | Class 100K | 5 to 10 spores per m2 |
| GI-24 | 132 | Polyester wipe | Spacecraft | 1 | Mars | Class 100K | 300 spores per m2 |
| Group IV: spacecraft surfaces (mission component-based determinations) (52 m2) | |||||||
| GI-38 | 151 | Polyester wipe | Spacecraft | 26 | Mars | Class 100K | Cruise stage (0.2 spores per m2) |
| GI-39 | 152 | Polyester wipe | Spacecraft | 9 | Mars | Class 100K | Descent stage (0.4 spores per m2) |
| GI-40 | 153 | Polyester wipe | Spacecraft | 16 | Mars | Class 100K | Rover (0.3 spores per m2) |
| GI-41 | 154 | Polyester wipe | Spacecraft | 1 | None | Class 100K | Nonflight samples (14 spores per m2) |
JPL-SAF, Jet Propulsion Laboratory Spacecraft Assembly Facility.
The elimination or minimization of cleanroom floor surface contamination resulting from deposition of moisture, particles, dirt, grease, oil, scale, corrosion, and nonvolatile residues introduced during fabrication, assembly, integration, testing, storage, and shipping operations was carried out via manual detergent-based cleaning. A protocol using an all-purpose cleaning and degreasing agent (Kleenol 30) (catalog. no. J-CC-00040; Accurate Industrial Supply, Inc., Cerritos, CA) was used to maintain cleanliness of the floor. Typically, cleanroom surface cleaning was performed twice a day when spacecraft were actively undergoing assembly (JPL-SAF; sample GI-36). In contrast to cleanroom floors, where detergents were routinely applied, GSE surfaces were cleaned solely by wiping with 70% alcohol. Both of the cleanroom facilities examined were maintained with stringent protocols pertaining to the replacement of tacky mats and to vacuuming and mopping of floors. In addition, staff members were required to follow protocols for donning cleanroom garments and minimizing the influx of particulate matter.
Sample categorization.
Table 1 summarizes the characteristics of the various spacecraft hardware, floor, and GSE surface samples examined in this study. The samples collected and analyzed over the course of this study were segregated into four distinct categories. Group I (8 sets; 33 m2) samples were analyzed together to examine the effect of various forms of cleanroom certification (i.e., cleanliness levels) on resulting pyrosequencing data. This group consisted of floor samples from a SAC, an ordinary room adjacent to this cleanroom, and a typical non-NASA cleanroom. Group II samples (4 sets; 38 m2) were analyzed together to assess the impact of cleaning procedures on pyrosequencing profiles. This group comprised SAC floor and GSE samples collected in a facility expected to receive spacecraft hardware from identical locations both (i) prior to cleaning and (ii) 1 day after cleaning. Group III samples (15 sets; 110 m2) were analyzed together to evaluate the extent of correlation between the endospore burden associated with spacecraft hardware surfaces and the resulting operational taxonomic unit (OTU) abundance. This group comprised spacecraft hardware surface samples whose bacterial endospore burden had been previously determined (4). Group IV samples (4 sets; 52 m2) were analyzed together to investigate how pyrosequencing results differed across various mission subsystem components (e.g., cruise stage, rover). This group consisted of spacecraft hardware surface samples collected from the various components employed in the mission. In total, 31 sample sets spanning a surface area of 233 m2 were collected and analyzed for pyrosequencing-derived microbial diversity.
Sample processing.
All BiSKit samples, and the vast majority of polyester wipes examined, were processed and analyzed immediately after collection. Only the group IV wipes collected at Kennedy Space Center (group IV samples) were stored at −20°C (14 days) prior to being transported on dry ice to JPL for further processing and analysis. To facilitate recovery of microorganisms and biomolecules from the polyester matrix, 200 ml of sterile rinse solution (85 mg of potassium dihydrogen phosphate, 200 mg of Tween 80 [per liter of water], pH 7.2) was added to each polyester wipe-containing Corning bottle prior to vigorous shaking and ultrasonic treatment, as previously described (4, 24). Previous studies have demonstrated that SAC samples contain extremely low biomass and seldom yield detectable PCR amplification products (34, 56). We therefore pooled multiple samples from each sampling event. All samples were filtered using Amicon Ultra-50 Ultracel centrifugal filter tubes (Millipore, Billerica, MA), which facilitated the concentration of microbial cells, endospores, and exogenous nucleic acid fragments greater than 100 bp in size into a final volume of 500 μl. Samples were divided into two aliquots, one of which was subjected to bead-beating protocols with a FastPrep instrument (MP Biomedicals, Solon, OH) (5 m/s, 60 s) to maximize the resulting detectable microbial diversity (29). After bead beating was performed, the two aliquots were combined and subjected to total DNA purification using the Maxwell MDx automated nucleic acid extraction system (Promega, Madison, WI), in accordance with the manufacturer's instructions. DNA extracts were then stored at −20°C.
454 tag-encoded pyrosequencing analysis.
Bacterium-biased primers 28F (5′-GAG TTT GAT CNT GGC TCA G-3′) and 519R (5′-GTN TTA CNG CGG CKG CTG-3′) were used to amplify ∼500-bp fragments spanning the V1 to V3 hypervariable regions of the bacterial 16S rRNA gene. Archaeon-biased primers 341F (5′-GYG CAS CAG KCG MGA AW-3′) and 958R (5′-GGA CTA CVS GGG TAT CTA AT-3′) were used to amplify ∼600-bp fragments spanning the V3 to V5 hypervariable regions of the archaeal 16S rRNA gene. A fungus-biased primer set consisting of ITS1F (5′-CTT GGT CAT TTA GAG GAA GTA A-3′) and ITS4R (5′-TCC TCC GCT TAT TGA TAT GC-3′) was employed to amplify ∼600-bp fragments of the fungal internal transcribed spacer (ITS) region. These primer pairs were tailored for pyrosequencing by adding a fusion linker and a proprietary 12-nucleotide (12-nt) barcode sequence at the 5′ end of the forward primer and a biotin and fusion linker sequence at the 5′ end of the reverse primer (10). A HotStarTaq Plus master mix kit (Qiagen, Valencia, CA) was used to catalyze the PCR under the following thermal cycling conditions: initial denaturing at 95°C for 5 min, followed by 35 cycles of denaturing at 95°C for 30 s, annealing at 54°C for 40 s, and extension at 72°C for 1 min, finalized by a 10-min elongation at 72°C. Resulting PCR products were purified using Diffinity RapidTip (Diffinity Genomics, Inc., West Henrietta, NY) chemistry and were then pooled according to the experimental plan. Small fragments were removed with Agencourt Ampure Beads (Beckman Coulter, Brea, CA).
In preparation for FLX-Titanium sequencing (Roche, Nutley, NJ), the resulting PCR amplicon fragment size and concentration were measured using DNA 1000 chips and a Bioanalyzer 2100 automated electrophoresis station (Agilent, Santa Clara, CA) and a TBS-380 Fluorometer (Turner Biosystems, Sunnyvale, CA). The total volumes of the initial PCR product used for the subsequent emulsion PCR were 2 μl for strong positives (>10 ng/μl), 5 μl for weak positives (5 to 10 ng/μl), and 20 μl for samples that failed to yield detectable PCR products (<5 ng/μl). This normalization step minimized bias in downstream amplification favoring initially strong PCR products. Approximately 9.6 × 106 molecules of ∼600 bp of double-stranded DNA were combined with 9.6 × 106 DNA capture beads and then subjected to emulsion PCR conditions. Following recovery and enrichment, bead-attached DNA molecules were denatured with NaOH and sequencing primers were annealed. A four-region 454 pyrosequencing run was performed on a GS PicoTiterPlate (PTP) housing a Genome Sequencer FLX system in accordance with manufacturer's instructions (Roche, Nutley, NJ). Twenty-four to 30 tagged samples were applied to each of the four regions of the PTP. All pyrosequencing procedures were performed at the Research and Testing Laboratory (Lubbock, TX) in accordance with established protocols (10). None of the negative controls, handling controls (sampling devices briefly exposed to the ambient sampling environment), or reagent blanks gave rise to measurable PCR products; thus, none of those controls were subjected to pyrosequencing analysis.
Bioinformatic analyses.
Bacterial and archaeal sequences were processed and analyzed using the MOTHUR software package (51), with the AmpliconNoise algorithm implemented. Previously described standard operating procedures were followed for the analysis of sequence data in this study (50). Sequences were removed from consideration if they (i) did not contain the primer sequence, (ii) contained an uncorrectable barcode, (iii) were <200 nt in length, (iv) had homopolymers longer than 8 nt, or (v) had a quality score of <25. Unique sequences were aligned using the Greengenes reference alignment (32, 51) and trimmed such that only regions of conserved overlapping sequence data were considered (∼85% of the overlapping sequence length). Filtered sequences were assigned to samples according to their 12-nt barcode. After chimeras were removed, sequences were classified in accordance with the new Greengenes training set and taxonomy (32, 59) and clustered into OTU at the 0.03 level (i.e., at 97% similarity) (17).
The full-length ITS1 subregion was extracted from the fungal ITS data set using ITS Extractor v. 1.1 (41), dismissing entries for which not both of the 3′ end of the SSU gene and the 5′ end of the 5.8S gene were found. The ITS1 sequences were clustered in CrunchClust v. 43 (http://code.google.com/p/crunchclust/) (16), with pyrosequencing homopolymer correction enabled at a Levenshtein distance of 7 (approximately 97% sequence similarity, a threshold value known to perform well for a wide range of fungi in terms of ITS distances) (40). A representative sequence from each OTU as designated by CrunchClust was annotated for taxonomic affiliation using an established ITS pyrosequence analysis pipeline (54) against the UNITE (1) and INSD (20) databases. The results were verified manually for each OTU. An entry was deemed identified as corresponding to an OTU if it produced a ≥97% match over the full length of the sequence to a fully identified reference sequence whose name was not contradicted by other, equally good matches. The corresponding values for tentative genus and order phylogenetic affiliation were 85% and 70%, respectively; they were deliberately set to be high to avoid false-positive inclusions. In the case of competing names for which neither synonymy nor anamorph-telemorph relationships could be established through MycoBank searches (7), the least inclusive parental nomenclatural level was used (e.g., Penicillium sp.).
Good's coverage values were calculated for OTU at the 0.03 level of definition (14).
Similarity and structure within memberships and across various samples were assessed according to Jaccard coefficients and thetayc calculations (51). A Newick-formatted tree that describes the dissimilarity (1 − similarity) among multiple groups was generated using the jclass calculators and visualized via the TreeView program (45, 51). The jclass calculator returns the traditional Jaccard index describing the dissimilarity between two communities. Parsimony analysis and the UniFrac weighted algorithm were used to determine whether the clustering within the resulting dendrogram was statistically significant (45). Visualization of beta-diversity information was achieved via ordination plotting with nonmetric multidimensional scaling (NMDS) in 2 dimensions, which was statistically examined via analysis of molecular variance (AMOVA) (51).
RESULTS
All microbial diversity results presented are based on DNA analysis, and any discussion of OTU presence thus addresses both living and deceased microbes.
Pyrosequencing-derived bacterial diversity.
In total, the pyrosequencing procedures carried out in this study yielded ∼200,000 bacterial 16S rRNA gene sequences >350 bp in length from the 31 sample sets examined. When these sequences were processed in MOTHUR, ∼70% of the sequences were omitted from consideration due to quality control criteria. The remaining 30% of sequences whose minimum length was 250 bp were aligned and subjected to cluster analyses to reveal their phylogenetic affiliations. The resulting pyrosequencing-derived sequence abundances and coinciding numbers of OTU associated with the various surface samples examined are given in Table 2 and Table 3, respectively. Collectively, group I and II samples, which had been collected from floor and GSE surfaces, gave rise to a greater number of OTU (2,686 OTU from 71 m2) than the spacecraft hardware surface samples of categories III and IV (194 OTU from 162 m2). The group I floor samples, represented by a surface area of ∼10 m2 from a typical but non-NASA cleanroom floor (sample 150), yielded two times the number of OTU seen with samples from the NASA cleanroom floor (sample 148; JPL-SAF). The JPL-SAF cleanroom GSE surfaces (sample 146) gave rise to 2.5 times more bacterial OTU than the floors that they rested on (sample 148; JPL-SAF). The floors of both NASA and non-NASA cleanrooms yielded 100 to 200 bacterial OTU, whereas cleanroom GSE surfaces harbored ∼500 bacterial OTU.
Table 2.
Bacterial pyrosequences retrieved from various spacecraft and associated surfaces
| Bacterial taxon | No. of pyrosequences from indicated sample (total no. of sequences) |
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I |
Group II |
Group III |
Group IV |
||||||||||||||||||||||||||||
| 150 (floor 70A) (8,598) | 143 (entrance floor) (8,107) | 141 (shoe cleaner) (2,622) | 144 (floor 1) (3037) | 145 (floor 2) (2,084) | 142 (air lock) (795) | 146 (JPL-SAF GSE) (4,695) | 148 (JPL-SAF floor) (1,783) | 155 (floor) (16,240) | 157 (GSE) (5,948) | 159 (floor) (1,511) | 161 (GSE) (1,583) | 124 (spacecraft) (13) | 125 (spacecraft) (3) | 133 (spacecraft) (112) | 134 (spacecraft) (275) | 135 (spacecraft) (63) | 136 (spacecraft) (171) | 137 (spacecraft) (1) | 126 (spacecraft) (59) | 127 (spacecraft) (219) | 128 (spacecraft) (339) | 129 (spacecraft) (698) | 130 (spacecraft) (33) | 138 (spacecraft) (21) | 140 (spacecraft) (139) | 132 (spacecraft) (459) | 151 (spacecraft) (69) | 152 (spacecraft) (92) | 153 (spacecraft) (79) | 154 (spacecraft) (15) | |
| Actinobacteria | 81 | 798 | 315 | 279 | 106 | 222 | 1,764 | 55 | 88 | 618 | 8 | 120 | 1 | 76 | 3 | 1 | 49 | 60 | 105 | 79 | 7 | 5 | |||||||||
| Armatimonadetes | 1 | 21 | 6 | 7 | 6 | 1 | 27 | ||||||||||||||||||||||||
| Bacteroidetes | 7 | 252 | 101 | 64 | 104 | 62 | 214 | 108 | 2,477 | 436 | 59 | 97 | 4 | 18 | 47 | ||||||||||||||||
| Chlorobi | 3 | ||||||||||||||||||||||||||||||
| Verrucomicrobia | 1 | 8 | 5 | ||||||||||||||||||||||||||||
| Chloroflexi | 1 | 10 | 15 | 7 | 17 | 5 | 26 | 4 | 27 | 25 | |||||||||||||||||||||
| Deinococcus-Thermus | 3 | 6 | 1 | 7 | 2 | 26 | 7 | 41 | 11 | 1 | |||||||||||||||||||||
| Acidobacteria | 4 | 23 | 10 | 11 | 16 | 1 | 113 | 350 | 1 | 37 | 11 | 3 | |||||||||||||||||||
| Firmicutes | 13 | 322 | 186 | 174 | 15 | 17 | 383 | 157 | 17 | 157 | 8 | 33 | 2 | 19 | 3 | 60 | |||||||||||||||
| Fusobacteria | 67 | 10 | 17 | 6 | 1 | 1 | 2 | 25 | |||||||||||||||||||||||
| Gemmatimonadetes | 1 | 13 | 4 | 17 | 3 | 2 | 2 | ||||||||||||||||||||||||
| Nitrospirae | 1 | 1 | |||||||||||||||||||||||||||||
| Planctomycetes | 11 | 2 | 3 | 1 | 2 | 4 | 3 | ||||||||||||||||||||||||
| Alphaproteobacteria | 5,776 | 2,465 | 693 | 1,065 | 785 | 196 | 1,120 | 606 | 7,495 | 2,517 | 692 | 73 | 1 | 23 | 22 | 26 | 129 | 15 | 127 | 54 | |||||||||||
| Betaproteobacteria | 1,478 | 431 | 215 | 154 | 215 | 104 | 523 | 100 | 1,031 | 565 | 291 | 271 | 9 | 2 | 42 | 101 | 33 | 139 | 32 | 77 | 139 | 329 | 26 | 3 | 103 | 37 | 2 | 5 | |||
| Deltaproteobacteria | 3 | 3 | 2 | 5 | 36 | 6 | 7 | 12 | |||||||||||||||||||||||
| Gammaproteobacteria | 1,211 | 3,374 | 962 | 1165 | 796 | 100 | 251 | 357 | 4,833 | 1,127 | 456 | 250 | 3 | 43 | 30 | 25 | 27 | 91 | 96 | 36 | 7 | 175 | 15 | 36 | 74 | 15 | |||||
| Unidentified | 13 | 6 | 24 | 12 | 7 | 104 | 13 | 4 | |||||||||||||||||||||||
| Spirochaetes | 1 | ||||||||||||||||||||||||||||||
| Tenericutes | 2 | 1 | |||||||||||||||||||||||||||||
| Unidentified division | |||||||||||||||||||||||||||||||
| SC4 | 24 | 6 | |||||||||||||||||||||||||||||
| TM7 | 6 | 3 | 3 | 12 | |||||||||||||||||||||||||||
| WPS-2 | 14 | 2 | |||||||||||||||||||||||||||||
| Unclassified bacteria | 20 | 277 | 89 | 61 | 10 | 78 | 224 | 8 | 159 | 384 | 737 | 1 | 29 | ||||||||||||||||||
Table 3.
Bacterial OTU retrieved from various spacecraft and associated surfaces
| Bacterial taxon | No. of OTU retrieved from indicated sample (total no. of OTU) |
||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I |
Group II |
Group III |
Group IV |
||||||||||||||||||||||||||||
| 150 (floor 70A) (227) | 143 (entrance floor) (654) | 141 (shoe cleaner) (486) | 144 (floor 1) (212) | 145 (floor 2) (159) | 142 (air lock) (145) | 146 (JPL-SAF GSE) (497) | 148 (JPL-SAF floor) (122) | 155 (floor) (513) | 157 (GSE) (662) | 159 (floor) (66) | 161 (GSE) (136) | 124 (spacecraft) (5) | 125 (spacecraft) (2) | 133 (spacecraft) (13) | 134 (spacecraft) (19) | 135 (spacecraft) (4) | 136 (spacecraft) (7) | 137 (spacecraft) (1) | 126 (spacecraft) (6) | 127 (spacecraft) (8) | 128 (spacecraft) (29) | 129 (spacecraft) (45) | 130 (spacecraft) (4) | 138 (spacecraft) (4) | 140 (spacecraft) (6) | 132 (spacecraft) (34) | 151 (spacecraft) (7) | 152 (spacecraft) (6) | 153 (spacecraft) (2) | 154 (spacecraft) (3) | |
| Actinobacteria | 31 | 169 | 122 | 50 | 31 | 54 | 185 | 25 | 51 | 186 | 3 | 30 | 1 | 6 | 1 | 1 | 3 | 10 | 15 | 5 | 1 | 1 | |||||||||
| Armatimonadetes | 1 | 6 | 4 | 1 | 3 | 1 | 1 | ||||||||||||||||||||||||
| Bacteroidetes | 6 | 65 | 45 | 9 | 21 | 10 | 43 | 8 | 39 | 75 | 4 | 22 | 1 | 1 | 4 | ||||||||||||||||
| Chlorobi | 1 | ||||||||||||||||||||||||||||||
| Verrucomicrobia | 1 | 4 | 2 | ||||||||||||||||||||||||||||
| Chloroflexi | 1 | 4 | 6 | 2 | 2 | 1 | 5 | 1 | 8 | 3 | |||||||||||||||||||||
| Deinococcus-Thermus | 2 | 3 | 1 | 3 | 2 | 8 | 5 | 15 | 3 | 1 | |||||||||||||||||||||
| Acidobacteria | 2 | 6 | 7 | 2 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 2 | |||||||||||||||||||
| Firmicutes | 7 | 49 | 50 | 15 | 4 | 3 | 27 | 11 | 10 | 33 | 2 | 1 | 1 | 4 | 2 | 1 | |||||||||||||||
| Fusobacteria | 2 | 2 | 3 | 1 | 1 | 1 | 2 | 1 | |||||||||||||||||||||||
| Gemmatimonadetes | 1 | 3 | 3 | 3 | 3 | 2 | 1 | ||||||||||||||||||||||||
| Nitrospirae | 1 | 1 | |||||||||||||||||||||||||||||
| Planctomycetes | 4 | 1 | 2 | 1 | 2 | 2 | 1 | ||||||||||||||||||||||||
| Alphaproteobacteria | 113 | 156 | 113 | 49 | 50 | 33 | 124 | 41 | 233 | 188 | 38 | 22 | 1 | 3 | 3 | 7 | 9 | 1 | 14 | 2 | |||||||||||
| Betaproteobacteria | 38 | 61 | 47 | 23 | 17 | 13 | 35 | 9 | 48 | 61 | 7 | 23 | 4 | 1 | 2 | 5 | 2 | 3 | 4 | 1 | 3 | 5 | 2 | 1 | 9 | 2 | 1 | 1 | |||
| Deltaproteobacteria | 1 | 3 | 1 | 1 | 1 | 3 | 3 | 1 | |||||||||||||||||||||||
| Gammaproteobacteria | 11 | 67 | 39 | 38 | 23 | 15 | 29 | 19 | 76 | 40 | 12 | 16 | 2 | 4 | 2 | 2 | 2 | 3 | 8 | 5 | 2 | 6 | 3 | 3 | 1 | 3 | |||||
| Unidentified | 3 | 4 | 4 | 3 | 1 | 4 | 3 | 1 | |||||||||||||||||||||||
| Spirochaetes | 1 | ||||||||||||||||||||||||||||||
| Tenericutes | 2 | 1 | |||||||||||||||||||||||||||||
| Unidentified division | |||||||||||||||||||||||||||||||
| SC4 | 3 | 3 | |||||||||||||||||||||||||||||
| TM7 | 2 | 3 | 1 | 1 | |||||||||||||||||||||||||||
| WPS-2 | 2 | 2 | |||||||||||||||||||||||||||||
| Unclassified bacteria | 12 | 50 | 35 | 12 | 4 | 12 | 27 | 4 | 28 | 33 | 15 | 1 | 2 | ||||||||||||||||||
The extremely clean nature of the JPL-SAF cleanroom floor samples required that 10 1-m2 surface areas be sampled and pooled to generate sufficient amounts of PCR product for downstream pyrosequencing. However, the floors of the ordinary room adjacent to the JPL-SAF cleanroom required only 1 m2 to be sampled and processed to amplify sufficient amounts of template DNA. With respect to the floors of this ordinary room, the entrance floor (sample 143) where staff ingress/egress in street clothes gave rise to ∼5.3 times more OTU (654 OTU) than the adjacent JPL-SAF cleanroom floor. The central floors of this ordinary room (samples 144 and 145), and the surfaces of a colocated shoe cleaner instrument (sample 141), yielded between 159 and 486 OTU, many of which were of proteobacterial and actinobacterial lineage (Table 3). The floors of the air lock chamber (sample 142) that connects the ordinary room to the JPL-SAF cleanroom gave rise to 145 distinct bacterial OTU.
The cleaning and maintenance involved in preparing the JPL building 144 cleanroom to receive mission-critical spacecraft hardware successfully reduced and (as made evident by the OTU not detected) even eliminated many bacterial lineages. Prior to cleaning, floor sample 155 gave rise to 513 OTU, whereas after cleaning, the same location (sample 159) showed a complete loss of 447 previously detected OTU. In general, the incidence of all bacterial lineages declined after cleaning of these cleanroom floors. In similar fashion, prior to cleaning procedures, 662 OTU were observed in samples from the JPL building 144 GSE surfaces (sample 157). This was ∼5 times more than the number of OTU detected after cleaning (136 OTU; sample 161). Several bacterial groups reported to be capable of surviving desiccation and UV radiation (e.g., actinobacteria, deinococci, firmicutes) were not detected after the cleaning of the floors, whereas the same bacterial types persisted even after cleaning on GSE surfaces. In contrast, the vast majority of purported human-associated bacteria (e.g., proteobacteria) was eliminated from both the cleanroom floors and colocated GSE surfaces after cleaning.
The results of this investigation showed a predominance of sequences typical of human skin-associated bacterial commensals in the group III and IV spacecraft hardware samples, whereas cleanroom floor samples yielded sequences from a greater variety of microbial sources. Even though firmicute sequences were generated from spacecraft hardware surface samples (group III), mission subsystem component surfaces (group IV) were devoid of these, as well as of sequences representing desiccation-resistant deinococci and actinobacteria.
Pyrosequencing-derived archaeal diversity.
Many fewer high-quality archaeal sequences were generated compared directly to the results of bacterial pyrosequencing for the very same samples. Although ∼30,000 sequences spanning >350 bp of the V3 to V5 region of archaeal 16S rRNA gene were generated, bioinformatic quality control measures demonstrated that only 151 sequences were truly of archaeal lineage. The remainder of these sequences were related to bacterial taxa, predominantly Verrucomicrobiae. Resulting archaeal sequence and OTU abundances from the various samples examined in this study are given in Table 4. None of the 31 sample sets examined yielded 1.1-kb archaeal 16S rRNA gene amplicons via traditional PCR techniques (35). In most cases, even following nested PCR amplification, there was insufficient DNA to perform successful pyrosequencing analysis. However, three samples (samples 142 to 144) collected from the ordinary room adjacent to the JPL-SAF cleanroom gave rise to measurable levels of archaeal 16S rRNA gene nested-PCR product (data not shown). Of the 12 samples composing groups I and II, no long-read (∼600-bp) archaeal sequences could be generated from any of the five cleanroom floor (∼10-m2 area) samples tested. In contrast, archaeal sequences were successfully obtained from all five of the ordinary room samples (1-m2 area) and two of the group II GSE samples (Table 4). In addition, archaeal sequences were generated from 6 of the 15 spacecraft hardware (group III) samples, whereas all four sets of mission subsystem component surface (group IV) samples failed to yield amplifiable archaeal DNA. Group I and II samples exclusively gave rise to sequences representing members of Nitrososphaeraceae (120 sequences; 3 OTU), while group III spacecraft hardware samples generated only Methanobacteriaceae OTU (31 sequences; 2 OTU).
Table 4.
Archaeal pyrosequences and OTU retrieved from various spacecraft and associated surfaces
| Archaeal taxon and species | No. of archaeal pyrosequences from indicated sample (no. of archaeal OTU) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I |
Group II |
Group III |
|||||||||||
| 150 (floor) (1) | 143 (entrance floor) (1) | 141 (shoe cleaner) (2) | 142 (air lock) (1) | 148 (JPL-SAF GSE) | 157 (GSE) (1) | 161 (GSE) (1) | 124 (spacecraft) (1) | 126 (spacecraft) (1) | 127 (spacecraft) (1) | 128 (spacecraft) (1) | 130 (spacecraft) (2) | 132 (spacecraft) (1) | |
| Nitrosphaeraceae SCA114-1 | 17 | 35 | 47 | 5 | 2 | ||||||||
| Nitrosphaeraceae SCA114-2 | 1 | ||||||||||||
| Nitrosphaeraceae SCA117 | 6 | ||||||||||||
| Methanobacteriaceae 1 | 1 | 2 | 7 | 3 | 16 | 1 | |||||||
| Methanobacteriaceae 3 | 1 | ||||||||||||
Pyrosequencing-derived fungal diversity.
Resulting fungal sequence abundances and the corresponding OTU associated with the various surface samples examined are given in Table 5. In total, 143,557 high-quality fungal sequences (∼50% of the total number generated and more than twice the number of high-quality bacterial sequences) comprising 456 distinct fungal OTU were generated in this investigation. Unlike the results seen with bacterial pyrosequencing, where at least some sequences were generated from each of the 31 sample sets examined, only 18 of these sample sets gave rise to fungal sequences. Fungal sequences were not generated from many of the spacecraft hardware samples (11 out of 19 sample sets), despite the fact that a large surface area was sampled. Group I samples gave rise to a high abundance of fungal OTU, compared to the samples of all other categories. Of these, the JPL-SAF cleanroom floor samples yielded only three OTU, whereas the non-NASA cleanroom floors (sample 150) gave rise to the most fungal OTU, followed by the entrance floor of the ordinary room adjacent to the JPL-SAF cleanroom (sample 141) and colocated shoe cleaner surfaces (sample 142). The central floors of this ordinary room did not present as much fungal diversity (12 and 3 OTU) as the entrance floor (133 OTU). Fungal communities were more diverse (44 OTU) about the floors of the air lock (sample 142) which connects this ordinary room to the JPL-SAF cleanroom.
Table 5.
Abundance and diversity of various fungal pyrosequences and OTU associated with spacecraft surfaces and assembly environments
| Fungal phylum, subphylum, and classa | No. of pyrosequences retrieved from indicated sample |
No. of OTU retrieved from indicated sample |
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I |
Group II |
Group III |
Group IV |
Group I |
Group II |
Group III |
Group IV |
|||||||||||||||||||||||||||||
| 150 (floor 70A) | 143 (entrance floor) | 141 (shoe cleaner) | 144 (floor 1) | 145 (floor 2) | 142 (air lock) | 146 (JPL-SAF GSE) | 148 (JPL-SAF floor) | 157 (GSE) | 161 (GSE) | 125 (spacecraft) | 126 (spacecraft) | 127 (spacecraft) | 128 (spacecraft) | 129 (spacecraft) | 130 (spacecraft) | 151 (spacecraft) | 154 (spacecraft) | 150 (floor 70A) | 143 (entrance floor) | 141 (shoe cleaner) | 144 (floor 1) | 145 (floor 2) | 142 (air lock) | 146 (JPL-SAF GSE) | 148 (JPL-SAF floor) | 157 (GSE) | 161 (GSE) | 125 (spacecraft) | 126 (spacecraft) | 127 (spacecraft) | 128 (spacecraft) | 129 (spacecraft) | 130 (spacecraft) | 151 (spacecraft) | 154 (spacecraft) | |
| Ascomycota | 844 | 76 | 357 | 4 | 467 | 44 | 535 | 28 | 12 | 7 | 3 | 1 | 4 | 4 | 3 | 1 | ||||||||||||||||||||
| Pezizomycotina | ||||||||||||||||||||||||||||||||||||
| Unclassified Pezizomycotina | 564 | 1,994 | 1,093 | 31 | 587 | 741 | 4,426 | 71 | 23 | 17 | 14 | 2 | 6 | 7 | 28 | 2 | ||||||||||||||||||||
| Dothideomycetes | 2,545 | 6,749 | 3,979 | 152 | 3 | 1,866 | 1,702 | 1,4204 | 2,129 | 59 | 2 | 492 | 8 | 59 | 36 | 35 | 3 | 1 | 13 | 17 | 56 | 16 | 2 | 1 | 1 | 1 | ||||||||||
| Eurotiomycetes | 803 | 898 | 498 | 3 | 163 | 87 | 54 | 857 | 95 | 1 | 18 | 69 | 27 | 11 | 5 | 1 | 3 | 2 | 2 | 12 | 1 | 1 | 1 | 1 | ||||||||||||
| Lecanoromycetes | 689 | 504 | 264 | 114 | 442 | 1,598 | 15 | 9 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||
| Leotiomycetes | 168 | 522 | 192 | 137 | 141 | 1,256 | 16 | 6 | 5 | 2 | 4 | 9 | ||||||||||||||||||||||||
| Pezizomycetes | 10 | 71 | 1 | 2 | ||||||||||||||||||||||||||||||||
| Sordariomycetes | 186 | 898 | 406 | 270 | 249 | 64 | 1,633 | 101 | 110 | 3 | 21 | 10 | 10 | 4 | 3 | 3 | 19 | 2 | 1 | 1 | ||||||||||||||||
| Saccharomycotina | ||||||||||||||||||||||||||||||||||||
| Saccharomycetes | 325 | 406 | 442 | 137 | 17 | 150 | 1 | 6 | 4 | 1 | 1 | 2 | ||||||||||||||||||||||||
| Taphrinomycotina | ||||||||||||||||||||||||||||||||||||
| Unclassified Taphrinomycotina | 2 | 44 | 1 | 1 | ||||||||||||||||||||||||||||||||
| Basidiomycota | 17 | 1 | ||||||||||||||||||||||||||||||||||
| Agaricomycotina | ||||||||||||||||||||||||||||||||||||
| Unclassified Agaricomycotina | 13 | 1 | ||||||||||||||||||||||||||||||||||
| Agaricomycetes | 82 | 251 | 110 | 341 | 5 | 234 | 39 | 1 | 11 | 4 | 3 | 2 | 1 | 3 | 1 | 1 | ||||||||||||||||||||
| Tremellomycetes | 62,584 | 3,053 | 1,366 | 864 | 248 | 7 | 2,805 | 526 | 28 | 17 | 16 | 5 | 9 | 1 | 23 | 3 | ||||||||||||||||||||
| Pucciniomycotina | 6 | 1 | ||||||||||||||||||||||||||||||||||
| Agaricostilbomycetes | 122 | 9 | 1 | 1 | ||||||||||||||||||||||||||||||||
| Cystobasidiomycetes | 76 | 1,109 | 355 | 18 | 175 | 301 | 45 | 4 | 5 | 4 | 1 | 1 | 3 | 1 | ||||||||||||||||||||||
| Microbotryomycetes | 12 | 50 | 1 | 2 | ||||||||||||||||||||||||||||||||
| Pucciniomycetes | 324 | 3,465 | 909 | 432 | 150 | 1,324 | 292 | 3 | 2 | 16 | 8 | 12 | 3 | 1 | 14 | 2 | 1 | 2 | ||||||||||||||||||
| Ustilaginomycotina | ||||||||||||||||||||||||||||||||||||
| Exobasidiomycetes | 255 | 108 | 93 | 71 | 133 | 166 | 4 | 1 | 1 | 2 | 1 | 3 | ||||||||||||||||||||||||
| Ustilaginomycetes | 4 | 2 | 15 | 1 | 1 | 4 | ||||||||||||||||||||||||||||||
| Chytridiomycota | ||||||||||||||||||||||||||||||||||||
| Chytridiomycetes | 36 | 52 | 30 | 1 | 3 | 1 | ||||||||||||||||||||||||||||||
| Former Zygomycota | ||||||||||||||||||||||||||||||||||||
| Mortierellomycotina | 3 | 1 | ||||||||||||||||||||||||||||||||||
| Total | 69,493 | 20,193 | 10,129 | 542 | 10 | 5,532 | 3,774 | 61 | 29,684 | 3,332 | 62 | 2 | 492 | 47 | 112 | 21 | 69 | 2 | 237 | 133 | 116 | 12 | 3 | 44 | 51 | 3 | 187 | 30 | 3 | 1 | 1 | 2 | 3 | 2 | 1 | 2 |
NCBI taxonomy.
Results show that the cleaning procedures applied to both the JPL-SAF and JPL-144 cleanroom floors effectively reduced a significant proportion of the fungal population, as 0 to 3 fungal OTU were observed in these samples postcleaning. In contrast, GSE surfaces that were sampled after cleaning still gave rise to 30 fungal OTU (compared to 186 fungal OTU observed in these samples before cleaning). The observed abundance of fungal OTU arising from GSE surfaces (sample 146) residing in the JPL-SAF cleanroom far exceeded (51 OTU) that corresponding to their floor counterparts (sample 148; 3 OTU). Fewer than four fungal OTU were observed in several (6 of 15) of the group III spacecraft hardware samples, and even fewer OTU (0 to 2) were detected in the group IV subsystem component samples.
With respect to diversity, the total fungal population encountered in this study spanned four distinct phyla (Table 5), and the majority of detected fungal sequences (99.9%) represented either Ascomycota (42.7%) or Basidiomycota (57.2%). With regard to OTU incidence and diversity, 71% and 28% of the 831 OTU observed in this study belonged to the phyla Ascomycota and Basidiomycota, respectively. Sequences representative of members of the Dothideomycetes were observed in the group III spacecraft hardware samples. Members of this class were encountered in high abundance (241 OTU) in most of the samples examined, strongly reminiscent of the results reported by Amend et al. (2). In addition, OTU representing species of the genus Penicillium (Eurotiomycetes) were found in the JPL-SAF cleanroom floor, spacecraft hardware, and mission subsystem component samples (Table 5).
Environmental clustering analyses.
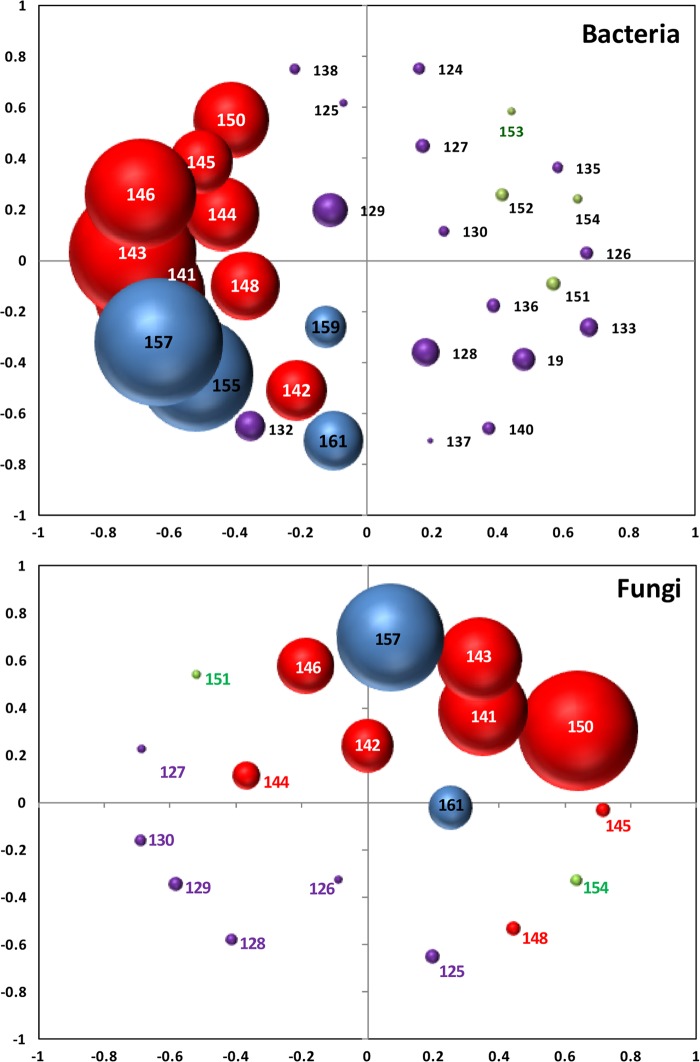
As Fig. 1 depicts, the non-NASA cleanroom floor samples (sample 148) were distinctly different from the NASA cleanroom floor samples (sample 148 and sample 159) examined. Samples collected from the JPL building 144 cleanroom (samples 155 and 159) and the ordinary room adjacent to the JPL-SAF cleanroom (samples 141 to 145) clustered together, indicative of the shared bacterial diversity present in these samples. Of particular note was the seemingly anomalous clustering of a group III spacecraft hardware sample (sample 132), whose spore count was very high (300 spores/m2), with samples collected from the ordinary support facility room (see Fig. S1 in the supplemental material). In a similar manner, distinct environmental clustering was observed among samples collected from the general surfaces of the spacecraft and mission subsystem component surface (see Fig. S1 in the supplemental material). Parsimony analysis for the bacterial tree showed a statistically significant difference for the data corresponding to the clustering of the four groups of samples considered in this study (P = <0.001). UniFrac weighted analysis, which determines whether two or more communities have statistically significant structural dissimilarity, indicated a statistically significant dissimilarity among all four of the sample categories (Wsig < 0.001) with respect to both bacterial and fungal diversity (observed OTU). AMOVA was used to assess the statistical significance of the spatial separation observed among the different groupings in NMDS ordination plots (i.e., where the centers of the clouds were representative of different categories more distantly separated than samples belonging to the same cloud). Statistically significant dissimilarities were observed across most categories with respect to both bacterial and fungal diversity (observed OTU; P < 0.05), with the exception of similarity between categories III and IV (0.064) with respect to bacteria and categories I and II (0.923) with respect to fungi.
Fig 1.
Nonmetric multidimensional scaling analysis depicting variations in bacterial (top) and fungal (bottom) diversity across samples. Stress, 0.38 and 0.35 for bacteria and fungi, respectively. Spheres are labeled according to numerical sample identification and sized according to relative OTU abundance. Red, cleanroom types; blue, surfaces prior to versus postcleaning; magenta, general spacecraft hardware surfaces; green, mission subsystem component samples. For a detailed explanation of these sample categories, see Table 1.
DISCUSSION
In previous works, 454 tag-encoded pyrosequencing methods targeting rRNA gene sequences as proxies for the presence of a given microbe estimated microbial diversity to be 100- to 1,000-fold greater than that determined solely on the basis of cultivation (18, 44, 53). A much greater understanding of the total biodiversity present was also unveiled when such methods were employed to elucidate the archaeal, bacterial, and fungal diversity of low-biomass spacecraft-associated settings. Compared directly, JPL-SAF cleanroom floors where spacecraft components were assembled did not display as rich a bacterial diversity as the floors of the ordinary room adjacent to this cleanroom (159 to 654 OTU). This is not to say that the bacterial populations about the JPL-SAF cleanroom floors were not diverse, as bacterial pyrosequencing analyses detected the presence of 122 distinct OTU from these samples. In contrast, a mere 1 to 45 bacterial OTU were detected in each of the 19 spacecraft and mission subsystem component samples (Table 3). The presence of OTU representative of actinobacteria, deinococci, acidobacteria, firmicutes, and proteobacteria on the spacecraft surfaces suggests that certain bacterial lineages persist even following rigorous quality control and cleaning practices. The majority of bacterial OTU observed as being recurrent belonged to actinobacteria (38 OTU) and alphaproteobacteria (35 OTU). Of the astrobiologically relevant species included in these groups, the presence of those belonging to the genera Arthrobacter (43), Modestobacter (15), Bacillus (39), Deinococcus (8, 58), and Acinetobacter (27) should not be taken lightly. The isolation of microbial species of each of these genera from spacecraft assembly environments has already been reported (13, 28, 43, 58). This is of consequence to NASA planetary protection practices and the validation of cleanroom maintenance. La Duc et al. (25) have gone as far as to posit that the measures of cleanliness exerted in SAC may inadvertently select for the hardiest organisms, which may in fact be the most fit to survive longer journeys in space (57, 58).
There were no archaeal sequences generated from any of the samples collected from the cleanroom floors. However, archaeal sequences were retrieved from the surfaces of GSE housed in these rooms (Table 4), which suggests that the different modes of maintenance of cleanliness about the floors and GSE in these cleanrooms have differential effects on the resident archaeal population. The retrieval of archaeal sequences belonging to the ammonia-oxidizing genus Nitrososphaeraceae of the recently proposed Thaumarchaeota phylum (3) in GSE samples collected both before (5 sequences) and after (2 sequences) cleaning was noteworthy. Members of this phylum have been shown to grow using only ammonia or urea as an energy source, and members of the Nitrososphaeraceae in particular have been reported to grow chemolithoautotrophically under oligotrophic conditions (small amounts of pyruvate) and when in coculture with bacteria. While Crenarchaeota and/or Euryarchaeota have previously been reported in cleanroom settings (34), this is the first account of Thaumarchaeota sequences having been detected in samples from these environments. Methanobacteriaceae sequences were also observed in the spacecraft hardware samples. This is particularly relevant for astrobiological issues, since members of this family have been reported to be obligate anaerobic, hydrogenotrophic, and methanogenic organisms and capable of utilizing carbon dioxide as their sole carbon source (22).
Ascomycetes dominate the mycobiota recovered from typical indoor environments (2), as humans, insects, and other animals contribute significantly to this population, both as vectors and as hosts (2, 46). The extent to which these observations carry over to cleanrooms and other facilities kept very clean is less clear, however. The various mechanisms employed to maintain a clean environment (e.g., chemical biocides, UV light) can be expected to erase many of the patterns observed for the mycobiota of regular indoor environments, notably, the taxonomic composition. High-efficiency particulate air (HEPA) filters, for example, are expected to favor the continued persistence of species with small spores or conidia (e.g., members of the genera Aspergillus and Cladosporium), whereas UV light would act as a selector for species with pigmented or thick-walled propagules (19, 52). Fungal diversity in cleanrooms and medical settings has primarily been addressed through culture-based methods and is known to feature a large proportion of fungi related to humans and human activities (5, 11, 21, 30, 48, 55). However, when Nagano et al. (37) applied Sanger sequencing-based approaches to examine fungal diversity, the mycobiota recovered from medical environments bore remarkable similarity to those now being detected in surfaces of the Kibo experimental module flown to the International Space Station, all of which are prime examples of a low-biomass environment (49). The Kibo module-associated dominant fungal community components included Alternaria, Aspergillus, Candida, Cladosporium, Penicillium, Trichoderma, and Malassezia (49).
Of heightened astrobiology relevance are studies that investigate fungal behavior in ionizing radiation. The explosion at the Chernobyl nuclear power plant in April 1986 renewed interest in the role fungi play in mediating radionuclide movement in ecosystems (9, 60–62). Radioresistance of some fungal species was linked to the presence of melanin, which was shown to have emerging properties of acting as an energy transporter for metabolism and has been implicated in enhancing hyphal growth and directed growth of sensitized hyphae toward sources of radiation. Up to 40% of all fungi isolated from the Chernobyl reactor room also contained melanin or other pigments (62). Furthermore, many plant-pathogenic fungi survive under highly desiccated conditions. By extension, this raises space exploration concerns of potential fungal forward contamination or sample return-based life detection. Among the limited number of fungal groups detected in this study, Capnodiales, Hypocreales, Pleosporales, and Cantharellales sequences were detected following cleaning. Among Hypocreales and Cantharellales, there are many species known for interactions with plants. The Pleosporales represent the largest order in the class Dothideomycetes, with a reported 23 families, 332 genera, and more than 4,700 species. In synonymy with Pleosporales is the order Melanommatales. Recent molecular studies have not supported separation of Melanommatales from Pleosporales (36). Dark-pigmented, double-wall-protected, often multicellular ascospores are typical of these fungi.
The findings of this study provide new and important insights into the benefits and limitations of innovative molecular approaches for assessing the microbial diversity associated with samples extremely low in total biomass which are of particular relevance to current and future NASA endeavors, as well as homeland security and medical, pharmaceutical, and semiconductor fabrication industries. Coupling the innovative pyrosequencing techniques employed in this study with other emerging molecular methodologies, such as generation 3 PhyloChip DNA microarrays (4, 28), microscopy-based viability assays (33), etc., could lead to significantly improved approaches for monitoring and mitigating the diverse microbial populations associated with ultraclean environments, without the biases that plague culture-dependent techniques. Ultimately, pyrosequencing analyses supported, and accentuated, the general trends observed with regard to environmental clustering where the greatest microbial diversity was encountered in ordinary room facilities, followed next by cleanroom floors, and, finally, spacecraft components (Fig. 1). The results of this study underscore that (i) continued monitoring and biosignature detection need to be extended throughout all three domains of life and (ii) a shift toward high-throughput, data-rich molecular assays with significant bioinformatic analysis is inevitable in microbial community analyses.
Supplementary Material
ACKNOWLEDGMENTS
Part of the research described in this study was carried out at the Jet Propulsion Laboratory, California Institute of Technology, under contract with the National Aeronautics and Space Administration.
We are largely indebted to P. Schloss for access to, and assistance with, MOTHUR software and extensive help with the interpretation of results. We are grateful to N. Hallenberg, A. Dahl, C. Conley, K. Buxbaum, and J. Andy Spry for advice and guidance. We also thank J. N. Benardini and E. Bargoma for assistance with sample collection and processing and Y. Sun, Research and Testing Laboratory, Lubbock, Texas, for improving the pyrosequencing procedures for low-biomass samples.
Footnotes
Published ahead of print 22 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Abarenkov K, et al. 2010. The UNITE database for molecular identification of fungi—recent updates and future perspectives. New Phytol. 186:281–285 [DOI] [PubMed] [Google Scholar]
- 2. Amend AS, Seifert KA, Samson R, Bruns TD. 2010. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc. Natl. Acad. Sci. U. S. A. 107:13748–13753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. 2008. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245–252 [DOI] [PubMed] [Google Scholar]
- 4. Cooper M, et al. 2011. Comparison of innovative molecular approaches and standard spore assays for assessment of surface cleanliness. Appl. Environ. Microbiol. 77:5438–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cordeiro RA, et al. 2010. Isolation of pathogenic yeasts in the air from hospital environments in the city of Fortaleza, northeast Brazil. Braz. J. Infect. Dis. 14:30–34 [DOI] [PubMed] [Google Scholar]
- 6. COSPAR 2002. Planetary protection policy, October 2002, as amended March 2008. COSPAR, Houston, TX [Google Scholar]
- 7. Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Stud. Mycol. 50:19–22 [Google Scholar]
- 8. Daly MJ, et al. 2004. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025–1028 [DOI] [PubMed] [Google Scholar]
- 9. Dighton J, Tugay T, Zhdanova N. 2008. Fungi and ionizing radiation from radionuclides. FEMS Microbiol. Lett. 281:109–120 [DOI] [PubMed] [Google Scholar]
- 10. Dowd SE, et al. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43 doi:10.1186/1471-2180-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ekhaise FO, Ighosewe OU, Ajakpovi OD. 2008. Hospital indoor airborne microflora in private and government owned hospitals in Benin City, Nigeria. World J. Med. Sci. 3:19–23 [Google Scholar]
- 12. Favero MS. 1971. Microbiologic assay of space hardware. Environ. Biol. Med. 1:27–36 [PubMed] [Google Scholar]
- 13. Ghosh S, Osman S, Vaishampayan P, Venkateswaran K. 2010. Recurrent isolation of extremotolerant bacteria from the clean room where Phoenix spacecraft components were assembled. Astrobiology 10:325–335 [DOI] [PubMed] [Google Scholar]
- 14. Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 15. Gtari M, et al. 2012. Contrasted resistance of stone-dwelling Geodermatophilaceae species to stresses known to give rise to reactive oxygen species. FEMS Microbiol. Ecol. 80:566–577 [DOI] [PubMed] [Google Scholar]
- 16. Hartmann M, et al. Significant and persistent impact of timber harvesting on soil microbial communities in northern coniferous forests. ISME J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hazen TC, et al. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208 [DOI] [PubMed] [Google Scholar]
- 18. Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jacobson ES. 2000. Pathogenic roles for fungal melanins. Clin. Microbiol. Rev. 13:708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karsch-Mizrachi I, Nakamura Y, Cochrane G. 2012. The international nucleotide sequence database collaboration. Nucleic Acids Res. 40:D33–D37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim KY, Kim YS, Kim D. 2010. Distribution characteristics of airborne bacteria and fungi in the general hospitals of Korea. Ind. Health 48:236–243 [DOI] [PubMed] [Google Scholar]
- 22. Kitamura K, Fujita T, Akada S, Tonouchi A. 2011. Methanobacterium kanagiense sp. nov., a hydrogenotrophic methanogen, isolated from rice-field soil. Int. J. Syst. Evol. Microbiol. 61:1246–1252 [DOI] [PubMed] [Google Scholar]
- 23. Krasnopolsky VA. 2006. Some problems related to the origin of methane on Mars. Icarus 180:359–367 [Google Scholar]
- 24. Kwan K, et al. 2011. Evaluation of procedures for the collection, processing, and analysis of biomolecules from low-biomass surfaces. Appl. Environ. Microbiol. 77:2943–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. La Duc MT, et al. 2007. Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean-room environments. Appl. Environ. Microbiol. 73:2600–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. La Duc MT, Kern RG, Venkateswaran K. 2004. Microbial monitoring of spacecraft and associated environments. Microb. Ecol. 47:150–158 [DOI] [PubMed] [Google Scholar]
- 27. La Duc MT, Nicholson W, Kern R, Venkateswaran K. 2003. Microbial characterization of the Mars Odyssey spacecraft and its encapsulation facility. Environ. Microbiol. 5:977–985 [DOI] [PubMed] [Google Scholar]
- 28. La Duc MT, et al. 2009. Comprehensive census of bacteria in clean rooms by using DNA microarray and cloning methods. Appl. Environ. Microbiol. 75:6559–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. La Duc MT, Osman S, Venkateswaran K. 2009. Comparative analysis of methods for the purification of DNA from low-biomass samples based on total yield and conserved microbial diversity. J. Rapid Methods Autom. Microbiol. 17:350–368 doi:10.1111/j.1745-4581.2009.00153.x [Google Scholar]
- 30. Lopolito P, Bartnett C, Polarine J. 2007. Control strategies for fungal contamination in cleanrooms. Control. Environ. 372:1–5 http://www.cemag.us/article/control-strategies-fungal-contamination-cleanrooms?page=0,4 [Google Scholar]
- 31. Margulies M, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McDonald D, et al. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mohapatra BR, La Duc MT. 2012. Evaluation of fluorescence in situ hybridization to detect encapsulated Bacillus pumilus SAFR-032 spores released from poly(methylmethacrylate). Microbiol. Immunol. 56:40–47 [DOI] [PubMed] [Google Scholar]
- 34. Moissl C, Bruckner J, Venkateswaran K. 2008. Archaeal diversity analysis of spacecraft assembly facilities. ISME J. 2:115–119 [DOI] [PubMed] [Google Scholar]
- 35. Moissl C, La Duc MT, Osman S, Dekas AE, Venkateswaran K. 2007. Molecular bacterial community analysis of clean rooms where spacecraft are assembled. FEMS Microbiol. Ecol. 61:509–521 [DOI] [PubMed] [Google Scholar]
- 36. Mugambi GK, Huhndorf SM. 2009. Molecular phylogenetics of Pleosporales: Melanommataceae and Lophiostomataceae re-circumscribed (Pleosporomycetidae, Dothideomycetes, Ascomycota). Stud. Mycol. 64:103-121S104 doi:10.3114/sim.2009.64.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nagano Y, et al. 2009. Identification of airborne bacterial and fungal species in the clinical microbiology laboratory of a university teaching hospital employing ribosomal DNA (rDNA) PCR and gene sequencing techniques. Int. J. Environ. Health Res. 19:187–199 [DOI] [PubMed] [Google Scholar]
- 38. Neefs JM, Van de Peer Y, Hendriks L, De Wachter R. 1990. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 18(Suppl):2237–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Newcombe DA, et al. 2005. Survival of spacecraft-associated microorganisms under simulated Martian UV irradiation. Appl. Environ. Microbiol. 71:8147–8156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH. 2008. Intraspecific ITS variability in the kingdom fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinform. Online 4:193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nilsson RH, et al. 2010. An open source software package for automated extraction of ITS1 and ITS2 from fungal ITS sequences for use in high-throughput community assays and molecular ecology. Fungal Ecol. 3:284–287 [Google Scholar]
- 42. Onofri S, et al. 2012. Survival of rock-colonizing organisms after 1.5 years in outer space. Astrobiology 12:508–516 [DOI] [PubMed] [Google Scholar]
- 43. Osman S, et al. 2008. Effect of shadowing on the survival of bacteria to conditions simulating Martian atmosphere and UV radiation. Appl. Environ. Microbiol. 74:959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pace NR. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734–740 [DOI] [PubMed] [Google Scholar]
- 45. Page RD. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 46. Pantoja LD, et al. 2009. Ants (Hymenoptera: Formicidae) as carriers of fungi in hospital environments: an emphasis on the genera Tapinoma and Pheidole. J. Med. Entomol. 46:895–899 [DOI] [PubMed] [Google Scholar]
- 47. Puleo JR, et al. 1977. Microbiological profiles of the Viking spacecraft. Appl. Environ. Microbiol. 33:379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saadoun I, Al Tayyar IA, Elnasser Z. 2008. Concentrations of airborne fungal contaminations in the medical surgery operation theaters (OT) of different hospitals in northern Jordan. Jordan J. Biol. Sci. 1:181–184 [Google Scholar]
- 49. Satoh K, et al. 2011. Microbe-I: fungal biota analyses of the Japanese experimental module KIBO of the International Space Station before launch and after being in orbit for about 460 days. Microbiol. Immunol. 55:823–829 [DOI] [PubMed] [Google Scholar]
- 50. Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310 doi:10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schloss PD, et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Selbmann L, de Hoog GS, Mazzaglia A, Friedmann EI, Onofri S. 2005. Fungi at the edge of life: cryptoendolithic black fungi from Antarctic deserts. Stud. Mycol. 51:1–32 [Google Scholar]
- 53. Sogin ML, et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tedersoo L, et al. 2010. 454 pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 188:291–301 [DOI] [PubMed] [Google Scholar]
- 55. Utescher CLDA, Franzolin MR, Trabulsi LR, Gambale V. 2007. Microbiological monitoring of clean rooms in development of vaccines. Braz. J. Microbiol. 38:710–716 [Google Scholar]
- 56. Vaishampayan P, Osman S, Andersen G, Venkateswaran K. 2010. High-density 16S microarray and clone library-based microbial community composition of the Phoenix spacecraft assembly clean room. Astrobiology 10:499–508 [DOI] [PubMed] [Google Scholar]
- 57. Vaishampayan P, Rabbow E, Horneck G, Venkateswaran K. 2012. Survival of Bacillus pumilus spores for a prolonged period of time in real space conditions. Astrobiology 12:487–497 http://online.liebertpub.com/doi/pdfplus/10.1089/ast.2011.0738 [DOI] [PubMed] [Google Scholar]
- 58. Vaishampayan P, et al. Deinococcus phoenicis sp. nov., an extreme ionizing radiation resistant bacterium isolated from the Phoenix Lander assembly facility. Int. J. Syst. Evol. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 59. Werner JJ, et al. 2012. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 6:94–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhdanova NN, et al. 1991. The interaction of soil micromycetes with “hot” particles in a model system. Mikrobiol. Zh. 53:9–17 (In Russian.) [PubMed] [Google Scholar]
- 61. Zhdanova NN, Zakharchenko VA, Haselwandter K. 2005. Radionuclides and fungal communities, p 759–768 In Dighton J, White JF, Oudemans P. (ed), The fungal community: its organization and role in the ecosystem. CRC Press, Baton Rouge, LA [Google Scholar]
- 62. Zhdanova NN, Zakharchenko VA, Vember VV, Nakonechnaya LT. 2000. Fungi from Chernobyl: mycobiota of the inner regions of the containment structures of the damaged nuclear reactor. Mycol. Res. 104:1421–1426 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.