Abstract
Epichloid endophytes provide protection from a variety of biotic and abiotic stresses for cool-season grasses, including tall fescue. A collection of 85 tall fescue lines from 15 locations in Greece, including both Continental and Mediterranean germplasm, was screened for the presence of native endophytes. A total of 37 endophyte-infected lines from 10 locations were identified, and the endophytes were classified into five distinct groups (G1 to G5) based on physical characteristics such as colony morphology, growth rate, and conidial morphology. These classifications were supported by phylogenetic analyses of housekeeping genes tefA and tubB, and the endophytes were further categorized as Neotyphodium coenophialum isolates (G1, G4, and G5) or Neotyphodium sp. FaTG-2 (Festuca arundinacea taxonomic group 2 isolates (G2 and G3). Analyses of the tall fescue matK chloroplast genes indicated a population-wide, host-specific association between N. coenophialum and Continental tall fescue and between FaTG-2 and Mediterranean tall fescue that was also reflected by differences in colonization of host tillers by the native endophytes. Genotypic analyses of alkaloid gene loci combined with chemotypic (chemical phenotype) profiles provided insight into the genetic basis of chemotype diversity. Variation in alkaloid gene content, specifically the presence and absence of genes, and copy number of gene clusters explained the alkaloid diversity observed in the endophyte-infected tall fescue, with one exception. The results from this study provide insight into endophyte germplasm diversity present in living tall fescue populations.
INTRODUCTION
Epichloid endophytes, comprised of sexual Epichloë and asexual Neotyphodium species, associate with cool-season grasses, including the agronomically important forage grass tall fescue [Lolium arundinaceum (Schreb.) Darbysh. syn Festuca arundinacea Schreb] (46, 47). Many Neotyphodium species arise through interspecific hybridization and contain genomic information from more than one progenitor species. Other Neotyphodium spp. are nonhybrids and appear to have evolved directly from sexual progenitors that have lost the ability to form stromata (47). These symbionts contribute to host persistence and tolerance to biotic and abiotic stresses (2, 13, 36). Collectively, these fungi are able to produce a range of bioactive alkaloids that deter herbivory, including the ergot alkaloids, pyrrolopyrazine (peramine), aminopyrrolizidines (lolines), and indole-diterpenes (including lolitrems and terpendoles) (7, 47, 53). Peramine and loline alkaloids confer anti-insect defenses to their host (7, 51, 57, 63), while ergot alkaloids and lolitrem B cause fescue toxicosis and ryegrass staggers in grazing mammals, respectively (4, 21, 22, 43).
Efforts to better understand the production of these compounds have led to the cloning and characterization of the genes required for the biosynthesis of the alkaloids from a number of epichloae, including hybrid and nonhybrid species (19, 40, 55, 57, 61, 68). The loci required for the production of lolines (LOL), indole-diterpenes (LTM or IDT), and ergot alkaloids (EAS) in Epichloë festucae and the related asexual nonhybrid Neotyphodium lolii are each found as complex gene clusters associated with AT-rich repetitive elements (18, 19, 34, 35, 52, 66–68). A single gene, perA, appears to be required for peramine production and is also associated with repeat sequences in some isolates (18).
Tall fescue is an important cool-season grass grown as forage in temperate climates. Three different races of tall fescue have been recognized (rhizomatous, Continental, and Mediterranean), each with distinctive physiological and morphological traits, geographical distributions, and population structures (27). Rhizomatous tall fescue originates from the Iberian Peninsula and is the target of turf breeding programs due to longer, more prevalent rhizomes (27, 56). Summer-active Continental tall fescue is prevalent in northern Europe and is the primary cultivated germplasm used in temperate regions for animal forage. Summer-dormant Mediterranean tall fescue has been collected from northern Africa and Mediterranean regions and has recently become the focus of grass breeders looking to expand the agronomic range of tall fescue to more arid regions (39, 60). Both Continental and Mediterranean tall fescue germplasm has been collected from Sardinia, indicating that the natural distributions of the two races overlap in the Mediterranean basin (27). However, crosses between Mediterranean germplasm and the other tall fescue races result in F1 hybrids with low fertility (30, 56), and phylogenetic analyses indicate that Mediterranean tall fescue is genetically distinct from Continental and rhizomatous races, possibly from an independent polyploid origin (26, 50).
Intriguingly, the genetic distinction between Continental and Mediterranean races of tall fescue appears to be reflected by the distribution of Neotyphodium endophytes within tall fescue populations. Three Neotyphodium species, i.e., N. coenophialum, Neotyphodium sp. FaTG-2 (Festuca arundinacea [tall fescue] endophyte taxonomic group 2), and Neotyphodium sp. FaTG-3, have been found to associate with tall fescue (12), two of which, Neotyphodium sp. FaTG-2 and Neotyphodium sp. FaTG-3, are found only in association with Mediterranean tall fescue. The most well-studied of these is N. coenophialum, which is commonly found in Continental tall fescue and thus prevalent throughout temperate-grown cultivars. Indeed, a large part of the success of this germplasm relates to the drought tolerance and antiherbivore properties imparted by the endophyte (2, 43, 53). All N. coenophialum isolates have been reported to produce peramine and loline alkaloids with known anti-insect properties, but unfortunately several strains also produce ergovaline, which causes fescue toxicosis in mammalian herbivores (12, 43, 48). Thus, the identification of “livestock-friendly” N. coenophialum isolates lacking ergovaline production is crucial to enhance the agronomic value of elite tall fescue lines without the risk of animal toxicity (6, 28, 29).
Neotyphodium sp. FaTG-2 and Neotyphodium sp. FaTG-3 have been isolated only from Mediterranean-type tall fescue from Italy, southern Spain, and North Africa (11, 13, 41). They are distinct from N. coenophialum in morphology, chemotype, isozyme profiles, alkaloid production, and microsatellite markers (12, 38). All FaTG-2 isolates thus far described produce ergovaline, but they are variable in the production of lolitrem B and peramine (12, 31, 42). However, no published studies have investigated the genetic diversity of these endophytes to understand the basis of this chemotype variation (41, 42).
Tall fescue germplasm was collected from Greece in an effort to incorporate summer dormancy traits into tall fescue breeding populations suitable for the south-central United States. We screened this tall fescue collection to assess endophyte incidence and to determine the likelihood of these lines causing livestock toxicity. Endophytes from infected tall fescue were characterized based on phylogenetic analyses and morphological, genetic, and biochemical traits. Furthermore, we investigated the diversity of the alkaloid genes within the fungal population to better understand the relationship between genetic diversity and chemotype variation of epichloid endophytes. This is the first comprehensive study of a tall fescue endophyte population to allow direct comparison of alkaloid gene profiling and chemotypes.
MATERIALS AND METHODS
Plant collection and fungal endophyte isolation.
In 2007, over 100 individual sites across Greece, Crete, and additional outlying islands were intensely surveyed for the presence of tall fescue. A total of 88 individual tall fescue plants were collected from 15 locations in Greece (Fig. 1; see Data Set S1 in the supplemental material) and are a random representation of tall fescue diversity from these regions. Seeds from 85 plants were produced in the Netherlands in 2008, and endophyte incidence was assessed via PCR screening of three seeds per line (see Fig. S1 in the supplemental material). The tall fescue race (Continental, Mediterranean, or rhizomatous) and ploidy status of each line were determined through sequence analysis of chloroplast matK as described by Hand et al. (27). Four endophyte-infected (E+) seedlings per line were maintained in the greenhouse (24°C during days [15 h] and 20°C during nights). KY31, a Continental-type tall fescue infected with N. coenophialum e19 was kindly provided by C. L. Schardl (University of Kentucky), and NFTF 1800, a Mediterranean-type tall fescue infected with a native Neotyphodium sp. FaTG-2 endophyte, were maintained under identical conditions and used as plant controls as indicated.
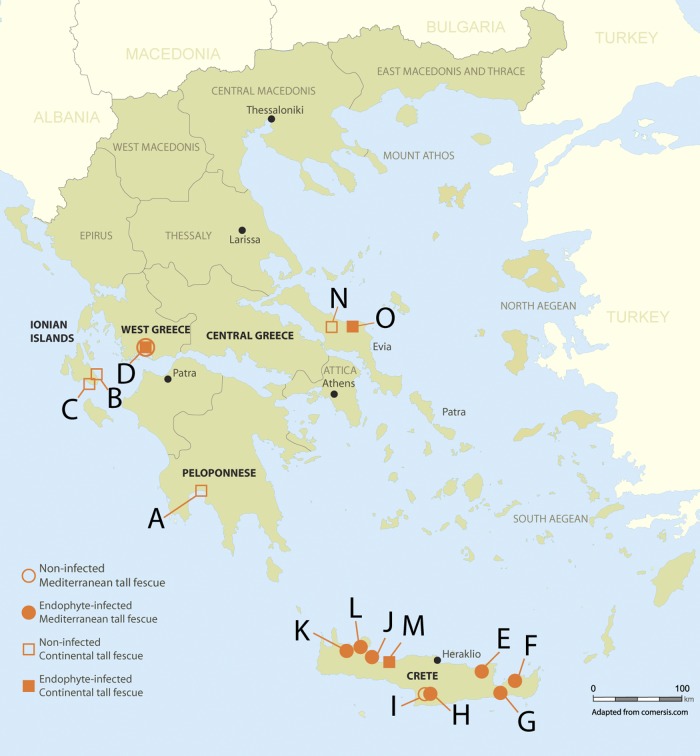
Fig 1.
Tall fescue collection locations in Greece. Open circles indicate locations where no endophyte-infected lines were collected. Filled shapes indicate locations where at least one endophyte-infected line was obtained. Squares represent N. coenophialum-infected lines, and circles represent Neotyphodium sp. FaTG-2-infected lines. (Adapted from comersis.com.)
Pure cultures of endophytes from each plant line were obtained from surface-sterilized pseudostem sections and purified by hyphal tipping (62). Growth rate measurements and colony morphology observations were made for all endophyte isolates grown on potato dextrose agar (PDA). Isolates were grown on 1% water agar plates to observe conidial morphology. Endophyte cultures are available at The Samuel Roberts Noble Foundation upon request.
Molecular biological techniques.
Genomic DNA from seeds and infected tillers was isolated using a MagAttract 96 DNA plant core kit (Qiagen, Valencia, CA). DNA extracted in this manner was not quantified prior to amplification. Genomic DNA from pure cultures of fungal isolates was isolated from freeze-dried mycelium using a ZR fungal/bacterial DNA Miniprep kit (Zymo Research, Irvine, CA). Fungal genomic DNA was quantified by Hoechst dye staining followed by fluorometry using a DyNA Quant 200 fluorometer (Hoefer, Inc., Holliston, MA).
Housekeeping genes tubB and tefA and alkaloid biosynthesis genes were amplified from total DNA extracted from seeds (6 μl), fresh tillers (3 μl), or fungal mycelia (5 ng) using primers listed in Table S1 in the supplemental material. Primers for the alkaloid biosynthesis genes were designed to bind to conserved gene regions as determined by available fungal gene and genome sequences. Alkaloid genes were initially amplified from DNA of infected tillers but were later confirmed with amplification of fungal DNA. PCR mixtures included 1× Green GoTaq reaction buffer (Promega, Madison, WI), 200 μM each deoxynucleoside triphosphate (dNTP), 200 nM each primer, and 1 U GoTaq DNA polymerase (Promega). PCR conditions for all amplifications were as follows unless indicated otherwise: 94°C for 2 min; 30 cycles of 94°C for 15 s, 56°C for 30 s, and 72°C for 1 min; and then 1 cycle of 72°C for 7 min. Amplification of tubB utilized a 50°C annealing temperature. PCR amplicons (10 μl) were observed using agarose gel electrophoresis, where the visualization of a band of the expected size indicated the presence of the endophyte or a copy of an alkaloid gene within the fungal genome.
The tefA, tubB, and perA (partial second adenylation domain) amplicons were TA ligated into a pGEM-T Easy vector (Promega) and subsequently used to transform One Shot TOP10 competent cells (Invitrogen). Individual clones were selected (12 to 48 clones per isolate per gene) and grown in 96-well plates in Terrific Broth (45) at 470 rpm for 20 h in the HiGro microwell plate growth system (Genomic Solutions, Inc., Ann Arbor, MI). Plasmid DNA isolation was performed using a Biomek FXP instrument (Beckman Coulter Inc., Brea, CA) and sequenced using primers SP6 and T7.
Purified plasmids and PCR amplicons of key alkaloid genes were sequenced using BigDye chemistry (v3.1; Applied Biosystems, Foster City, CA) and analyzed with an Applied Biosystems 3730 DNA Analyzer. Sequences were viewed, edited, and managed using Sequencher v5.0 (Gene Codes, Ann Arbor, MI). Additional sequences were obtained from the GenBank database at the National Center for Biotechnology Information (NCBI) for comparative analyses.
Phylogenetic analyses.
Sequences from all Greek isolates were aligned using the platform at www.phylogeny.fr (15, 16) with the following steps: sequences were aligned using MUSCLE 3.7 (17) and refined using Gblocks 0.9b (8), the phylogenetic trees were inferred with PhyML 3.0 (25), branch support was estimated by the approximate likelihood-ratio test (1) with the SH-like option, and TreeDyn 198.3 rendered the phylogenetic trees (10). Phylogenies of the housekeeping genes tubB and tefA included sequences from representative Epichloë and Neotyphodium species, specifically, E. baconii, E. festucae, E. typhina, N. coenophialum, and Neotyphodium sp. FaTG-2 (see Table S2 in the supplemental material). For the sake of clarity, where necessary, gene copies are amended with a prefix referring to the isolate or species, followed by the gene name, with a suffix that reflects the phylogenetic heritance of the gene; e.g., Nco-tefA-Efe refers to the N. coenophialum tefA gene copy inherited from the E. festucae progenitor (14).
Chemical analyses of secondary metabolites.
Tillers from 3 or 4 plants per E+ line were harvested in May 2011 and subdivided into blade and pseudostem samples. Plant tissues were ground to a powder and stored at −20°C until used for further analyses.
Ergovaline concentrations in pseudostems and blades of all endophyte-infected tall fescue lines were measured in duplicate using 10 ± 0.10 mg of tissue. Tissue samples were extracted in 200 μl methanol solution containing 0.005 mg/ml dihydroergotamine tartrate salt (Sigma-Aldrich) for 2.5 to 3 h and then centrifuged at 1,800 × g for 5 min. For each sample, 2.5 μl of the supernatant was analyzed for the presence of ergovaline using an Acquity ultra-performance liquid chromatography (UPLC) system (Waters Corporation, Milford, MA) fitted with a fluorescence detector. Separations were achieved using a 2.1- by 150-mm BEH C18 column (Waters) with a flow rate of 0.35 ml/min at 30°C. The mobile phase consisted of a linear gradient of 80:20 to 5:95 (0.1 M ammonium acetate-acetonitrile) in 6.0 min followed by reequilibration for 1.5 min at 80:20. Detection was achieved using a fluorescence detector with excitation at 310 nm and emission at 410 nm with a photomultiplier (PMT) gain setting of 1.00 and data rate of 5 scans/s. Data were processed using Empower 2 software (Waters). As ergovaline is not commercially available, seed extracts in which the ergovaline content was previously quantified (A. M. Craig, Endophyte Testing Laboratory, Oregon State University) were used as standards (0, 50, 97, 500, and 1000 ppb) for quantification. Standards and samples were analyzed in duplicate and values averaged.
Peramine and indole-diterpene analyses of pseudostem samples from representative tall fescue lines were performed by AgResearch (Palmerston North, New Zealand) as described previously (66, 67) using 50.0 mg lyophilized tissue. Loline alkaloid analyses of blade and pseudostem samples from selected tall fescue lines were performed at the University of Kentucky (Lexington, KY) from 100.0 mg lyophilized tissue using the method as described by Blankenship et al. (5) except that sodium hydroxide was substituted for sodium bicarbonate to basify tissue samples. Quinoline was used as an internal standard, and the lolines were quantified by gas chromatography-ion trap mass spectrometry (GC/ITMS) using a Saturn 2200 machine (Varian Inc., Santa Clara, CA).
Nucleotide sequence accession numbers.
The unique tubB and tefA sequences have been submitted to GenBank under accession numbers JX028244 to JX028269 (see Table S2 in the supplemental material).
RESULTS
Tall fescue classification and endophyte incidence.
The chloroplast matK gene was amplified and partially sequenced to determine the ploidy and race of each plant line from Greece. All lines were determined to be hexaploid tall fescue, with 38 lines from 7 locations (A, B, C, D, M, N, and O) classified as Continental and 47 lines from 9 locations (D, E, F, G, H, I, J, K, and L) as Mediterranean (Fig. 1; see Data Set S1 in the supplemental material).
Endophyte-specific tefA primers were used to amplify total DNA isolated from seed of each line (see Fig. S1 and Data Set S1 in the supplemental material), followed by PCR analysis of eight seedlings per plant line to evaluate percent infection and transmissibility. A total of 37 lines were identified as endophyte infected, with the endophyte incidence ranging from 37 to 100%. Although the number of tall fescue plants collected was almost evenly split between southern Greece (43 lines from 6 locations) and Crete (42 lines from 9 locations), 92% (34/37) of endophyte-positive tall fescue lines were collected from Crete (Fig. 1). Overall, tall fescue collected from Crete had an infection rate of 81% (34/42), whereas that obtained from mainland Greece had an overall infection rate of 6.9% (3/43).
Endophyte characterization.
Pure endophyte cultures were isolated and observed for physical characteristics such as colony morphology, growth rate, and conidial morphology (see Fig. S2 and Data Set S1 in the supplemental material). Colony morphology was consistent among isolates from the same location, although some variation in growth rate was observed. The conidial shape was consistent among isolates from the same location and also among isolates that share the same colony morphology. Five distinct morphotypes (Greek types 1 to 5 [G1 to G5]) were observed across the 37 individual isolates.
The colonization pattern in planta of the Greek tall fescue endophytes was assessed by a PCR screen using DNA extracted from the pseudostem, collar, and blade tissues of endophyte-infected plants (Fig. 2A). Colonization of G1, G4, and G5 endophytes was readily detected in the pseudostem and collar, but little colonization was observed in the leaf blades of their native Continental tall fescue hosts (Fig. 2B). However, in all G2 and G3 endophyte/Mediterranean tall fescue associations, the endophyte could be readily detected in pseudostem, collar, and blades, indicating fungal colonization and accumulation of significant biomass within all aerial tissues. To assess whether the pseudostem/blade colonization pattern of the Greek tall fescue endophytes is consistent with other N. coenophialum and FaTG-2/tall fescue associations, we observed endophyte-infected Continental and Mediterranean tall fescue tillers for the growth pattern of the endophyte (Fig. 2B). Endophyte presence could easily be detected in the blade tissue of E+ NFTF 1800, a Mediterranean-type tall fescue, but was not readily observed in the blade tissue of E+ KY31, a Continental-type tall fescue.
Fig 2.
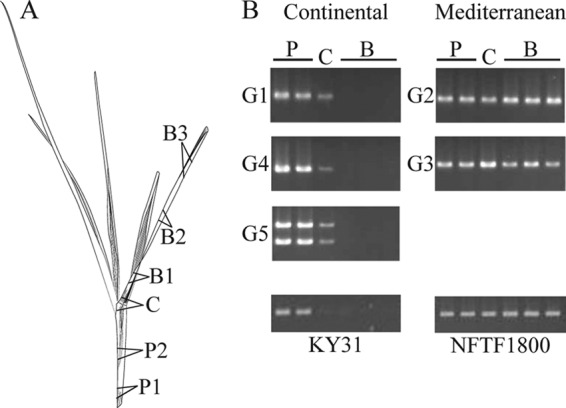
Colonization pattern of endophytes within tall fescue lines. (A) Diagram of a tall fescue tiller, with locations of tissue samples marked accordingly. (B) Amplification of endophyte DNA in pseudostem (P), collar (C), and leaf blade (B) tissues of Greek tall fescue plants and plants previously characterized as E+ Continental type (KY31) or E+ Mediterranean type (NFTF 1800). Total DNA extracted from the specified tissues of infected tillers was amplified using endophyte-specific tefA primers. Amplicon detection indicates the presence of fungal biomass within the plant tissue.
Phylogenetic analyses of all the Greek isolates were conducted using tubB and tefA intron sequences. The sequences and copy number for each gene were identical within each of the Greek-type groupings and have been collapsed to represent G1 to G5. Three tubB and tefA copies were identified in G1, G4, and G5, but only two copies were identified in G2 and G3 (see Table S2 in the supplemental material). Phylogenetic trees for the two genes are shown in Fig. 3 and in Fig. S3 in the supplemental material, respectively.
Fig 3.
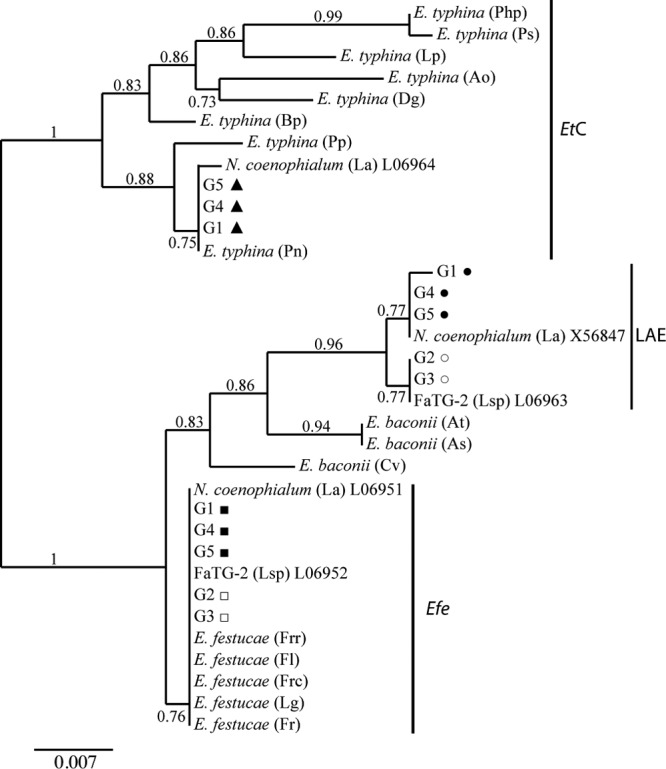
Phylogeny derived from maximum-likelihood (ML) analysis of introns 1 to 4 of tubB genes from representative haploid Epichloë species, hybrid Neotyphodium species, and copies obtained from the Greek tall fescue endophytes. The midpoint root is at the left edge. Numbers at branches are the branch support values; branches with less than 50% support were collapsed. Letters in parentheses after the Epichloë and Neotyphodium isolates refer to host designations as follows: Ao, Anthoxanthum odoratum; As, Agrostis stolonifera; At, Agrostis tenuis; Bp, Brachypodium pinnatum; Cv, Calamagrostis villosa; Fl, Festuca longifolia; Frc, Festuca rubra subsp. commutata; Frr, Festuca rubra subsp. rubra; La, Lolium arundinaceum = Schedonorus phoenix; Lp, Lolium perenne subsp perenne; Lsp, Lolium sp.; Php, Phleum pratense; Pn, Poa nemoralis; Pp, Poa pratensis; Ps, Poa silvicola. Multiple alleles from N. coenophialum and FaTG-2 isolates are designated by GenBank accession number, and G1, G4, and G5 copies are designated by ▲ (copy 1), ■ (copy 2), or ● (copy 3); alleles from G2 and G3 isolates are designated by □ (copy 1) or ○ (copy 2). EtC, Epichloë typhina complex; Efe, E. festucae; LAE, Lolium-associated endophyte clade.
Phylogenetic analyses of the tubB copies representing G1, G4, and G5 reveal that these isolates are closely related to N. coenophialum and its progenitor species E. typhina, E. festucae, and a proposed, but currently unidentified, Lolium-associated endophyte (LAE) (Fig. 3). The G1, G4, and G5 tubB1 copies are identical to that of E. typhina from Poa nemoralis but have one base difference from the N. coenophialum copy from the E. typhina progenitor (Nco-tubB-Ety). The G1, G4, and G5 tubB2 sequences were identical to the Nco-tubB-Efe copy, and the G4 and G5 tubB3 copies were identical to Nco-tubB-LAE, but G1-tubB3 has a single base difference. Similar results were observed with the tefA sequences, with three copies of tefA present in the genome G1, G4, and G5 isolates that cluster with the known N. coenophialum gene copies. Of particular interest, the G5 tefA2 copy contains a 279-bp deletion that correlates with the smaller band observed during PCR amplification of G5 isolates (see Fig. S1 and S3 in the supplemental material) that has not been previously described for epichloid endophytes (37, 58).
Phylogenetic analyses of tubB sequences from the G2 and G3 isolates indicate that they are closely related to Neotyphodium sp. FaTG-2, its progenitor species E. festucae, and the LAE clade representative (Fig. 3; see Fig. S3 in the supplemental material). G2 and G3 tubB1 are identical to FaTG2-tubB-LAE, but G2 and G3 tubB2 have a single base difference from FaTG2-tubB-Efe (Fig. 3). The tefA copies in the G3 isolates are both identical to sequences reported for the FaTG-2 representative isolate 4078 (= Tf14) (12, 37). The G2 tefA copies are more variable, as G2-tefA1 contains a seven-base deletion not present in FaTG-2-tefA-Efe and the G2-tefA2 copy has a single nucleotide variation from FaTG-2-tefA-LAE.
Alkaloid genotype and chemotype diversity.
A PCR-based approach was utilized to detect the presence of genes required for alkaloid biosynthesis from endophyte-infected plants and cultured isolates representing each line (Table 1; see Data Set S1 in the supplemental material). Each independent plant per line produced results identical to the data generated from the pure cultures, indicating that a single endophyte was present in each plant line. All isolates produced PCR amplicons for the five representative gene markers spanning the EAS locus (predicting the presence of dmaW, cloA, easH, lpsA, and lpsB) (Table 1) and thus have the genetic capacity to produce ergovaline, the causal agent of fescue toxicosis (3, 19, 40). Similarly, all isolates were predicted to have the ability to produce peramine due to the presence of PCR amplicons from three perA gene regions, i.e., the second adenylation (A2), second thiolation (T2), and reductase (R) domains (Table 1). The reductase domain encoded by the 3′ end of the 8.5-kb perA gene was specifically targeted as a marker because a deletion in this region has been linked to the inability to produce peramine in N. lolii Lp14 and an identical deletion is found in E. festucae E2368 (18). Amplification of the A2 domain resulted in an additional, smaller amplicon observed only in G2 and G3 isolates (see Fig. S1 in the supplemental material).
Table 1.
Alkaloid profiles of endophytes isolated from Greek tall fescue
| Groupa | Site(s) | E+ incidence | Peramineb |
Ergot alkaloidsc |
Lolines (lol) |
Indole-diterpenes (ltm) |
Expected chemotyped | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PerA-A2 | PerA-red | dmaW | lpsA | C | F | D | T | A | U | O | E | P | G | M | C | B | P | Q | F | K | J | E | ||||
| G1 | D | 1/10 | + | + | + | + | + | + | + | + | + | + | + | + | + | PER, EV, NFL | ||||||||||
| G2 | E, F, G | 15/18 | ++ | + | + | + | + | + | + | + | + | + | + | + | + | + | PER, EV, LB | |||||||||
| G3 | H, J, K, L | 16/19 | ++ | + | + | + | + | + | + | + | + | + | + | + | PER, EV, TER | |||||||||||
| G4 | M | 3/4 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | PER, EV, NFL | |||||
| G5 | O | 2/5 | + | + | + | + | + | + | + | + | + | + | + | + | + | PER, EV, NFL | ||||||||||
The 37 tall fescue endophytes described in this study have been categorized into five morphotypes/genotypes, Greek types 1 through 5 (G1 through G5).
+, one PCR amplicon; ++, two PCR amplicons.
dmaW and lpsA are genes encoding enzymes involved early and late in the ergot alkaloid pathway, respectively.
PER, peramine; EV, ergovaline; NFL, N1-formylloline; LB, lolitrem B; TER, terpendoles.
Genomic diversity was more apparent across the LOL and LTM loci. All nine loline biosynthesis (LOL) genes (51, 55) were detected only in the G1, G4, and G5 isolates (Table 1). This gene profile predicts that these isolates will likely contain the complete LOL locus and have the ability to produce N1-formylloline (NFL) in planta, the final compound synthesized in the loline biosynthetic pathway (51, 55). However, no LOL genes were observed in the G2 and G3 isolates. Ten indole-diterpene/lolitrem biosynthesis (LTM) genes across the LTM locus were evaluated, with only the G2, G3, and G4 isolates observed to contain LTM genes (Table 1). Only G2 isolates appeared to contain all the genes from the LTM locus, and as such they appear capable of producing lolitrem B in planta, an end product of the indole-diterpene pathway (67). G3 isolates contain all LTM genes except ltmE and ltmJ, and based on this gene profile, they would be unlikely to produce lolitrem B but could potentially produce terpendole intermediates, which was recently demonstrated with isolates that have a similar LTM profile (67). G4 isolates contained only five of the LTM genes but lacked genes encoding the early pathway steps, such as ltmG and ltmM (Table 1), indicating that these isolates would be unable to produce any indole-diterpenes.
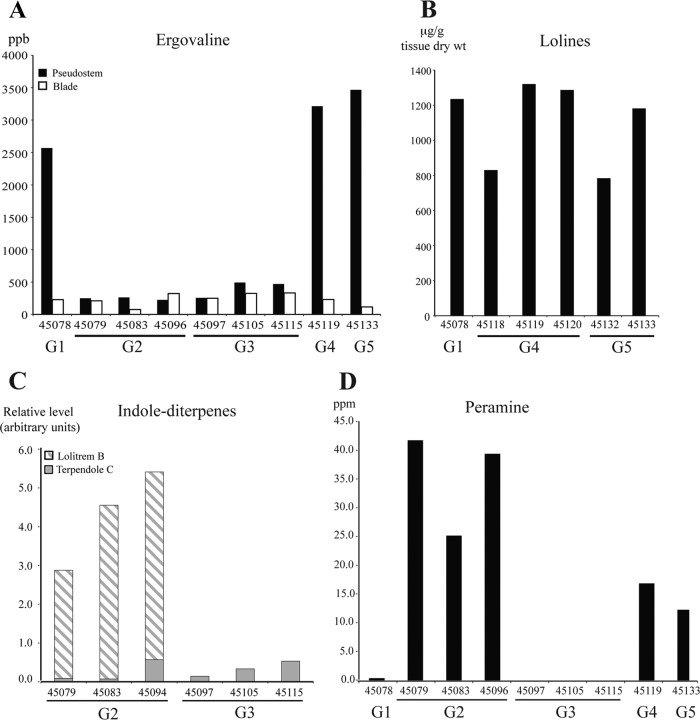
The alkaloid chemotypes predicted by gene profiling were confirmed by chemical analysis of endophyte-infected plant material (Table 1; Fig. 4). All chemotype predictions except for peramine production were validated. Ergovaline was detected in all endophyte-infected tall fescue, with N. coenophialum (G1, G4, and G5)-infected lines producing higher concentrations of ergovaline (average of 1,732 ppm) than FaTG-2 (G2 and G3)-infected lines (average of 276 ppm) (Fig. 4A). However, the ergovaline concentration also varied by plant tissue type. In tall fescue infected with N. coenophialum (G1, G4, and G5), ergovaline in pseudostems was on average 19.5 times higher than that in the leaf blades. Plants infected with FaTG-2 isolates had a more consistent concentration in both tissues, with pseudostems containing ergovaline at levels only 1.3 times higher than in those leaf blades.
Fig 4.
Alkaloid concentrations of selected representative endophyte-infected Greek tall fescue accessions. (A) Ergovaline; (B) lolines; (C) indole-diterpenes; (D) peramine. Genotype designations are listed below the tall fescue lines in each graph. Unless otherwise noted, samples from pseudostem tissue samples of endophyte-infected tillers were analyzed. Analyses of representative samples are shown.
As predicted by the LOL gene profiles (Table 1), tall fescue plants infected with G1, G4, and G5 isolates all produced NFL (Fig. 4B) as well as pathway intermediates N1-acetylnorloline (NANL), N1-acetylloline (NAL), N1-methylloline (NML). The total loline concentrations detected for all lines were comparable to levels reported previously for other Neotyphodium species (69). Consistent with the LTM gene profiles of the G2 and G3 isolates (Table 1), lolitrem B and terpendole intermediates were detected in G2-infected plant lines, whereas only terpendole intermediates, such as terpendole C, were observed in the G3-infected plants (Fig. 4C).
All isolates were predicted to have a complete copy of perA as determined by amplification of three regions within the 8.5-kb gene (Table 1), but not all chemotype predictions were supported by chemical analyses. Representative plants infected with G2, G4, and G5 endophytes contained 12.4 to 43.7 ppm peramine (Fig. 4D), consistent with previously published results for epichloid-infected plants (32, 44, 57, 66). However, none of the G3-infected lines produced peramine, and the single G1 line, 45078, produced less than 0.5 ppm peramine, a value near the level of detection (Fig. 4D).
Copy number and origin of alkaloid genes.
To better understand the genetic and chemotype diversities observed in the Greek tall fescue endophyte population, the copy number and progenitor origin of the alkaloid genes and/or loci were determined by directly sequencing PCR amplicons (see Fig. S5). Although we amplified regions that span at least one intron (except for perA) to increase the likelihood of identifying multiple gene copies, we cannot discount the possibility that additional gene copies may be overlooked.
The copy number of perA was determined by sequencing fragments of the second adenylation and reductase domains. Due to the amplicon size variation noted in the FaTG-2 isolates (G2 and G3) in the second adenylation domain of perA (see Fig. S1 in the supplemental material), fragments from representative isolates were cloned, and 24 clones from each were sequenced. Phylogenetic analysis of the cloned fragments revealed that the genomes of G1, G4, G5, and N. coenophialum e19 contained three copies of perA, where copies 1, 2, and 3 are likely inherited from the E. typhina, E. festucae, and LAE progenitors, respectively (Fig. 5). Two copies of perA were identified in the G2 and G3 isolates, where perA1 and perA2 are likely inherited from the E. festucae and LAE progenitors, respectively, but the perA2 copy contains a 328-bp deletion within the coding region, rendering it nonfunctional. Phylogenetic analysis across the second adenylation domain suggests that G2-perA1 was more closely related to perA from E. festucae Fl1, an isolate capable of producing peramine, and G3-perA1 was more similar to the genes from E. festucae 2368 and N. lolii Lp14 (Fig. 5), isolates with a deletion in the 3′ region of perA linked to the inability to produce peramine (18). Direct sequencing of the perA reductase PCR amplicons revealed polymorphic peaks, which indicates that at least two copies of the perA reductase domain are present in all isolates.
Fig 5.
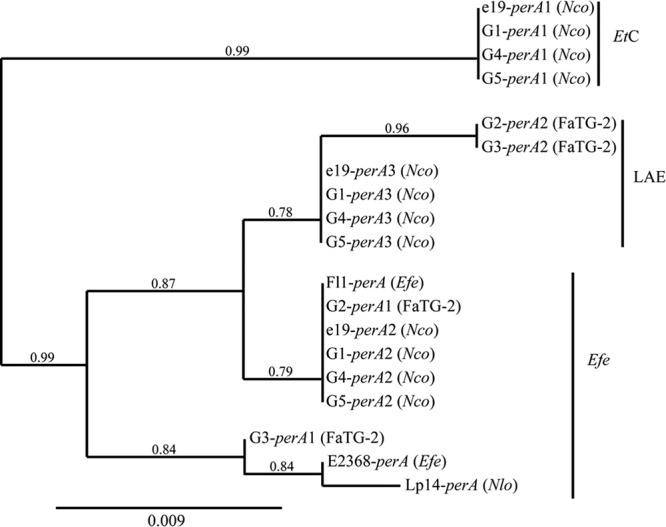
Phylogeny derived from maximum-likelihood (ML) analysis of sequence fragments of perA genes from representative Epichloë and Neotyphodium species and copies obtained from the Greek tall fescue endophytes. The midpoint root is at the left edge. Numbers at branches are the branch support values; branches with less than 50% support were collapsed. EtC, Epichloë typhina complex; Efe, E. festucae; LAE, Lolium-associated endophyte clade.
The copy number of the EAS loci was determined by directly sequencing PCR amplicons of dmaW and intron-rich cloA from representative isolates. Polymorphic peaks, identified in less than 7% of the sequence, were present in both dmaW and cloA sequences from G1, G2, G4, and G5 isolates, suggesting the presence of at least two copies of each gene. No sequence ambiguities were observed in amplicons from the G3 isolates, indicating the likelihood that only one copy of these genes is present in these isolates. Comparison of the single G3-dmaW with published dmaW sequences (see Fig. S4 in the supplemental material) revealed a higher (99%) identity to N. coenophialum e19 dmaW1 than to Nco-dmaW2 (95% identity) and other copies originating from E. festucae (61). Consistent with these data, G3-cloA has 99% identity to Nco-cloA1 (C. A. Young and C. L. Schardl, unpublished data) but only 93% identity to cloA originating from E. festucae.
The copy number of the LOL loci was determined by direct sequencing of the lolC and lolP amplicons from isolates G1, G4, and G5. No polymorphic peaks were observed in the amplicons, indicating that all isolates contained a single copy of the LOL genes, which were identical to those from N. coenophialum (see Fig. S4 in the supplemental material). Previous analyses of the N. coenophialum LOL genes determined that the genes were inherited from the E. typhina progenitor, despite the fact that no current member of the E. typhina complex (ETC) contains LOL genes (34).
PCR amplicons from five genes spanning the LTM locus (ltmM, ltmK, ltmQ, ltmP, and ltmE) were observed for polymorphic peaks in representative isolates. G2 isolates, which contain all LTM genes (Table 1), were revealed to contain two copies of ltmK, ltmM, ltmP, and ltmQ but only one copy of ltmE. No polymorphic peaks were detected in G3 or G4 ltmM, ltmK, ltmQ, or ltmP amplicons, suggesting that these isolates have only one copy of the LTM genes in their respective genomes. The G2, G3, and G4 gene sequences were aligned to the LTM locus of N. lolii, a known lolitrem B producer, to estimate the percent similarity of the amplified regions. The ltmP and ltmQ sequences from G4 were found to be highly similar to Nlo-ltmP and Nlo-ltmQ (98% and 99% identity, respectively), but all the LTM genes observed in the G3 isolates shared between 95 and 96% identity to the corresponding N. lolii genes. The G3 ltmK, ltmM, and ltmQ sequences were used as a reference to predict each multicopy LTM gene present in G2 isolates. Of the two deconvoluted sequences from each gene, one copy shared 99% sequence identity with N. lolii with only 95 to 96% identity to the G3 sequences, while the second copy was 99% identical to G3 with only 95 to 96% identity to N. lolii.
DISCUSSION
Previous studies of endophytes from tall fescue populations have relied heavily on phenotypic data such as conidium length, isozyme analyses, and alkaloid production to classify individual isolates as N. coenophialum or FaTG-2 or -3 (Festuca arundinaceum endophyte taxonomic groups 2 and 3, respectively) (12, 14, 31, 41, 42). In this study, we combined phylogenetic analyses of housekeeping genes tubB and tefA with a PCR-based alkaloid gene profiling approach and chemical analyses to comprehensively characterize five distinct endophyte morphotypes from a representative tall fescue collection from Greece.
Phylogenetic analyses indicate that three of the unique Greek-type endophytes (G1, G4, and G5) are N. coenophialum isolates, while the G2 and G3 isolates are FaTG-2 (Fig. 3; see Fig. S3 in the supplemental material). Phylogenetic classifications were supported by observations of culture and conidial morphology (see Fig. S2 in the supplemental material). N. coenophialum isolates were obtained only from Continental tall fescue lines, whereas FaTG-2 isolates were isolated only from Mediterranean tall fescue. Host-specific associations between N. coenophialum isolates and Continental tall fescue and between Neotyphodium sp. FaTG-2 isolates and Mediterranean-type tall fescue have been reported previously (12, 14, 42, 49), but this is the first tall fescue endophyte population study to observe this correlation on a larger scale. Our findings, together with tall fescue interfertility data and cladogenic segregation based on grass gene sequences (26, 27, 30, 50), suggest that these tall fescue races are likely different species. Furthermore, the mixture of both tall fescue races identified in this study suggests that Greece, particularly Crete, is a region with overlapping Continental and Mediterranean tall fescue populations and may be a region of considerable biodiversity for both plant and fungal germplasms.
The production of bioactive alkaloids by epichloid endophytes within tall fescue and other cool-season grasses has been demonstrated to provide host protection from herbivory, resulting in substantial ecological and agricultural impacts (3, 6, 7, 43, 44, 48). Traditionally, the alkaloid chemotype of a given endophyte was tested for the presence of in planta pathway end products, as production in culture (in the absence of the host plant) is most often limited or nonexistent. More recently, alkaloid gene profiling of the epichloae has become a critical component of endophyte characterization because it not only facilitates a better understanding of the genetic and metabolic diversity of these plant symbionts but also accurately and rapidly elucidates their agronomic potential (9, 24, 48, 54, 67).
In this study, the alkaloid gene profiles of the EAS, LOL, LTM, and PER loci correctly predicted the chemotypes of the G1, G4, and G5 isolates (ergovaline, N1-formylloline [NFL], and peramine) and the G2 isolates (ergovaline, lolitrem B, and peramine), suggesting that all the alkaloid genes characterized by PCR profiling were functional. Although the G1-infected plant line contained a very low concentration (<0.5 ppm) of peramine, this result may have been influenced by having only three plants of a single line available for analysis or could be due to a host plant effect. In contrast, the alkaloid gene profile of the G3 isolates was only partially correct. These endophytes were predicted to produce, and did in fact produce, ergovaline and terpendoles. However, the presence of all three perA gene markers in the G3 isolates did not correlate with the production of peramine (Fig. 4D), suggesting that G3-perA may contain sequence variation that results in a nonfunctional gene. Variation in peramine and lolitrem B (an indole-diterpene) alkaloid production by FaTG-2 isolates has been reported previously, resulting in three reported chemotypes: (i) ergovaline, (ii) ergovaline and peramine, and (iii) ergovaline, peramine, and lolitrem B, where production of indole-diterpenes was limited to detection of lolitrem B (12). More recently, using liquid chromatography/mass spectrometry (LC/MS) analyses, the terpendoles, intermediates in lolitrem B biosynthesis, have been identified in endophyte-infected plant material containing AR1 (N. lolii) and Lp1 (Neotyphodium sp. LpTG-2), which were previously described as unable to produce lolitrem B (67). The terpendole chemotype has now been equated with isolates that contain a functional LTM locus that lacks the two genes ltmE and ltmJ, which are required for prenylation of the indole moiety (67), similar to the case for G3-infected tall fescue characterized in this study.
Phylogenetic analyses have determined that N. coenophialum is a triparental hybrid consisting of E. festucae, E. typhina, and LAE progenitors, while FaTG-2 is a biparental hybrid of E. festucae and LAE (37, 58). Our phylogenetic analyses confirm that all the endophytes isolated from this Greek tall fescue collection are interspecific hybrids (Fig. 3), which allows for a greater chance of accumulating multiple alkaloid genes from different ancestral parents. Partial sequence analysis of key alkaloid genes indicates that N. coenophialum G1, G4, and G5 isolates contain three copies of perA, two copies of the EAS genes, and a single copy of the LOL locus. Similar results have been reported for the N. coenophialum isolate e19 (20, 34). The alkaloid gene sequences from G1, G4, and G5 were highly similar (99% identity) to sequences from N. coenophialum e19, suggesting a shared pattern of inheritance for all the N. coenophialum isolates. Intriguingly, G4 isolates also contain five LTM genes, a pattern observed in some E. festucae isolates (67), but are unable to produce indole-diterpenes because they lack early pathway genes ltmG and ltmM, which are essential for indole-diterpene production (66, 68). The presence of multiple LTM genes in the G4 isolates (and their absence in G1 and G5 isolates) (Table 1) and of ltmP in N. coenophialum e19 suggests that the E. festucae ancestor contained an LTM locus that subsequently lost one or more genes in lolitrem biosynthesis during the evolution of G1, G5, and other N. coenophialum isolates.
Unlike the N. coenophialum isolates, the FaTG-2 G2 and G3 isolates have different gene copy numbers and inheritance patterns that are supported by the observed chemotypes (Fig. 4). G2 isolates have two copies of EAS genes, one inherited from each progenitor. Two copies of the LTM locus are also present in G2 isolates, with a complete copy derived from the E. festucae progenitor and a second partial copy that lacks ltmE (and likely ltmJ since it is linked with ltmE). In contrast, G3 isolates have only a single copy of the EAS and LTM loci. Alkaloid gene profiling (Table 1) and chemotype analyses (Fig. 4) of the G3-infected tall fescue lines indicate that both loci are functional and provide evidence of a complete EAS locus and a partial LTM locus in the G3 isolates. The G3 EAS and LTM gene sequences are identical to copies present in the G2 isolates but less similar (<97%) to E. festucae genes, suggesting that the G3 isolates inherited both these loci from the LAE progenitor (see Fig. S4 in the supplemental material). Although both G2 and G3 isolates contain two copies of perA, sequence comparison of the G2-perA1 and G3-perA1 adenylation domains identified some variation between the genes. Subsequent phylogenetic analysis of this domain indicates that G2-perA1 was likely derived from an E. festucae Fl1 type, whereas G3-perA1 is more similar to E. festucae E2368-perA (Fig. 5). Interestingly, although Fl1 is a known peramine producer, while E2368 is not (48), we cannot discount that additional sequence variation exists between G2-perA1 and G3-perA1 that explains the different peramine chemotype between the isolate groups. G3-perA1 is the only alkaloid gene in the G3 isolates inherited from an E. festucae ancestor, whereas G2 isolates contain copies of perA and the EAS and LTM loci that were likely provided by the E. festucae progenitor.
We speculate that the G2 and G3 isolates shared a common LAE progenitor but have different E. festucae ancestors. This conclusion is drawn from phylogenetic data, represented by the identity between the G2 and G3 LAE tefA and tubB sequences and alkaloid gene differences that present as both copy number differences (LTM and EAS) and sequence differences (perA). However, we cannot discount the possibility that the LTM and EAS genes were lost from the E. festucae progenitor during or after hybridization. Interestingly, the chemotype variation observed between the G2 and G3 isolates also correlates with a distinct geographical distribution; all G2 isolates were collected from the eastern portion of Crete (locations E, F, and G), whereas all G3 isolates were collected from western Crete (locations H, J, K, and L). This suggests that chemotype data can discriminate some populations, and perhaps even subpopulations, of conspecific endophytes.
Alkaloid gene profiling and subsequent analyses for ergot alkaloids and indole-diterpenes indicate that all endophyte-infected lines would produce ergovaline at levels toxic to grazing livestock and that the G2-infected lines can also produce lolitrem B, the causative agent of ryegrass staggers. Although the FaTG-2-infected Mediterranean tall fescue produces less ergovaline than the N. coenophialum-infected Continental tall fescue (Fig. 4A), livestock toxicity may be exacerbated by ergovaline accumulation in the leaf blades, which correlates with the endophyte colonization patterns observed in tillers in this study (Fig. 2) and previously (11). Our results indicate that the tall fescue lines described in this study, which are currently being evaluated for agronomic performance for future cultivar development (M. A. Trammell, unpublished data), will require the replacement of the endemic endophytes by “livestock-friendly” isolates. Although endophyte growth patterns may partially explain the higher accumulation of ergovaline in pseudostems of N. coenophialum-infected tall fescue lines, it is possible that in conjunction with plant genotype effects, gene copy and/or expression may also play a role in alkaloid production, thus explaining the concentration differences observed between N. coenophialum and FaTG-2.
This study describes the incidence, genetic diversity, and chemotypic diversity of 37 epichloid endophytes identified from 85 representative tall fescue lines collected from Greece. The comprehensive analysis of this endophyte population provides insight into the relationship between genetic diversity at the alkaloid loci and the resulting chemotypic diversity of epichloid endophytes. Our results also indicate that although N. coenophialum G1, G4, and G5 isolates all share common ancestry with the model isolate e19, the Neotyphodium sp. FaTG-2 G2 and G3 isolates likely arise from independent hybridization events. Additional large-scale population studies of tall fescue endophytes, particularly those from Mediterranean tall fescue, are necessary to better understand the evolutionary history of these species.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nikki Charlton and Kelly Craven for valuable discussion on grass endophyte associations and phylogeny. We are grateful to Summer Houghton for providing the tall fescue tiller illustration utilized in this work. The peramine and indole-diterpene analyses were completed by Wade Mace (AgResearch, New Zealand). Loline analyses were completed by Padmaja Nagabhyru and Christopher L. Schardl (University of Kentucky). We thank Pieter Den Haan of Steenbergen, the Netherlands, for his assistance in material collection. We thank Lark Trammell (Forage Analysis Core Facility), David Huhman (Metabolomics Core Facility), and the Genomic Core Facility and the Greenhouse Core Facility at The Samuel Roberts Noble Foundation for technical support.
Support for loline analyses was provided by USDA-ARS Specific Cooperative Agreement 200911131030. We thank The Samuel Roberts Noble Foundation for financial support.
Footnotes
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 2. Arachevaleta M, Bacon CW, Hoveland CS, Radcliffe DE. 1989. Effect of the tall fescue endophyte on plant response to environmental stress. Agron. J. 81:83–90 [Google Scholar]
- 3. Bacon CW. 1995. Toxic endophyte-infected tall fescue and range grasses: historic perspectives. J. Anim. Sci. 73:861–870 [DOI] [PubMed] [Google Scholar]
- 4. Bacon CW, Porter JK, Robbins JD, Luttrell ES. 1977. Epichloë typhina from toxic tall fescue grasses. Appl. Environ. Microbiol. 34:576–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blankenship JD, et al. 2001. Production of loline alkaloids by the grass endophyte, Neotyphodium uncinatum, in defined media. Phytochemistry 58:395–401 [DOI] [PubMed] [Google Scholar]
- 6. Bouton JH, et al. 2002. Reinfection of tall fescue cultivars with non-ergot alkaloid-producing endophytes. Agron. J. 94:567–574 [Google Scholar]
- 7. Bush LP, Wilkinson HH, Schardl CL. 1997. Bioprotective alkaloids of grass-fungal endophyte symbioses. Plant Physiol. 114:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 9. Charlton ND, Craven KD, Mittal S, Hopkins AA, Young CA. 6 June 2012. Epichloë canadensis, a new interspecific epichloid hybrid symbiotic with Canada wildrye (Elymus canadensis). Mycologia [Epub ahead of print.] doi:10.3852/11-403 [DOI] [PubMed] [Google Scholar]
- 10. Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439 doi:10.1186/1471-2105-7-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christensen MJ, Easton HS, Simpson WR, Tapper BA. 1998. Occurrence of the fungal endophyte Neotyphodium coenophialum in leaf blades of tall fescue and implications for stock health. N. Z. J. Agric. Res. 41:595–602 [Google Scholar]
- 12. Christensen MJ, Leuchtmann A, Rowan DD, Tapper BA. 1993. Taxonomy of Acremonium endophytes of tall fescue (Festuca arundinacea), meadow fescue (F. pratensis) and perennial rye-grass (Lolium perenne). Mycol. Res. 97:1083–1092 [Google Scholar]
- 13. Clay K, Schardl CL. 2002. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 160:S99–S127 [DOI] [PubMed] [Google Scholar]
- 14. Clement SL, Elberson LR, Youssef NN, Davitt CM, Doss RP. 2001. Incidence and diversity of Neotyphodium fungal endophytes in tall fescue from Morocco, Tunisia, and Sardinia. Crop Sci. 41:570–576 [Google Scholar]
- 15. Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8 doi:10.1186/1471-2148-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fleetwood DJ, et al. 2011. Abundant degenerate miniature inverted-repeat transposable elements in genomes of epichloid fungal endophytes of grasses. Genome Biol. Evol. 3:1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fleetwood DJ, Scott B, Lane GA, Tanaka A, Johnson RD. 2007. A complex ergovaline gene cluster in Epichloë endophytes of grasses. Appl. Environ. Microbiol. 73:2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Florea S, Andreeva K, Machado C, Mirabito PM, Schardl CL. 2009. Elimination of marker genes from transformed filamentous fungi by unselected transient transfection with a Cre-expressing plasmid. Fungal Genet. Biol. 46:721–730 [DOI] [PubMed] [Google Scholar]
- 21. Gallagher RT, et al. 1982. Ryegrass staggers: the presence of lolitrem neurotoxins in perennial ryegrass seed. N. Z. Vet. J. 30:183–184 [DOI] [PubMed] [Google Scholar]
- 22. Gallagher RT, Hawkes AD, Steyn PS, Vleggaar R. 1984. Tremorgenic neurotoxins from perennial ryegrass causing ryegrass staggers disorder of livestock: structure elucidation of lolitrem B. J. Chem. Soc. Chem. Commun. (Camb.) 1984:614–616 [Google Scholar]
- 23. Reference deleted.
- 24. Ghimire SR, Rudgers JA, Charlton ND, Young CA, Craven KD. 2011. Prevalence of an intraspecific Neotyphodium hybrid in natural populations of stout wood reed (Cinna arundinacea L.) from eastern North America. Mycologia 103:75–84 [DOI] [PubMed] [Google Scholar]
- 25. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 26. Hand ML, Cogan NOI, Stewart AV, Forster JW. 2010. Evolutionary history of tall fescue morphotypes inferred from molecular phylogenetics of the Lolium-Festuca species complex. BMC Evol. Biol. 10:303 doi:10.1186/1471-2148-10-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hand ML, Cogan NOI, Forster JW. 2012. Molecular characterisation and interpretation of genetic diversity within globally distributed germplasm collections of tall fescue (Festuca arundinacea Schreb.) and meadow fescue (F. pratensis Huds.). Theor. Appl. Genet. 124:1127–1137 [DOI] [PubMed] [Google Scholar]
- 28. Hopkins AA, et al. 2010. Agronomic performance and lamb health among several tall fescue novel endophyte combinations in the south-central USA. Crop Sci. 50:1552–1561 [Google Scholar]
- 29. Hopkins AA, Young CA, Bouton JH, Butler TJ. 2011. Registration of ‘Texoma’ MaxQ II tall fescue. J. Plant Reg. 5:14–18 [Google Scholar]
- 30. Hunt KL. 1981. Fertility of hybrids between two geographic races of tall fescue. Crop Sci. 21:400–404 [Google Scholar]
- 31. Jensen AMDM, Mikkelsen L, Roulund N. 2007. Variation in genetic markers and ergovaline production in endophyte (Neotyphodium)-infected fescue species collected in Italy, Spain, and Denmark. Crop Sci. 47:139–147 [Google Scholar]
- 32. Koulman A, Lane GA, Christensen MJ, Fraser K, Tapper BA. 2007. Peramine and other fungal alkaloids are exuded in the guttation fluid of endophyte-infected grasses. Phytochemistry 68:355–360 [DOI] [PubMed] [Google Scholar]
- 33. Reference deleted.
- 34. Kutil BL, et al. 2007. Comparison of loline alkaloid gene clusters across fungal endophytes: predicting the co-regulatory sequence motifs and the evolutionary history. Fungal Genet. Biol. 44:1002–1010 [DOI] [PubMed] [Google Scholar]
- 35. Kutil BL, Liu G, Vrebalov J, Wilkinson HH. 2004. Contig assembly and microsynteny analysis using a bacterial artificial chromosome library for Epichloë festucae, a mutualistic fungal endophyte of grasses. Fungal Genet. Biol. 41:23–32 [DOI] [PubMed] [Google Scholar]
- 36. Malinowski DP, Belesky DP. 2000. Adaptations of endophyte-infected cool-season grasses to environmental stresses: mechanisms of drought and mineral stress tolerance. Crop Sci. 40:923–940 [Google Scholar]
- 37. Moon CD, Craven KD, Leuchtmann A, Clement SL, Schardl CL. 2004. Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Mol. Ecol. 13:1455–1467 [DOI] [PubMed] [Google Scholar]
- 38. Moon CD, Tapper BA, Scott B. 1999. Identification of Epichloë endophytes in planta by a microsatellite-based PCR fingerprinting assay with automated analysis. Appl. Environ. Microbiol. 65:1268–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Norton MR, Volaire F, Lelièvre F, Fukai S. 2009. Identification and measurement of summer dormancy in temperate perennial grasses. Crop Sci. 49:2347–2352 [Google Scholar]
- 40. Panaccione DG, et al. 2001. Elimination of ergovaline from a grass—Neotyphodium endophyte symbiosis by genetic modification of the endophyte. Proc. Natl. Acad. Sci. U. S. A. 98:12820–12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pecetti L, Romani M, Carroni AM, Annicchiarico P, Piano EB. 2007. The effect of endophyte infection on persistence of tall fescue (Festuca arundinacea Schreb.) populations in two climatically contrasting Italian locations. Aust. J. Agric. Res. 58:893–899 [Google Scholar]
- 42. Piano EB, et al. 2005. Specificity of host-endophyte association in tall fescue populations from Sardinia, Italy. Crop Sci. 45:1456–1463 [Google Scholar]
- 43. Porter JK. 1995. Analysis of endophyte toxins: fescue and other grasses toxic to livestock. J. Anim. Sci. 73:871–880 [DOI] [PubMed] [Google Scholar]
- 44. Rowan DD, Dymock JJ, Brimble MA. 1990. Effect of fungal metabolite peramine and analogs on feeding development of Argentine stem weevil (Listronotus bonariensis). J. Chem. Ecol. 16:1683–1695 [DOI] [PubMed] [Google Scholar]
- 45. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 46. Schardl CL. 2010. The Epichloae, symbionts of the grass subfamily Poöideae. Ann. Mo. Bot. Gard. 97:646–665 [Google Scholar]
- 47. Schardl CL, Leuchtmann A, Spiering MJ. 2004. Symbioses of grasses with seedborne fungal endophytes. Annu. Rev. Plant Biol. 55:315–340 [DOI] [PubMed] [Google Scholar]
- 48. Schardl CL, Young CA, Faulkner JR, Florea S, Pan J. 2011. Chemotypic diversity of epichloae, fungal symbionts of grasses. Fungal Ecol. 5:331–344 [Google Scholar]
- 49. Schardl CL. 2001. Epichloë festucae and related mutualistic symbionts of grasses. Fungal Genet. Biol. 33:69–82 [DOI] [PubMed] [Google Scholar]
- 50. Schardl CL, et al. 2008. A novel test for host-symbiont codivergence indicates ancient origin of fungal endophytes in grasses. Syst. Biol. 57:483–498 [DOI] [PubMed] [Google Scholar]
- 51. Schardl CL, Grossman RB, Nagabhyru P, Faulkner JR, Mallik UP. 2007. Loline alkaloids: currencies of mutualism. Phytochemistry 68:980–996 [DOI] [PubMed] [Google Scholar]
- 52. Scott B, et al. 2009. Regulation and functional analysis of bioprotective metabolite genes from the grass symbiont Epichloë festucae, p 199–216 In Gisi U, Chet I, Gullino ML, Cheung EY. (ed), Recent developments in disease management, vol 1 Springer, Berlin, Germany [Google Scholar]
- 53. Siegel MR, et al. 1990. Fungal endophyte-infected grasses: alkaloid accumulation and aphid response. J. Chem. Ecol. 16:3301–3315 [DOI] [PubMed] [Google Scholar]
- 54. Spiering MJ, Wilkinson HH, Blankenship JD, Schardl CL. 2002. Expressed sequence tags and genes associated with loline alkaloid expression by the fungal endophyte Neotyphodium uncinatum. Fungal Genet. Biol. 36:242–254 [DOI] [PubMed] [Google Scholar]
- 55. Spiering MJ, Moon CD, Wilkinson HH, Schardl CL. 2005. Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics 169:1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stewart A. 1997. The development of a rhizomatous tall fescue (Festuca arundinacea) cultivar, p 136–138 In Proceedings of the 8th International Turfgrass Research Conference Australian Turfgrass Research Institute, Ltd, Concord West, New South Wales, Australia [Google Scholar]
- 57. Tanaka A, Tapper BA, Popay A, Parker EJ, Scott B. 2005. A symbiosis expressed non ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol. Microbiol. 57:1036–1050 [DOI] [PubMed] [Google Scholar]
- 58. Tsai HF, et al. 1994. Evolutionary diversification of fungal endophytes of tall fescue grass by hybridization with Epichloë species. Proc. Natl. Acad. Sci. U. S. A. 91:2542–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reference deleted.
- 60. Volaire F, Norton M. 2006. Summer dormancy in perennial temperate grasses. Ann. Bot. 98:927–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang J, Machado C, Panaccione DG, Tsai HF, Schardl CL. 2004. The determinant step in ergot alkaloid biosynthesis by an endophyte of perennial ryegrass. Fungal Genet. Biol. 41:189–198 [DOI] [PubMed] [Google Scholar]
- 62. Whitney HS, Parmeter JR. 1963. Synthesis of heterokaryons in Rhizoctonia solani Kühn. Can. J. Bot. 41:879–886 [Google Scholar]
- 63. Wilkinson HH, et al. 2000. Contribution of fungal loline alkaloids to protection from aphids in a grass-endophyte mutualism. Mol. Plant Microbe Interact. 13:1027–1033 [DOI] [PubMed] [Google Scholar]
- 64. Reference deleted.
- 65. Reference deleted.
- 66. Young CA, et al. 2005. Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Genet. Genomics 274:13–29 [DOI] [PubMed] [Google Scholar]
- 67. Young CA, et al. 2009. Indole-diterpene biosynthetic capability of Epichloë endophytes as predicted by ltm gene analysis. Appl. Environ. Microbiol. 75:2200–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Young CA, et al. 2006. A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fungal Genet. Biol. 43:679–693 [DOI] [PubMed] [Google Scholar]
- 69. Zhang DX, Nagabhyru P, Schardl CL. 2009. Regulation of a chemical defense against herbivory produced by symbiotic fungi in grass plants. Plant Physiol. 150:1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.