Abstract
Aerobic biological ammonia oxidation is carried out by two groups of microorganisms, ammonia-oxidizing bacteria (AOB) and the recently discovered ammonia-oxidizing archaea (AOA). Here we present a study using cultivation-based methods to investigate the differences in growth of three AOA cultures and one AOB culture enriched from freshwater environments. The strain in the enriched AOA culture belong to thaumarchaeal group I.1a, with the strain in one enrichment culture having the highest identity with “Candidatus Nitrosoarchaeum koreensis” and the strains in the other two representing a new genus of AOA. The AOB strain in the enrichment culture was also obtained from freshwater and had the highest identity to AOB from the Nitrosomonas oligotropha group (Nitrosomonas cluster 6a). We investigated the influence of ammonium, oxygen, pH, and light on the growth of AOA and AOB. The growth rates of the AOB increased with increasing ammonium concentrations, while the growth rates of the AOA decreased slightly. Increasing oxygen concentrations led to an increase in the growth rate of the AOB, while the growth rates of AOA were almost oxygen insensitive. Light exposure (white and blue wavelengths) inhibited the growth of AOA completely, and the AOA did not recover when transferred to the dark. AOB were also inhibited by blue light; however, growth recovered immediately after transfer to the dark. Our results show that the tested AOB have a competitive advantage over the tested AOA under most conditions investigated. Further experiments will elucidate the niches of AOA and AOB in more detail.
INTRODUCTION
Nitrification, the microbial oxidation of NH3 (ammonia) to NO3− (nitrate), is one of the key processes of the global nitrogen cycle. The first and rate-limiting step of nitrification is the oxidation of NH3 to NO2− (nitrite). Until recently, aerobic ammonia oxidation was attributed to only a small subset of the Proteobacteria; most freshwater and terrestrial ammonia-oxidizing bacteria (AOB) belong to a distinct group in the Betaproteobacteria, while a few marine AOB species belong to the Gammaproteobacteria (29, 32, 33). The AOB have a chemolithoautotrophic metabolism, oxidizing NH3 to NO2− via the intermediate NH2OH (hydroxylamine) and fixing carbon from CO2 (carbon dioxide) via the Calvin cycle (1).
Recently, genes encoding ammonia monooxygenase (amoA), the first enzyme in the process of ammonia oxidation, were discovered together with archaeal 16S rRNA genes in a metagenomic study (60) and a soil fosmid library (57). At the same time, Nitrosopumilus maritimus, the first archaeal ammonia oxidizer, was isolated in pure culture from a saltwater aquarium (30). Ammonia-oxidizing archaea (AOA) in pure and enrichment cultures have essentially the same metabolism as AOB; they oxidize NH3 stoichiometrically to NO2− and fix carbon from bicarbonate (HCO3−) (15, 20, 30, 36, 43, 56). However, the genomes of N. maritimus and “Candidatus Nitrosoarchaeum limnia” revealed differences between AOA and AOB, such as the use of the 3-hydroxypropionate/4-hydroxybutyrate pathway for HCO3− fixation, the absence of hydroxylamine oxidoreductase, and the presence of many copper-containing enzymes (5, 63).
AOA and AOB often co-occur in the same environment, but the contributions of AOA and AOB to the total ammonia oxidation still need to be elucidated. Many previous studies focused on the influence of environmental factors on niche differentiation between AOA and AOB using cultivation-independent molecular methods. From those studies, it can be concluded that AOA are frequently found in environments with lower substrate (NH4+ and O2) availability and AOB are frequently found in environments with higher substrate availability (see references 4, 13, 17, 24, 42, and 55, among others). However, most of these studies were conducted using methods that target the abundance and/or expression of the archaeal and bacterial amoA genes. Unfortunately, it is not possible to draw direct conclusions about the activity of the AOA and AOB on the basis of the abundance and expression of the amoA gene, because amoA mRNA has been detected in AOB for weeks and 16S rRNA (ribosomes) has been detected for up to a year after the onset of starvation (8, 25, 26). The response of AOA toward starvation and resuscitation has not yet been investigated. In addition, it has been shown that not all amoA-encoding Thaumarchaeota are autotrophic ammonia oxidizers (39, 64). While studies focusing on the analysis of abundance and activity of microbes using molecular methods give very valuable insights, it is also necessary to investigate the response of microbes to environmental factors using cultivation-based approaches, because these experiments will demonstrate changes in physiological activity more conclusively.
Here we present a study that used a cultivation-dependent approach to investigate the responses of AOA and AOB to environmental factors. Cultures of three phylogenetically distinct AOA from freshwater sediments in Ohio were enriched, and the growth of the AOA was characterized under different conditions and compared with that of AOB from freshwater in an enrichment culture. Factors of interest include the NH4+ concentration, pH, O2 concentration, and light wavelength and intensity. These factors have strong effects on the physiology and niche differentiation of AOB (7, 23, 44, 54) and are, therefore, also very likely to influence the physiology and niche differentiation between AOA and AOB.
MATERIALS AND METHODS
Sampling.
Near-shore sediment samples were taken from Lakes Acton (AC; 39°57′N, 84°74′W) and Delaware (DW; 40°39′N, 83°05′W) in fall 2008. Additional sediment core samples were collected from Lake Acton in summer 2009.
Medium.
The mineral salts (MS) medium used to enrich and cultivate AOA and AOB contained 10 mM NaCl, 1 mM KCl, 1 mM CaCl2 · 2H2O, 0.2 mM MgSO4 · 7H2O, and 1 ml liter−1 trace elements solution (9, 61). HEPES buffer was added in a 4-fold molar ratio to the NH4+ concentration, and the pH was adjusted to 7.5 before autoclaving. After autoclaving, sterile KH2PO4 solution was added to obtain a final concentration of 0.4 mM (9, 61).
Enrichment of AOA (AOA-AC2, AOA-AC5, and AOA-DW enrichment cultures).
Sediment samples (1 g) were inoculated into 50 ml MS medium with 0.25 mM NH4+ immediately upon arrival in the laboratory. The enrichments were incubated at 27°C in the dark. NH4+ levels were monitored weekly using a colorimetric assay (9, 28). When the cultures reached late logarithmic growth phase (depletion of about 80% of the initial NH4+ concentration), they were transferred to fresh medium using a 10% (vol/vol) inoculum. The cultures were passed through 0.45-μm-pore-size filters for the first five to six transfers to exclude AOB (9; Annika Mosier, personal communication). In addition to filtration, the enrichment cultures from DW were also treated with 100 μg ml−1 streptomycin to eliminate AOB. After several transfers, when the cultures depleted NH4+ at regular intervals, 20 ml was collected on 0.1-μm-pore-size nitrocellulose filters for molecular characterization. The filters were stored at −20°C.
AOB culture.
We used the previously described AOB strain from a freshwater enrichment culture G5-7 (AOB-G5-7) to compare the growth of AOA to that of AOB (6, 7). The strain belongs to the Nitrosomonas oligotropha cluster and is adapted to low NH4+ concentrations (6, 7). Members of this AOB cluster have been found in many freshwater environments around the world (11, 12, 14, 22, 53; E. French and A. Bollmann, unpublished data).
Growth experiments.
All growth experiments were conducted in MS medium with 0.5 mM NH4+ at pH 7.5 in 125-ml Erlenmeyer flasks with cotton stoppers unless otherwise noted. We tested the influence of different factors (NH4+ concentration, O2 concentration, pH, and light) on the rate of NO2−/NO3− production of the three AOA enrichment cultures (AOA-AC2, AOA-AC5, and AOA-DW) and the AOB enrichment culture (AOB-G5-7). All cultures were inoculated with 10% (vol/vol) conditioned late-log-phase cells and incubated in the dark at 27°C. Samples (1 ml) were taken at regular intervals and centrifuged at 16,000 rpm for 20 min. The supernatant was stored at −20°C for further chemical analysis. To investigate the influence of different NH4+ concentrations, medium with 15 μM to 5 mM NH4+ was prepared with the corresponding HEPES concentrations. The influence of pH was investigated by adjusting the initial pH in the medium to values between 6 and 9. The influence of the O2 concentration was investigated by equilibrating the medium in serum bottles under anaerobic conditions overnight. After equilibration, the bottles were sealed with rubber stoppers. Different calculated O2 concentrations in the headspace were achieved by exchanging the corresponding volume of the headspace with sterile filtered air. The influence of light was investigated by incubating the cultures 18 cm above light-emitting diode panels emitting 30 μmol photons m−2 s−1 at the wavelengths 5,000 to 7,000K (white light), 623 ± 3 nm (red light), and 470 ± 5 nm (blue light) and 3 μmol photons m−2 s−1 at the wavelength 470 ± 5 nm (blue light). The light intensity inside the glass bottles was 25 μmol photons m−2 s−1 (high light conditions) and 2.5 μmol photons m−2 s−1 (low light conditions), as measured with a LI-250A light meter (LI-COR Biosciences, Lincoln, NE), indicating that the glass filtered approximately 15% of the light. To investigate the influence of light-to-dark and dark-to-light transitions on the growth of AOA and AOB, cultures were incubated in the dark until 50% of the NH4+ was consumed and then transferred to the light. At the same time, cultures that were incubated in the light were transferred from the light to the dark. Controls were incubated for the complete cycle in the dark.
Evaluation of growth experiments.
NO2− and NO3− concentrations were determined in the supernatants using colorimetric assays (9, 49). NO2−/NO3− concentrations were log transformed and plotted against time (see Fig. S1 in the supplemental material). Growth rates were calculated from the linear increase (slope) of the log-transformed NO2−/NO3− concentrations over time, assuming that NO2−/NO3− production in the cultures is correlated with the growth of AOA and AOB (3, 9, 30). The increase in NO2−/NO3− production was linear for several days to 1 week, and the correlation coefficients were always ≥0.97 but in most cases were even ≥0.99.
Molecular analysis. (i) DNA isolation from AOA enrichment cultures.
DNA was isolated from the nitrocellulose filters using a Qiagen DNeasy blood and tissue kit (Valencia, CA) with the following modifications. Acid-washed zirconium beads (1 g) and 500 μl high-salt buffer (1 M NaCl, 5 mM MgCl2 · 2H2O, 10 mM Tris, pH 8) (2) were added to the nitrocellulose filters. The filters were homogenized using a bead beater (Biospec Products, Bartlesville, OK) at 4,800 rpm for 30 s. This was repeated three times, and the samples were stored in between cycles on ice for 10 min. After bead beating, 500 μl Qiagen buffer AL and 50 μl proteinase K were added and the mixture was incubated at 56°C for 30 min. The reaction mixture was spun down at 8,000 rpm for 1 min and transferred to spin columns supplied by the manufacturer. The spin columns were treated according to the manufacturer's recommendations, and the DNA was eluted with 100 μl elution buffer AE (Qiagen).
(ii) PCR.
GoTaq green master mix (Promega, Madison WI) was used for all standard PCRs, according to the manufacturer's recommendations, using the primers and protocols summarized in Table S1 in the supplemental material.
(iii) Cloning and sequencing.
PCR products were cleaned using a Wizard SV gel and PCR product cleanup system (Promega, Madison, WI) and cloned into the pGEM-T Easy vector system (Promega, Madison, WI). Transformants were screened for inserts using PCR with M13 primers, and the PCR products were cleaned up and sequenced using a BigDye Terminator (version 3.1) cycle sequencing kit (Life Technology Corporation, Carlsbad, CA) on an Applied Biosystems 3730xl DNA analyzer (Life Technology Corporation).
(iv) DNA sequence analysis.
All sequences were edited with the 4Peaks program (A. Griekspoor and T. Groothuis, The Netherlands Cancer Institute). The sequences were aligned using ARB software (35). Phylogenetic trees were constructed using the neighbor-joining algorithm in ARB, and parsimony and maximum likelihood methods were performed using the PHYLIP program (16). Trees constructed with all three methods showed the same overall grouping; therefore, only the tree constructed with the neighbor-joining method has been presented.
(v) FISH.
The catalyzed reporter deposition (CARD)-fluorescence in situ hybridization (FISH) protocol (45, 48) was used with the following modifications: the hybridization temperature was 46°C, and the first wash was performed at 48°C, followed by an amplification step at 46°C. All probes (see Table S2 in the supplemental material) were labeled at their 5′ ends with horseradish peroxidase and used at a final concentration of 50 ng μl−1. All filters were counterstained with DAPI (4′,6-diamidino-2-phenylindole) for total cell counts. Direct microscopic counts were performed by fluorescence microscopy (Zeiss Axiophot HB0100; Carl Zeiss Inc., North America) at ×1,000 magnification.
Nucleotide sequence accession numbers.
All sequences were deposited in GenBank under the numbers JQ669389 to JQ669394.
RESULTS
Enrichment of AOA.
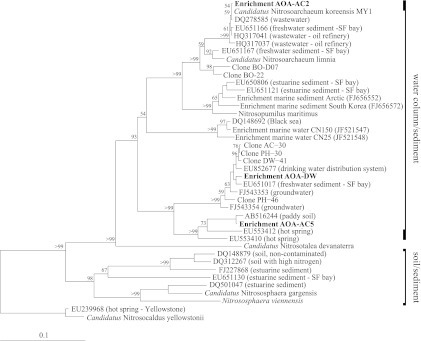
AOA were enriched from the sediment of Lakes Acton (AOA-AC2 and AOA-AC5 cultures) and Delaware (AOA-DW culture) under autotrophic conditions with NH4+ as the sole electron donor in the medium. On the basis of the AOA amoA sequences, all enrichment cultures belong to water column/sediment group I.1a of the Thaumarchaeota (Fig. 1). The strain in the AOA-AC2 culture was 81 to 81.7% (amoA) and 92.8 to 93.1% (16S rRNA gene) identical to the strains in the other two enrichment cultures, while the strains in AOA-AC5 and AOA-DW were 87.1% (amoA) and 97.9% (16S rRNA gene) identical to each other. The amoA sequences of the strain in AOA-DW were 98.2 to 98.5% identical to those of clones from the sediment of Lakes Acton, Delaware, and Pleasant Hill (C. Li and A. Bollmann, unpublished), 98.5% identical to those of clones from the freshwater sediment in the San Francisco Bay (38), and 98.1% identical to those of clones from a drinking water distribution system in The Netherlands (59). The amoA sequences of the strain in AOA-AC5 were 99% identical to the amoA sequence of a clone from a paddy soil in Japan (18). The strain in the third enrichment culture, AOA-AC2, is closely related to “Ca. Nitrosoarchaeum koreensis” (99.8% identity for amoA and 99.6% identity for 16S rRNA gene) and “Ca. Nitrosoarchaeum limnia” (94.3% identity for amoA and 98.5% identity for the 16S rRNA gene) (5, 27). In contrast to the strain in AOA-AC2, the strains in AOA-AC5 and AOA-DW were not closely related to described AOA isolates or strains in enrichment cultures, such as N. maritimus and Nitrososphaera viennensis, among others (70 to 82% identity for amoA and 81 to 93% identity for the 16S rRNA gene) (Table 1).
Fig 1.
Neighbor-joining phylogenetic tree of the AOA in enrichment cultures based on amoA gene sequences (595bp). Bootstrap values of >50 of 100 replicates are shown at the nodes. SF, San Francisco.
Table 1.
Identities of AOA in the AOA-AC2, AOA-AC5, and AOA-DW enrichment cultures in comparison with previously cultivated AOA
| Species (reference) | % identitya |
|||||
|---|---|---|---|---|---|---|
| AOA-AC2 |
AOA-AC5 |
AOA-DW |
||||
| amoA | 16S rRNA | amoA | 16S rRNA | amoA | 16S rRNA | |
| Nitrosopumilus maritimus (30) | 88.6 | 96.2 | 79.8 | 92.9 | 78.8 | 92.9 |
| Nitrososphaera viennensis (56) | 69.6 | 82.7 | 70.4 | 83.7 | 71.1 | 83.7 |
| “Ca. Nitrososphaera gargensis” (20) | 70.9 | 81.9 | 72.7 | 82.7 | 72.1 | 82.2 |
| “Ca. Nitrosocaldus yellowstonii” (15) | 71.1 | 80.4 | 71.1 | 81.8 | 70.0 | 81.0 |
| “Ca. Nitrosoarchaeum limnia” (5) | 94.3 | 98.4 | 81.9 | 92.6 | 81.5 | 92.9 |
| “Ca. Nitrosoarchaeum koreensis” (27) | 99.8 | 99.6 | 81.6 | 92.9 | 81.2 | 92.8 |
| “Ca. Nitrosotalea devanaterra” (34) | 77.9 | 88.5 | 76.7 | 89.7 | 76.1 | 89.3 |
Comparisons are based on 16S rRNA genes (794 bp; corresponding to 109 to 915 in Escherichia coli numbering) and amoA genes (595 bp).
CARD-FISH was used to determine the proportion of AOA in the enrichment cultures at the end of the logarithmic growth phase. AOA-DW contained 85% AOA, AOA-AC2 contained 91% AOA, and AOA-AC5 contained 81% AOA (Table 2). AOB and nitrite-oxidizing bacteria (NOB) were not detected, as tested by PCR amplification with AOB-specific 16S rRNA and amoA primers (see Table S1 in the supplemental material) (results not shown) and FISH with AOB- and NOB-specific 16S rRNA probes (Table 2; see Table S2 in the supplemental material).
Table 2.
Quantitative analysis of compositions of AOA-AC2, AOA-AC5, and AOA-DW enrichment cultures
| Organism | Composition (%)a |
||
|---|---|---|---|
| AOA-AC2 | AOA-AC5 | AOA-DW | |
| Crenarchaeota | 91.0 | 81.2 | 85.4 |
| Bacteria | 9.5 | 3.3 | 9.2 |
| Nitrospira (NOB) | ND | ND | ND |
| AOB | ND | ND | ND |
The cell numbers were determined using CARD-FISH (percentage of DAPI counts) (n = 1). Samples were taken at the end of the logarithmic phase. ND, not detected.
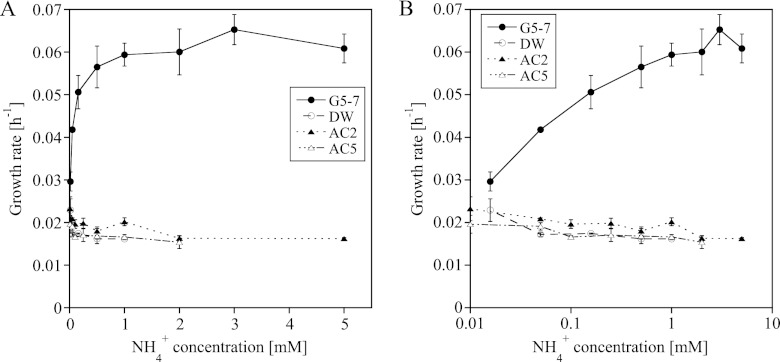
Influence of NH4+ concentration on growth rates of AOA and AOB.
During stratification in the summer, the NH4+ concentration in Lake Acton increases to up to 400 μM (41), which falls within the tested range of NH4+ concentrations of 15 μM and 5 mM NH4+. Increasing NH4+ concentrations up to 1 mM NH4+ doubled the growth rate of the AOB in AOB-G5-7, while the growth rates of the AOA in the enrichment cultures decreased or remained constant (Fig. 2). The growth rate of the AOA in AOA-DW at the lowest NH4+ concentration (15 μM) was significantly higher than the growth rate at higher NH4+ concentrations (see Table S3 in the supplemental material). The same tendency was observed for the other two cultures, although the statistical support was less strong (Fig. 2; see Table S3 in the supplemental material). The AOA strains in the enrichment cultures exhibited different tolerances to high NH4+ concentrations; the strain in AOA-DW grew at NH4+ concentrations up to 1 mM, the strain in AOA-AC2 grew at NH4+ concentrations up to 2 mM, and the strain in AOA-AC5 grew at NH4+ concentrations up to 5 mM (Fig. 2). The lag phase of AOA and AOB differed; the strain in AOB-G5-7 became active 1 to 3 days after inoculation at all tested NH4+ concentrations, whereas the lag phase of the AOA increased with increasing initial NH4+ concentrations up to more than 2 weeks before logarithmic growth could be detected at NH4+ concentrations between 1 mM and 5 mM NH4+ (see Fig. S2 and Table S4 in the supplemental material).
Fig 2.
Influence of NH4+ concentration on the growth rates of the strains in enrichment cultures AOA-AC2, AOA-AC5, AOA-DW, and AOB-G5-7 (mean ± SD; n = 3). NH4+ concentrations are shown on a linear scale (A) and on a logarithmic scale (B).
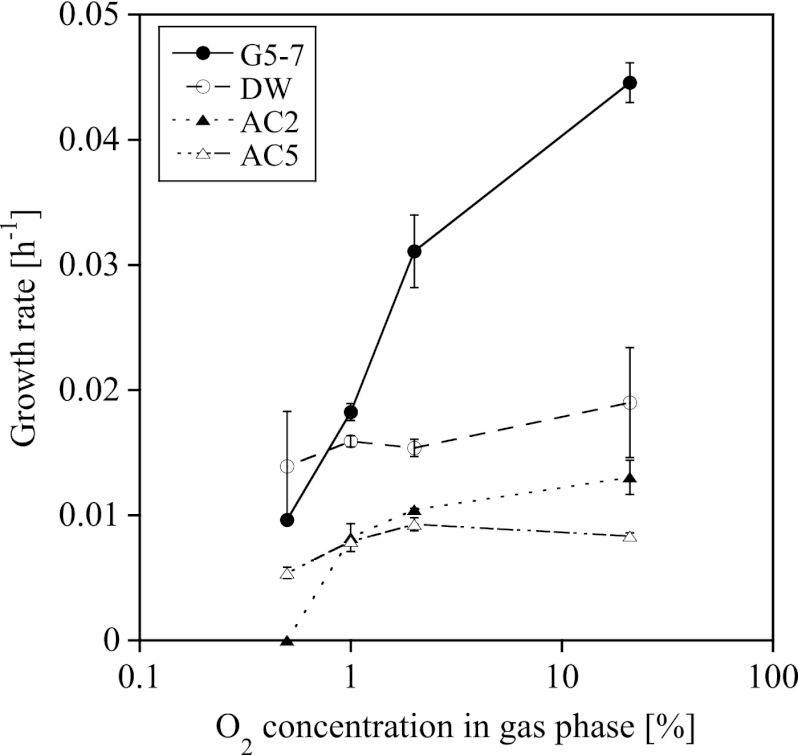
Influence of O2 concentration on growth of AOA and AOB.
Lake Acton stratifies during the summer and has an anaerobic zone as well as a zone with low oxygen availability (1 mg liter−1 O2) (41). We therefore investigated the response of the strains in our enrichment cultures to 0.5 to 2% O2 (calculated) in the headspace, which corresponded to 0.2 to 0.8 mg liter−1 O2 in the medium. The growth rate of the AOB in AOB-G5-7 decreased with decreasing O2 concentration, and the growth rates at all different O2 concentrations were significantly different from each other (Fig. 3; see Table S5 in the supplemental material). The strains in the AOA enrichment cultures grew at all O2 concentrations in the headspace, with the exception of that in the AOA-AC2 enrichment culture at 0.5% O2. The decrease of the growth rates with decreasing O2 concentration in the AOA cultures was less steep than the decrease of the growth rates in the AOB enrichment culture. However, at low O2 concentrations the growth rates in the AOA-AC2 and AOA-AC5 enrichment cultures were significantly lower than the growth rates at 21% O2 (Fig. 3; see Table S5 in the supplemental material).
Fig 3.
Influence of the calculated O2 concentration in the headspace of the bottle on the growth rates of the strains in enrichment cultures AOA-AC2, AOA-AC5, AOA-DW, and AOB-G5-7 (mean ± SD; n = 3).
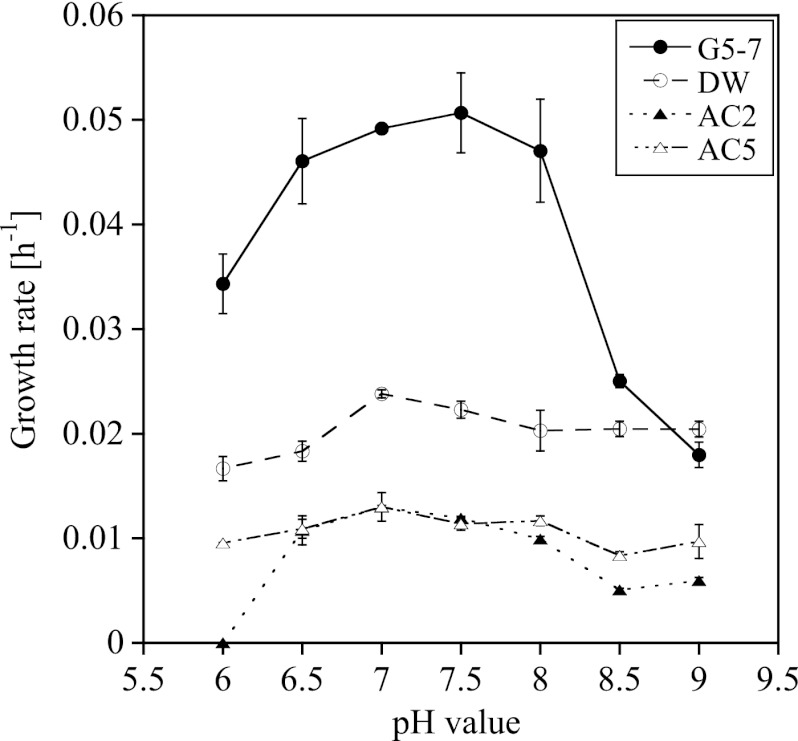
Influence of pH on growth of AOA and AOB.
We investigated the growth of the strains in all cultures at pH 6 to 9, the range at which nonacidophilic ammonia oxidizers grow (31, 32, 33). The growth rates of the strains in all cultures showed bell-shaped curves in relation to the pH, with maximum growth rates at pH 7 to 7.5 (Fig. 4). The strain in the AOA-AC2 culture did not grow at pH 6, while the other AOA strains and the AOB strain did. The growth rates of the strains in AOA-AC5 and AOA-DW cultures at pH 9 were similar to the growth rates at pH 7.5, while the growth rates of the strains in AOA-AC2 and AOB-G5-7 cultures differed significantly from their respective rates at pH 7.5 (Fig. 4; see Table S6 in the supplemental material).
Fig 4.
Influence of the pH of the medium on the growth rates of the strains in enrichment cultures AOA-AC2, AOA-AC5, AOA-DW, and AOB-G5-7 (mean ± SD; n = 3).
Influence of light on growth of AOA and AOB.
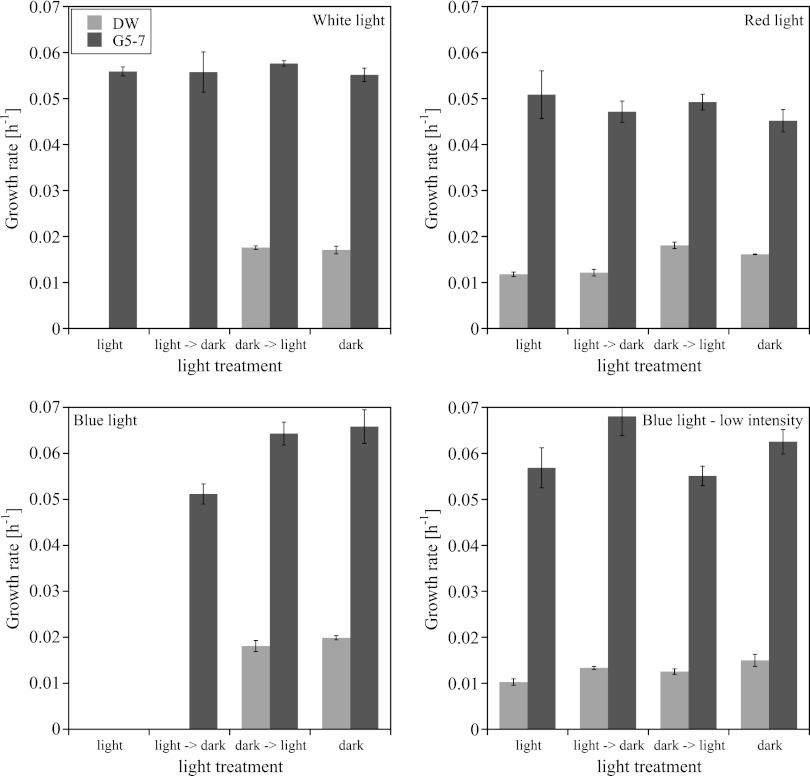
The investigated intensities represent a range of light, but below light saturation, at which phytoplankton in freshwater systems are able to grow (51). White light (30 μmol photons m−2 s−1) strongly inhibited the growth of the AOA in the AOA-DW culture but had no effect on the AOB in the AOB-G5-7 culture (Fig. 5). The AOA did not grow in white light and did not begin to grow after being transferred from the light to the dark. However, growth continued when the AOA cultures were transferred from the dark to the light. To get a better insight into which wavelength of light had the strongest influence on the growth of AOA and AOB, we conducted similar experiments with red (623 ± 3 nm) and blue (470 ± 5 nm) light. Strains in both cultures grew in the red light, but while the growth of the AOB in the AOB-G5-7 culture was not influenced by the red light, the growth rate of the AOA in the AOA-DW culture was significantly lower in the red light and after transfer from the light to the dark (Fig. 5; see Tables S7 and S8 in the supplemental material). Blue light at 30 μmol photons m−2 s−1 had the strongest effect on the growth of strains in both cultures (Fig. 5). In the blue light, strains did not grow in any of the cultures, and growth of the AOA in the AOA-DW culture did not recover after transfer from the light to the dark. In contrast, the AOB in the AOB-G5-7 culture recovered immediately after transfer from the light to the dark, but the growth rate was significantly lower than the growth rate in the continuous dark (see Table S7 in the supplemental material). Transfer of the cultures from the dark into blue light stopped growth immediately. Strains in both cultures grew in the less intense blue light (3 μmol photons m−2 s−1), but the growth rate of strain in the AOA-DW culture was significantly lower in the low blue light than in the dark (Fig. 5; see Table S8 in the supplemental material).
Fig 5.
Influence of white, red, and blue light with an intensity of 30 μmol photons m−2 s−1 and blue light with an intensity of 3 μmol photons m−2 s−1 on the growth rates of the strains in enrichment cultures AOA-DW and AOB-G5-7 (mean ± SD; n = 3).
DISCUSSION
Enrichment of AOA cultures AOA-DW, AOA-AC2, and AOA-AC5.
We enriched and characterized the growth of three different freshwater AOA belonging to thaumarchaeal group I.1a within the newly described phylum Thaumarchaeota (10, 52). The AOA in one of the cultures, AOA-AC2, is closely related to “Ca. Nitrosoarchaeum koreensis,” while the strains in the other two cultures, AOA-AC5 and AOA-DW, are only 70 to 82% (amoA) and 81 to 93% (16S rRNA gene) identical to other cultivated isolates and enrichment culture strains, such as N. maritimus and Nitrososphaera viennensis (Table 1). This finding indicates that the AOA in these two enriched AOA cultures belong to a new genus of the ammonia-oxidizing Thaumarchaeota, assuming that the identity between the two genera is, on average, 96.4% on the basis of the 16S rRNA gene sequence (66). This new genus/group includes many ribotypes from non-salt water systems, such as freshwater systems (38; Li and Bollmann, unpublished) and drinking water systems (59), as well as soil and hot spring environments (67), as indicated by highly identical clones (Fig. 1).
Pure cultures.
In this study, no pure cultures of the AOA were obtained. It is safe to assume that the heterotrophic satellite community is providing some compound that enabled the AOA to grow in the enrichment culture. Similar observations have been made with other AOA as well as with AOB. Potential compounds that positively influence the growth of AOA could be small organic compounds such as pyruvate, which improved growth and enabled isolation of N. viennensis (56). However, the addition of pyruvate during serial dilution has not led to isolation of any of these strains to date, indicating that different compounds might be important for different AOA. Further research will be necessary to elucidate the interactions between AOA (and AOB) and the heterotrophic satellite bacteria in ammonia-oxidizing enrichment cultures.
Growth of AOA and AOB.
Overall, the growth experiments showed that the growth rates of the AOA were almost always lower than the growth rates of the AOB. All our experiments have been conducted under strict chemolithoautotrophic conditions. The results indicate that the strain in AOB-G5-7 had an advantage over the strains in the three tested AOA cultures under the conditions investigated. In nature, however, conditions are often less defined with respect to energy-generating processes. It has been suggested that not all Thaumarchaeota are chemolithoautotrophic ammonia oxidizers; some carry the amoA gene but are not actively oxidizing NH4+, and others utilize mixotrophic or heterotrophic lifestyles in pure and enrichment cultures (39, 56, 65). On the basis of these observations and our data, one could speculate that AOA in natural samples utilize a mixotrophic and/or heterotrophic lifestyle rather than a completely autotrophic lifestyle, which could explain their success in nature compared to that in the laboratory.
Increasing NH4+ concentrations had different influences on the growth rates and lag phases of AOA and AOB, with AOB growing faster and having shorter lag phases than AOA (Fig. 2; see Fig. S2 and Tables S3 and S4 in the supplemental material). After comparing these results with data provided by other studies that determined the Km of AOA for NH3/NH4+ to be approximately 1,000 times lower than the Km of AOB (27, 36, 43), we suggest that AOB have an advantage over AOA at higher NH4+ concentrations (>10 μM). This assumption is supported by the detection of high abundances of AOB in environments with higher NH4+ input due to fertilization and other processes, while AOA are more abundant in low-NH4+ and unfertilized environments (17, 21, 24, 62, 64).
The AOA in enrichment cultures AOA-DW and AOA-AC5 showed lower tolerance to high NH4+ concentrations than the strain in the AOA-AC2 culture, with the highest concentrations supporting growth at 1 mM NH4+ (AOA-DW) and 2 mM NH4+ (AOA-AC5). These concentrations are lower than the highest tolerances toward NH4+ observed for N. viennensis (15 mM), “Ca. Nitrosoarchaeum koreensis” (10 mM), and the strain in the AOA-AC2 enrichment culture (5 mM), which is closely related to “Ca. Nitrosoarchaeum koreensis” (27, 56). These results indicate that the strains in the AOA-DW and AOA-AC5 cultures are less tolerant to high NH4+ concentrations than other AOA isolates and isolates in other enrichment cultures. Similar observations have been made for AOB; members of the Nitrosomonas oligotropha cluster, which are also commonly found in freshwater environments, are less tolerant to high NH4+ concentrations and better adapted to low NH4+ concentrations, while members of the Nitrosomonas europaea/N. eutropha cluster are primarily found in environments with high NH4+ concentrations (6, 7, 31, 32, 33).
AOA and AOB responded differently when cultured over a range of O2 concentrations. The strains in the AOA-AC5 and AOA-DW cultures grew at all tested O2 concentrations at the same rate, while the strain in the AOA-AC2 culture did not grow at 0.5% O2, and the growth rate of the strain in the AOB-G5-7 culture decreased with decreasing O2 concentrations (Fig. 3; see Table S5 in the supplemental material). Environmental surveys often detected AOA at the oxic-anoxic interface (4, 13, 17, 47), indicating an adaptation to low oxygen conditions. The low Km for O2 found for N. maritimus as well as other AOA (27, 36, 43) and the environmental data support the hypothesis that AOA are very likely better adapted to low O2 than AOB and may therefore have a competitive advantage at the oxic-anoxic interface, while AOB are active under more aerobic conditions.
AOA and AOB grew at most of the tested pH values, with AOA growing at almost the same rate over a wide pH range and AOB showing a more bell-shaped curve, with the highest growth rate occurring at pH 7 to 7.5 (Fig. 4; see Table S6 in the supplemental material). AOA are found over a wide pH range in different environments, such as soils and hot springs (15, 19, 21, 40, 46), but most cultivated AOA, such as N. maritimus, N. viennensis, “Ca. Nitrosoarchaeum koreensis,” and “Ca. Nitrosotalea devanaterra,” have rather narrow pH ranges for growth and activity compared with the AOA strains tested in enrichment cultures (27, 30, 56, 58).
The strain in AOB-G5-7 was more tolerant to light than the strain in AOA-DW and also recovered faster after exposure, while the strain in AOA-DW did not fully recover from light exposure (Fig. 5; see Tables S7 and S8 in the supplemental material). In the environment, maximum numbers of thaumarchaeal amoA and 16S rRNA copies have been detected at levels below where photosynthetically active radiation (PAR) in the water column dropped to 0, indicating that no light was penetrating to this depth (4, 47). In the same study, AOB and AOA were detected in low abundance in more shallow waters of the Pacific, indicating that AOB as well as some AOA strains could be more tolerant to light than those that are the most abundant in the lower parts of the water column (47). The light response of AOA and AOB could be due to differences in the reaction of the copper-containing enzymes to light. AOB are very sensitive to blue near-UV light (23, 50). The authors discussed that this inhibition could be attributed to the absorption of light by the oxygenated state of the copper-containing ammonia monooxygenase, which leads to inactivation of the enzyme (50). Genome studies of AOA showed a large number of copper-containing enzymes such as multicopper oxidases and blue copper proteins (5, 63), suggesting that some of the copper-containing enzymes in AOA could be sensitive to light as well, leading to inhibition of overall metabolism in AOA by light. During the preparation of the manuscript, Merbt et al. (2012) published a study investigating the response of two AOB (Nitrosomonas europaea and Nitrosospira multiformis) and two AOA (N. maritimus and “Ca. Nitrosotalea devanaterra”) to white light (37). The study confirmed our findings.
Conclusion.
The results of this study show that AOB are able to outcompete AOA under almost all conditions tested. These findings are in accordance with those of other cultivation-based studies, as well as observations made in the environment using molecular approaches. Further investigation must be done using other cultivation-based experiments, such as continuous cultures, which enable us to cultivate AOA and AOB under more stringently controlled conditions, and in situ incubations, which enable us to investigate the response of AOA and AOB to environmental changes under conditions which allow AOA and AOB to utilize metabolic functions as they would naturally in the environment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Annika Mosier (University of California, Berkeley) for helpful discussions at the beginning of the project, Michael Vanni and Beth Mette (Department of Zoology, Miami University) for support with sampling, Lynn Johnson (Instrumentation Laboratory, Miami University) for construction of the light installations, Anne Bernhard (Connecticut College, New London, CT) for providing her AOA amoA ARB alignment file, and Anne Morris Hooke (Department of Microbiology, Miami University) for critical reading of the manuscript.
This work was supported by startup funds of Miami University and by National Science Foundation grants DEB-1120443 to A.B. and OCE-0927277 to G.B.
Footnotes
Published ahead of print 8 June 2012
This paper is dedicated to the memory of John W. Hawes (Center for Bioinformatics and Functional Genomics and Department of Chemistry and Biochemistry, Miami University).
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Arp DJ, Sayavedra-Soto LA, Hommes NG. 2002. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 178:250–255 [DOI] [PubMed] [Google Scholar]
- 2. Bateson MM, Ward DM. 2008. Methods for extracting DNA from microbial mats and cultivated micro-organisms: high molecular weight DNA from French press lysis, p 53–60 In Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD. (ed), Molecular microbial ecology manual, vol 1 Springer, Dordrecht, The Netherlands [Google Scholar]
- 3. Belser LW, Schmidt EL. 1980. Growth and oxidation kinetics of 3 genera of ammonia-oxidizing nitrifiers. FEMS Microbiol. Lett. 7:213–216 [Google Scholar]
- 4. Beman JM, Popp BN, Francis CA. 2008. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2:429–441 [DOI] [PubMed] [Google Scholar]
- 5. Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. 2011. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6:e16626 doi:10.1371/journal.pone.0016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bollmann A, Laanbroek HJ. 2001. Continuous culture enrichments of ammonia-oxidizing bacteria at low ammonium concentrations. FEMS Microbiol. Ecol. 37:211–221 [Google Scholar]
- 7. Bollmann A, Bär-Gilissen MJ, Laanbroek HJ. 2002. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68:4751–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bollmann A, Schmidt I, Saunders A, Nicolaisen M. 2005. Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl. Environ. Microbiol. 71:1276–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bollmann A, French E, Laanbroek HJ. 2011. Isolation, cultivation, and characterization of ammonia-oxidizing bacteria and archaea adapted to low ammonium concentrations. Methods Enzymol. 486:55–88 [DOI] [PubMed] [Google Scholar]
- 10. Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. 2008. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245–252 [DOI] [PubMed] [Google Scholar]
- 11. Chen GY, Qui SL, Zhou YY. 2009. Diversity and abundance of ammonia-oxidizing bacteria in eutrophic and oligotrophic basins of a shallow Chinese lake (Lake Donghu). Res. Microbiol. 160:173–178 [DOI] [PubMed] [Google Scholar]
- 12. Coci M, Bodelier PLE, Laanbroek HJ. 2008. Epiphyton as a niche for ammonia-oxidizing bacteria: detailed comparison with benthic and pelagic compartments in shallow freshwater lakes. Appl. Environ. Microbiol. 74:1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coolen MJL, et al. 2007. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ. Microbiol. 9:1001–1016 [DOI] [PubMed] [Google Scholar]
- 14. De Bie MJM, et al. 2001. Shifts in the dominant populations of ammonia-oxidizing β-subclass proteobacteria along the eutrophic Schelde estuary. Aquat. Microb. Ecol. 23:225–236 [Google Scholar]
- 15. de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810–818 [DOI] [PubMed] [Google Scholar]
- 16. Felsenstein J. 2005. PHYLIP (Phylogenetic Inference Package), version 3.6 Department of Genome Science, University of Washington, Seattle, WA [Google Scholar]
- 17. Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujii C, et al. 2010. Succession and community composition of ammonia-oxidizing archaea and bacteria in bulk soil of a Japanese paddy field. Soil Sci. Plant Nutr. 56:212–219 [Google Scholar]
- 19. Hansel CM, Fendorf S, Jardine PM, Francis CA. 2008. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 74:1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hatzenpichler R, et al. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He JZ, et al. 2007. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ. Microbiol. 9:2364–2374 [DOI] [PubMed] [Google Scholar]
- 22. Herrmann M, Saunders AM, Schramm A. 2009. Effect of lake trophic status and rooted macrophytes on community composition and abundance of ammonia-oxidizing prokaryotes in freshwater sediments. Appl. Environ. Microbiol. 75:3127–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hooper AB, Terry KR. 1974. Photoinactivation of ammonia oxidation in Nitrosomonas. J. Bacteriol. 119:899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jia Z, Conrad R. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658–1671 [DOI] [PubMed] [Google Scholar]
- 25. Johnstone BH, Jones RD. 1988. Recovery of a marine chemolithotrophic ammonium-oxidizing bacterium from long-term energy-source deprivation. Can. J. Microbiol. 34:1347–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones RD, Morita RY. 1985. Survival of a marine ammonium oxidizer under energy-source deprivation. Mar. Ecol. Prog. Ser. 26:175–179 [Google Scholar]
- 27. Jung M-Y, et al. 2011. Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl. Environ. Microbiol. 77:8635–8647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kandeler E, Gerber H. 1988. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fertil. Soils 6:68–72 [Google Scholar]
- 29. Klotz MG, et al. 2006. Complete genome sequence of the marine, chemolithoautotrophic, ammonia-oxidizing bacterium Nitrosococcus oceani ATCC 19707. Appl. Environ. Microbiol. 72:6299–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Könneke M, et al. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed] [Google Scholar]
- 31. Koops HP, Pommerening-Roeser A. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1–9 [Google Scholar]
- 32. Koops HP, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M. 2007. The lithoautotrophic ammonia-oxidizing bacteria, p 778–811 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes 5. Springer, New York, NY [Google Scholar]
- 33. Kowalchuk GA, Stephen JR. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485–529 [DOI] [PubMed] [Google Scholar]
- 34. Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. 2011. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. U. S. A. 108:15892–15897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ludwig W, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–981 [DOI] [PubMed] [Google Scholar]
- 37. Merbt SN, et al. 2012. Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol. Lett. 327:41–46 [DOI] [PubMed] [Google Scholar]
- 38. Mosier AC, Francis CA. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002–3016 [DOI] [PubMed] [Google Scholar]
- 39. Mussmann M, et al. 2011. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108:16771–16776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966–2978 [DOI] [PubMed] [Google Scholar]
- 41. Nowlin WH, Evarts JL, Vanni MJ. 2005. Release rates and potential fates of nitrogen and phosphorus from sediments in a eutrophic reservoir. Freshw. Biol. 50:301–322 [Google Scholar]
- 42. Offre P, Prosser JI, Nicol GW. 2009. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol. Ecol. 70:99–108 [DOI] [PubMed] [Google Scholar]
- 43. Park BJ, et al. 2010. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl. Environ. Microbiol. 76:7575–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park HD, Noguera DR. 2007. Characterization of two ammonia-oxidizing bacteria isolated from reactors operated with low dissolved oxygen concentrations. J. Appl. Microbiol. 102:1401–1417 [DOI] [PubMed] [Google Scholar]
- 45. Pernthaler A, Pernthaler J, Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reigstad LJ, et al. 2008. Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol. Ecol. 64:167–174 [DOI] [PubMed] [Google Scholar]
- 47. Santoro AE, Casciotti KL, Francis CA. 2010. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ. Microbiol. 12:1989–2006 [DOI] [PubMed] [Google Scholar]
- 48. Sekar R, et al. 2003. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shand CA, Williams BL, Coutts G. 2008. Determination of N-species in soil extracts using microplate techniques. Talanta 74:648–654 [DOI] [PubMed] [Google Scholar]
- 50. Shears JH, Wood PM. 1985. Spectroscopic evidence for a photosensitive oxygenated state of ammonia monooxygenase. Biochem. J. 226:499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sigee DC. 2005. Freshwater microbiology: biodiversity and dynamic interactions of microorganisms in the aquatic environment. John Wiley & Sons, Chichester, Great Britain [Google Scholar]
- 52. Spang A, et al. 2010. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 18:331–340 [DOI] [PubMed] [Google Scholar]
- 53. Speksnijder AGCL, Kowalchuk GA, Roest K, Laanbroek HJ. 1998. Recovery of a Nitrosomonas-like 16S rDNA sequence group from freshwater habitats. Syst. Appl. Microbiol. 21:321–330 [DOI] [PubMed] [Google Scholar]
- 54. Suzuki I, Dular U, Kwok SC. 1974. Ammonia or ammonium ion as substrate for oxidation by Nitrosomonas europaea cells and extracts. J. Bacteriol. 120:556–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tourna M, Freitag TE, Nicol GW, Prosser JI. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357–1364 [DOI] [PubMed] [Google Scholar]
- 56. Tourna M, et al. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U. S. A. 108:8420–8425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Treusch AH, et al. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985–1995 [DOI] [PubMed] [Google Scholar]
- 58. Urakawa H, Martens-Habbena W, Stahl DA. 2011. Physiology and genomics of ammonia-oxidizing archaea, p 117–155 In Ward BB, Arp DJ, Klotz MG. (ed), Nitrification. ASM Press, Washington, DC [Google Scholar]
- 59. van der Wielen PWJJ, Voost S, van der Kooij D. 2009. Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl. Environ. Microbiol. 75:4687–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 61. Verhagen FJM, Laanbroek HJ. 1991. Competition for ammonium between nitrifying and heterotrophic bacteria in dual energy-limited chemostats. Appl. Environ. Microbiol. 57:3255–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Verhamme DT, Prosser JI, Nicol GW. 2011. Ammonia concentration determines differential growth of ammonia-oxidizing archaea and bacteria in soil microcosms. ISME J. 5:1067–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Walker CB, et al. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:8818–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wells GF, et al. 2009. Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ. Microbiol. 11:2310–2328 [DOI] [PubMed] [Google Scholar]
- 65. Xu M, Schnorr J, Keibler B, Simon HM. 2012. Comparative analysis of 16S rRNA and amoA genes from archaea selected with organic and inorganic amendments in enrichment culture. Appl. Environ. Microbiol. 78:2137–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yarza P, et al. 2008. The all-species living tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31:241–250 [DOI] [PubMed] [Google Scholar]
- 67. Zhang CL, et al. 2008. Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl. Environ. Microbiol. 74:6417–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.