Abstract
Sphingomonas sp. strain Fr1 has recently been shown to protect Arabidopsis thaliana against the bacterial leaf pathogen Pseudomonas syringae DC3000. Here, we describe a forward genetic in planta screen to identify genes in Sphingomonas sp. Fr1 necessary for this effect. About 5,000 Sphingomonas sp. Fr1 mini-Tn5 mutants were assayed for a defect in plant protection against a luxCDABE-tagged P. syringae DC3000 derivative in a space-saving 24-well plate system. The bioluminescence of the pathogen was used as the indicator of pathogen proliferation and allowed for the identification of Sphingomonas sp. Fr1 mutants that had lost the ability to restrict pathogen growth before disease symptoms were visible. Potential candidates were validated using the same miniaturized experimental system. Of these mutants, 10 were confirmed as plant protection defective yet colonization competent. The mutants were subsequently evaluated in a previously described standard microbox system, and plants showed enhanced disease phenotypes after pathogen infection relative to those inoculated with the parental strain as a control. However, the disease severities were lower than those observed for control plants that were grown axenically prior to pathogen challenge, which suggests that several traits may contribute to plant protection. Transposon insertion sites of validated mutants with defects in plant protection were determined and mapped to 7 distinct genomic regions. In conclusion, the established screening protocol allowed us to identify mutations that affect plant protection, and it opens the possibility to uncover traits important for in planta microbe-microbe interactions.
INTRODUCTION
Plants are colonized by a variety of microorganisms, including bacteria, fungi, and yeasts. In the above-ground parts of plants, known as the phyllosphere, bacteria are the most abundant inhabitants (31), and their populations in the phyllosphere are estimated to number up to 1026 cells globally (34). Members of the genus Sphingomonas are frequently found in the phyllospheres of different plants (17, 29). It was recently shown that various Sphingomonas strains protect Arabidopsis thaliana against bacterial pathogens: upon pathogen application, Sphingomonas-inoculated plants exhibited reduced pathogen cell numbers compared to control plants, and plant phenotypes were similar to those of uninfected plants (25). Different mechanisms by which biological control agents can confer protection against plant pathogens have been proposed (3, 26). These either may be on the level of bacterium-bacterium antagonism through competition for space and nutrients and the production of antimicrobial compounds or may be plant mediated with the induction of systemic resistance. These modes of action are not mutually exclusive, and plant-protective microorganisms may employ more than one mechanism (26).
The mechanism by which Sphingomonas spp. suppress disease is not yet known. The high overlap in carbon source utilization between beneficial Sphingomonas spp. and Pseudomonas syringae is consistent with substrate competition playing a role in the antagonistic effect; however, direct evidence is lacking (25), and additional mechanisms cannot be excluded. Two general strategies can be used to decipher the mode of protection: (i) targeted approaches to test presumed mechanisms individually or (ii) a forward genetic screen. The screening of randomly generated mutants for loss-of-function mutations is a commonly applied method. For example, this approach was used to identify root symbiosis-defective mutants of Bradyrhizobium sp. strain ORS278 (11), virulence-defective mutants of P. syringae (14), and antibiosis-negative mutants of Erwinia herbicola (28). Screenings aimed at the detection of plant-protective effects of presumed antibiotics have often been performed by cultivating bacteria on agar plates (28). However, it is well known that these plant-protective phenotypes, such as antibiosis, do not necessarily correlate with in planta phenotypes (4, 12, 20, 26). Therefore, in planta analysis of mutations for the loss of protective effects represents a more straightforward method of screening.
The assessment of the beneficial effects of plant-colonizing bacteria usually requires long-term monitoring of plant growth and disease development in combination with quantification of bacterial populations (6, 12). Most methods used for the quantification of plant-associated microbial populations are time-consuming and invasive, thereby restricting dynamic observations or high-throughput applications (27). Bioluminescent reporter strains offer an alternative, nondisruptive approach for measuring bacterial growth on plants by detecting photons emitted by the labeled bacteria (21, 42).
Here, our aim was to establish a space- and time-saving system that allows for the screening of a library of Sphingomonas sp. strain Fr1 mutants for defects in plant protection. The screen is based on a chromosomally luxCDABE-tagged P. syringae strain (18) whose infection and subsequent amplification are monitored in A. thaliana.
MATERIALS AND METHODS
Strains and culture conditions.
The plant-protective strain Sphingomonas sp. Fr1 (25) was grown on nutrient broth without additional NaCl (NB; Fluka) or on minimal medium (MM) (37) containing 0.5% glucose as a carbon source at 28°C. Escherichia coli S17-1(λpir) carrying the suicide vector pAG408 (46) was cultivated at 37°C on LB medium supplemented with 25 μg/μl kanamycin (10). The lux-tagged P. syringae DC3000 (P. syringae DC3000 lx) (18) was grown on King's B medium (KB) (30) at 28°C.
Transposon mutagenesis.
Sphingomonas sp. Fr1 mutants were constructed by mini-Tn5 transposon mutagenesis. Biparental mating was carried out overnight on MM plates without a carbon source at 28°C with the conjugal donor E. coli S17-1(λpir) containing the plasmid pAG408 (46) and the acceptor Sphingomonas sp. Fr1. Cells were then scraped off and suspended in 0.9% NaCl. Serial dilutions were plated onto selective MM plates supplemented with glucose. To suppress the growth of cells that had acquired a weak kanamycin resistance but did not insert the transposon into the chromosome, kanamycin was used at a high concentration (200 μg/μl). The donor E. coli cells were counterselected by the addition of colistin (10 μg/μl) to the medium. Transconjugants were selected after 5 days of incubation and were further maintained on MM supplemented with glucose and kanamycin (50 μg/μl).
Growth conditions for Arabidopsis thaliana plants in 24-well plates.
A. thaliana Col-0 seeds were grown on Murashige-Skoog medium (MS) (including vitamins [Duchefa]) (36) supplemented with 3% sucrose and 0.55% plant agar (Duchefa) in 24-well plates, with each well containing 1.5 ml of medium. Seeds were surface sterilized as described by Schlesier et al. (41), placed in 24-well plates (1 seed/well), and then stratified at 4°C for 2 days. After inoculation (see below), plates were sealed with Parafilm and placed in a growth chamber at 22°C under long-day conditions (16-h photoperiod) for 1 week and then under short-day conditions (9-h photoperiod). One day before infection with P. syringae DC3000 lx, the Parafilm was removed from the plates to reduce the relative humidity within the plates and the variation between plates.
Inoculation and infection of plants in 24-well plates.
Cells of 2-day-old Sphingomonas sp. Fr1 mutants grown on MM (supplemented with glucose and kanamycin [50 μg/μl]) agar plates were resuspended in 120 μl of 10 mM MgCl2 in 96-well microtiter plates to obtain an optical density at 600 nm (OD600) of approximately 0.5. Seeds were inoculated with 4 μl of bacterial suspension. As controls, seeds inoculated with a Sphingomonas sp. Fr1 wild-type suspension or 10 mM MgCl2 (axenic) were included. Ten to 14 days after seed inoculation, the plants were infected with P. syringae DC3000 lx. The infection inoculum was prepared as described previously (25), except that the concentration was adjusted to an OD600 of 3 × 10−6, corresponding to approximately 1,500 CFU per ml. Plants were infected by pipetting 15 to 20 μl of the suspension to the aerial parts of the plants, distributing it to the leaves and the center of the plant.
Throughout this paper, the term inoculation is used for the application of commensal Sphingomonas, whereas the term infection refers to the application of the pathogenic Pseudomonas.
Luminescence detection.
Luminescence signals (photon counts per seconds [cps]) were quantified with a plate reader (Victor3; Perkin-Elmer). Preliminary tests revealed 6 days postinfection (dpi) with P. syringae DC3000 lx as a suitable time point to monitor luminescence of protected and nonprotected plants; as a visual aid, luminescence images were taken at 6 dpi with the IVIS Spectrum imaging system (Xenogen) using the Living Image software 3.2. For luminescence imaging, a 20-s exposure and a 500-nm emission filter were used. The filter allowed for the detection of photons emitted during the luciferase reaction that peaked at 490 nm (49) but blocked the detection of plant phosphorescence. A Tn5 mutant was considered a putative candidate with a defect in plant protection when the photons exceeded a threshold level of 600 to 2,000 cps (corresponding to at least a 20-fold increase in cps relative to background level). The adjustment of the absolute luminescence intensity threshold was set for each screening round due to variations in the baseline luminescence, which may have been caused by slightly varying concentrations of the initial P. syringae DC3000 lx inoculum and differing environmental conditions.
Validation of candidate mutants from the initial screen in 24-well plates.
Each candidate was validated in the 24-well assay with 12 replicates distributed on three 24-well plates (at different positions to exclude local site effects). Luminescence values for wells with ungerminated seeds or plants with a strong growth delay were not considered for statistical analysis. For mutants that showed significantly higher log-transformed cps than wild type controls (one-way analysis of variance [ANOVA] with Tukey's post hoc test, P < 0.05), Sphingomonas sp. Fr1 colonization was determined on 6 plants at 7 dpi as described previously (25). Aerial parts of plants were separated from the roots and placed into 2-ml tubes containing 1.3 ml phosphate buffer (100 mM, pH 7) amended with 0.2% Silwet L-77 (GE Bayer Silicones). The tubes were shaken for 15 min at 25 Hz in a Retsch tissue lyser (Retsch) and sonicated in a water bath for 5 min. Dilution series were spotted onto MM (with glucose and streptomycin [20 μg/μl]) agar plates; for mutants, 50 μg/μl kanamycin was added. After incubation at 28°C for 3 days, plates were inspected for CFU. The remaining plants were incubated to monitor disease phenotypes of plants after 3 weeks postinfection. Mutants were considered tentatively validated candidates when their bioluminescence signals were elevated (determined by the test described above) and/or when disease phenotypes (determined by visual inspection) were observed, and the mutants could be recovered from the phyllosphere of the tested plants.
Microbox in planta assay.
For further evaluation of the mutants identified in the 24-well plate system, the effect of the mutants was analyzed in a previously established microbox system as described previously (25), except that the bacterial cultures used for seed inoculation were grown in MM supplemented with glucose instead of in NB. Bacterial cell numbers on aerial plant parts at the day of infection and at 21 days postinfection were determined as described previously (25). Dilution series were spotted onto different selective media. For Sphingomonas sp. Fr1, MM with glucose and streptomycin (20 μg/μl) was used, and for mutants, kanamycin (50 μg/μl) was added. P. syringae DC3000 lx was selected on KB supplemented with rifampin (50 μg/μl). CFU were counted after 2 to 3 days of incubation at 28°C. Additionally, disease phenotypes of plants were scored at 20 dpi with a disease severity index from 1 (no visible disease symptoms) to 5 (dead plant), as described by Whalen et al. (51). The weighted arithmetic means were calculated per mutant and then normalized to results for the wild-type (0) and noninoculated (1) controls.
Identification of transposon insertion sites.
The regions flanking the transposon insertion sites were amplified by arbitrarily primed PCR using primer sequences (Table 1) modified from those described by Das et al. (15). Genomic DNA was extracted from freshly grown plate cultures with the MasterPure DNA purification kit (Epicentre Biotechnologies). Transposon-specific primers, Iseq (inner end) or Oseq (outer end), were paired with the arbitrary primer Arb1, Arb2, or Arb3 in a first-round PCR with 100 ng chromosomal template DNA. PCR products were purified using the Gel and PCR Cleanup kit (Macherey-Nagel), and 1/10 was used as the template for the second-round PCR with the primer PCR2 paired with pagI (inner end) or pagO (outer end). Primer concentrations and PCR conditions were as described elsewhere (15). The PCR products were separated on 1.5% agarose gels, and single bands were excised, extracted with the Gel and PCR Cleanup kit (Macherey-Nagel), and sequenced (Microsynth) with the corresponding transposon primer, pagO or pagI. The obtained DNA sequences were compared, using BLAST (1), to the automatically annotated Sphingomonas sp. Fr1 draft genome sequence on RAST (5) to identify transposon insertion sites.
Table 1.
Oligonucleotides used for arbitrary primed PCR to identify transposon insertion sites
| Primer name | Sequence (5′ → 3′) | Reference |
|---|---|---|
| Arb1 | GGCCACGCGTCGACTAGTCANNNNNNNNNNGCTCGa | This study |
| Arb2 | GGCCACGCGTCGACTAGTCANNNNNNNNNNGACTCa | This study |
| Arb3 | GGCCACGCGTCGACTAGTCANNNNNNNNNNGATACa | This study |
| Iseq | TACGTGCAAGCAGATTACGG | 35 |
| Oseq | TTGTGCCCATTAACATCACC | 35 |
| PCR2 | GGCCACGCGTCGACTAGTCA | 15 |
| pagI | CAATTCGTTCAAGCCGAGAT | 35 |
| pagO | TGGGACAACTCCAGTGAAAA | 35 |
As described by Das et al. (15) but with a modified 3′-pentamer varying in GC content. N, ATCG.
RESULTS
Establishment of an in planta screening assay.
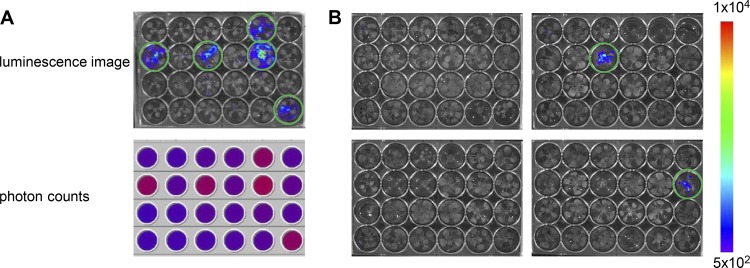
For the successful implementation of an in planta forward genetic screen, medium- to high-throughput procedures are needed. Because the identification of Sphingomonas sp. Fr1 mutants with defects in plant protection requires the testing of mutants one by one, we established an experimental setup that allowed for a reduction in the time and space required for such a screen. This screen was based on a previously established microbox assay (25) with the following adjustments: (i) a 24-well microtiter plate system was used, (ii) plant infection was carried out at an earlier time point, and (iii) a previously described lux-tagged Pseudomonas syringae DC3000 derivative (P. syringae DC3000 lx) was used as reporter strain for pathogen proliferation because it was shown that luminescence and pathogen population size are positively correlated in planta (18). The luminescence of the strain could be measured in a nondestructive way (Fig. 1A), and it allowed for the prediction of disease incidence at 6 days postinfection rather than 2 to 3 weeks postinfection, as is required for disease symptoms to be easily scored.
Fig 1.
Detection of photons emitted by the lux-tagged pathogen P. syringae DC3000 on plants. (A) Luminescence image and microplate reader photon counts of a 24-well plate with A. thaliana plants at 3 days postinfection (dpi) with P. syringae DC3000 lx. Plants were seed inoculated with wild-type Sphingomonas sp. Fr1 (unmarked) or kept axenically (marked with a circle on the luminescence image) prior to P. syringae DC3000 lx application. The photon counts (cps) measured with the microplate reader were converted to a color image, where red indicates high values and blue low values. (B) Exemplary luminescence images of 24-well plates used for screening at 6 dpi. Each well harbors a plant inoculated with a different Sphingomonas sp. Fr1 mutant. The luminescence signals are represented in colors as indicated by the scale bar (in arbitrary units). Sphingomonas mutants unable to restrict P. syringae DC3000 lx proliferation showed higher luminescence signals and are circled in green.
Screening and validation.
A library of Sphingomonas sp. Fr1 mutants was obtained by mini-Tn5 random mutagenesis. To circumvent the selection of auxotrophic mutants that are likely to be colonization deficient, a minimal medium containing glucose as the carbon source was used to grow cells after transformation and prior to plant inoculation. The genome size of Sphingomonas sp. Fr1 is predicted to be in the range of 4 Mbp based on a draft genome sequence generated by 454 shotgun sequencing (unpublished data). In total, 5,088 mutants were screened in 11 screening rounds, and controls (wild-type Sphingomonas sp. Fr1-inoculated plants infected with the pathogen and plants grown axenically prior to pathogen infection) were included in each of the screens. Exemplary luminescence images are shown in Fig. 1B. Mutants for which light intensity exceeded the threshold level were selected for validation. To account for the possibility of several mechanisms acting in concert, the luminescence threshold was set below intensities measured for axenic controls. This was due to the assumption that potential candidates were expected to show phenotypes intermediate between those of wild-type and axenic controls. Low threshold levels thus reduced the risk of missing mutants mildly affected in plant protection. In total, 121 mutants passed the threshold criteria and were subsequently validated further.
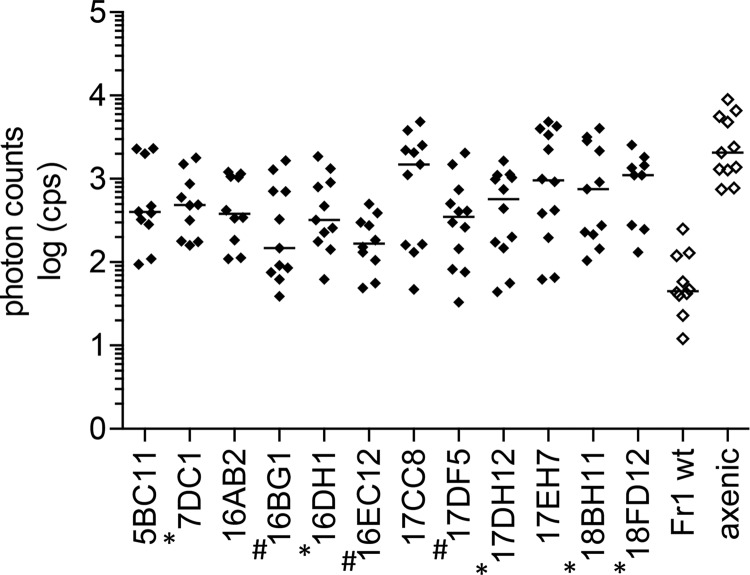
Mutants with a potential defect in plant protection were validated in replicates (n = 12) using the in planta screening protocol described above. Mutants either allowing pathogen growth on plants and/or failing to prevent disease symptoms were further investigated (exemplary validation results are shown in Fig. 2 and 3). To ensure that the elevated luminescence signals or enhanced disease symptom development did not result from a loss in phyllosphere colonization, the presence of Sphingomonas mutants on the aerial plant parts was checked by plating assays. Of the 15 mutants investigated, five did not show consistent plant colonization; therefore, they likely had a negative plant-protective phenotype. Overall, of the 121 validated mutants, 10 were further considered plant-protective-negative and five were considered plant colonization-negative candidates.
Fig 2.
Exemplary luminescence data of mutants validated in replicates of 12 in 24-well plates. Shown are log-transformed photon counts per second (cps) at 6 days postinfection with P. syringae DC3000 lx of mutant-inoculated plants that showed significantly higher log cps (Tukey's posttest, P < 0.05) than plants inoculated with wild-type Sphingomonas sp. Fr1 or allowed disease symptom formation (closed symbols). Values for control plants that were inoculated with wild-type Sphingomonas sp. Fr1 (Fr1 wt) or grown axenically (axenic) prior to pathogen infection are included (open symbols). Horizontal bars indicate the median. Sphingomonas sp. Fr1 mutants that were colonization defective on plants harvested are marked with an asterisk. Sphingomonas sp. Fr1 mutants that allowed disease symptom formation but did not show significantly higher log cps than the wild-type control are marked with a number sign.
Fig 3.
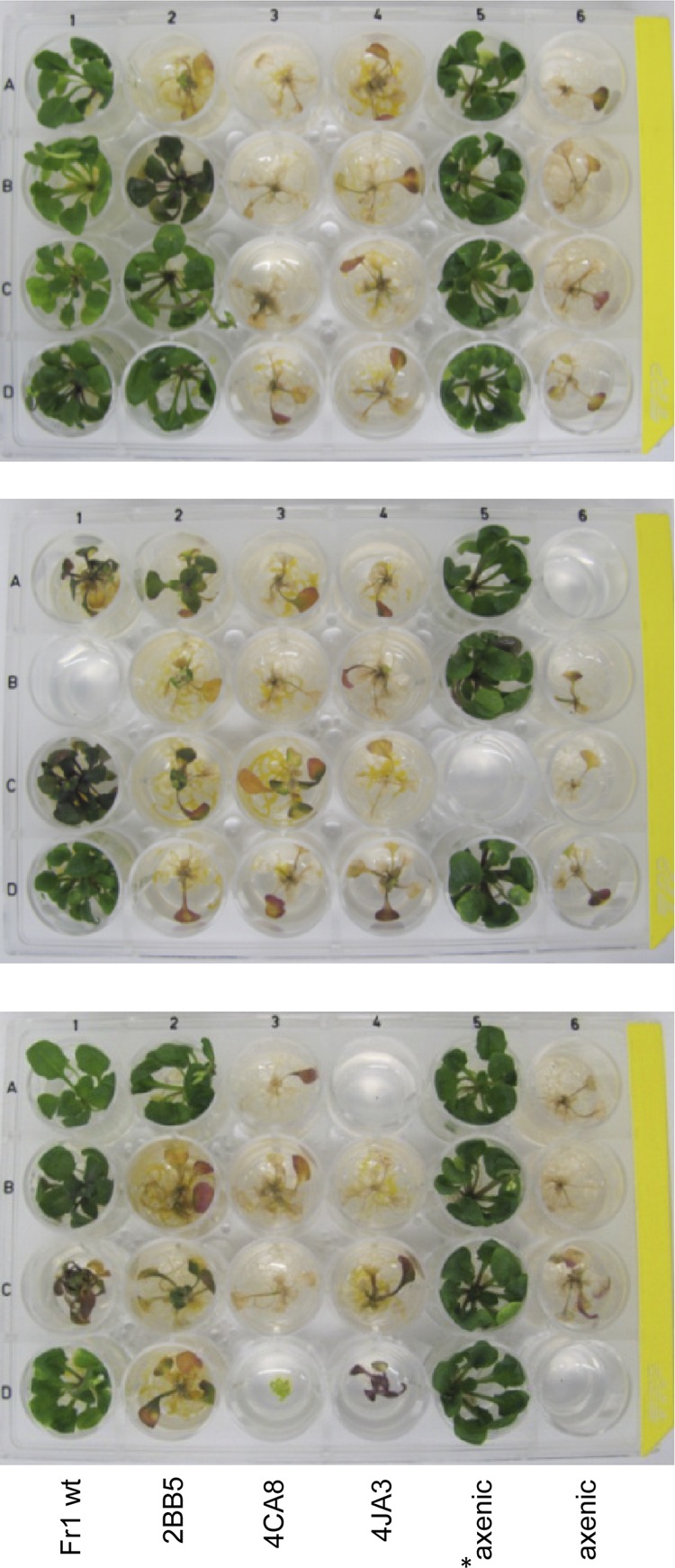
Exemplary photographs of three replicate validation plates at 21 days postinfection. A. thaliana plants were seed inoculated with wild-type Sphingomonas sp. Fr1 (Fr1 wt) or the plant protection-deficient mutants 2BB5, 4CA8, and 4JA3 or grown axenically prior to pathogen application (axenic). Ten-day-old plants were infected with the lux-tagged pathogen P. syringae DC3000 lx or mock treated with 10 mM MgCl2 (indicated by an asterisk). Note that this figure is for illustrative purposes; in standard validation plates, candidates were inoculated at different positions to avoid site effects.
Evaluation of mutants in microboxes.
To test whether the plant-protective-deficient phenotype was also observable in the previously established microbox system, the 10 mutants showing plant colonization and reduced plant protection in the 24-well plate system were further evaluated as described previously (25). Cell numbers of Sphingomonas were determined at 0 and 21 dpi and those of Pseudomonas at 21 dpi. Sphingomonas population sizes were in the range of 106 to 108 CFU per gram of leaf fresh weight, at both 0 and 21 dpi (Table 2). All mutants were able to colonize the plants; however, most mutants showed colonization levels lower than that of the wild type, which showed colonization in the range of 108 CFU per gram of leaf fresh weight, as previously shown (25). Exceptions were the mutants 17EH7 and 5BC11, which showed population sizes at 0 dpi that were similar to that of the wild type. Pathogen cell numbers at 21 dpi determined on plants inoculated with different Sphingomonas mutants were in the range of 108 to 1010 CFU per gram of leaf fresh weight and were thus elevated for most of the mutants compared to the wild type. Not only were pathogen cell numbers different, but plants inoculated with the different mutants also showed different levels of disease symptom formation, ranging from mildly diseased with a few small lesions to a strongly diseased phenotype comparable to that of plants that were grown axenically prior to pathogen infection (see Fig. S1 and S2 in the supplemental material). Disease severity scores at 21 dpi normalized to wild-type (0) and noninoculated (1) control phenotypes were determined. This was based on the disease severity index as described by Whalen et al. (51). The normalized index ranged from 0.3 to 0.8 for the different mutants (Table 2). For some mutants, such as mutant 16BG1, a pronounced plant-to-plant variation was observed, even for plants present in the same microbox. For others, such as mutant 16AB2, some phenotypic differences were apparent between microboxes, but nevertheless, most plants showed an enhanced disease phenotype compared to that of the wild-type-inoculated plants (see Fig. S1 and S2 in the supplemental material).
Table 2.
Colonization levels of and disease severity caused by Sphingomonas sp. Fr1 mutants and controls with genes disrupted by transposon insertion and the corresponding gene in Sphingomonas sp. S17
| Inoculation | Population in the phyllosphere, log(CFU/g)a |
Disease indexb | Gene annotation | Identifierc | Corresponding gene in Sphingomonas sp. S17d | ||
|---|---|---|---|---|---|---|---|
|
Sphingomonas sp. Fr1 |
P. syringae DC3000, 21 dpi | ||||||
| 0 dpi | 21 dpi | ||||||
| Fr1 (wild type) | 8.0 ± 0.2 | 8.3 ± 0.4 | 8.1 ± 1.1 | 0.0 | |||
| 17DF5 | 7.6 ± 0.6 | 7.0 ± 0.8 | 8.6 ± 0.7 | 0.3 | Cytochrome o ubiquinol oxidase subunit II | 2,537 | SUS17_373 |
| 17CC8 | 7.0 ± 0.3 | 6.4 ± 0.7 | 8.6 ± 0.9 | 0.4 | Intergenic | 1,536/1,537 | —e |
| 16BG1 | 7.3 ± 0.5 | 8.1 ± 0.9 | 8.3 ± 1.4 | 0.4 | Mo cofactor biosynthesis protein C | 990 | SUS17_830 |
| 4JA3 | 5.7 ± 0.7 | 6.3 ± 1.0 | 9.1 ± 0.6 | 0.5 | Sensor histidine kinase | 1,140 | SUS17_272 |
| 4CA8 | 7.4 ± 0.3 | 6.9 ± 0.6 | 9.5 ± 0.4 | 0.6 | Putative membrane protein | 1,490 | SUS17_2491 |
| 17EH7 | 7.5 ± 1.0 | 7.8 ± 0.7 | 8.9 ± 0.8 | 0.7 | Cytochrome o ubiquinol oxidase subunit IV | 2,534 | SUS17_370 |
| 2BB5 | 7.0 ± 1.2 | 7.5 ± 0.7 | 9.5 ± 0.7 | 0.7 | GTP pyrophosphokinase (SpoT) | 155 | SUS17_3068 |
| 16EC12 | 7.7 ± 0.3 | 6.7 ± 0.6 | 9.4 ± 0.7 | 0.7 | Putative membrane protein | 1,490 | SUS17_2491 |
| 5BC11 | 8.4 ± 0.4 | 8.2 ± 0.5 | 9.3 ± 0.5 | 0.7 | Cytochrome o ubiquinol oxidase subunit II | 2,537 | SUS17_373 |
| 16AB2 | 6.5 ± 0.7 | 7.4 ± 0.7 | 9.1 ± 0.6 | 0.8 | Multimodular transpeptidase-transglycosylase | 869 | SUS17_3528 |
| Axenicf | 9.4 ± 0.4 | 1.0 | |||||
Sphingomonas sp. Fr1 and P. syringae DC3000 populations in the phyllosphere at 0 and 21 days postinfection. Shown are means ± standard deviations of log-transformed CFU per gram of leaf fresh weight (n = 12).
Disease severity at 20 dpi. Each plant was given a rank of 1 to 5 (healthy to diseased). Shown are normalized data for the weighted arithmetic means (n = 16 to 24).
Gene identifier according to a RAST annotation of a Sphingomonas sp. Fr1 draft genome (unpublished).
From reference 19.
—, no microsynteny in Sphingomonas sp. S17.
Control plants that were grown axenically prior to pathogen application.
Identification of transposon insertion sites.
For all mutants that were evaluated in the microbox system, transposon insertion sites were identified (Table 2). Insertions were found within the following genes: spoT (2BB5); genes encoding a sensor histidine kinase (4JA3), a multimodular transpeptidase-transglycosylase (16AB2), and Mo cofactor biosynthesis protein C (16BG1); genes belonging to an operon encoding a cytochrome o ubiquinol oxidase (17DF5, 17EH7, and 5BC11); and a gene encoding a putative membrane protein (4CA8 and 16EC12). In one mutant, the transposon insertion was intergenic (17CC8) and upstream of genes encoding a sugar transporter and an acyl coenzyme A (acyl-CoA) dehydrogenase.
DISCUSSION
The identification of traits involved in plant protection requires genotype-phenotype correlations and causality. In this study, we established and applied a forward genetic screen to identify genes affecting plant protection by Sphingomonas sp. Fr1 against the bacterial pathogen P. syringae DC3000. The in planta screening of a mutant library provides a number of advantages: (i) it does not rely on in vitro culture conditions, which are usually not adequate to reflect in situ conditions, and (ii) an unbiased approach has the potential to uncover novel mechanisms. The presented procedure corresponds to a miniaturized setup of a previously described standardized microbox system with similar cultivation, inoculation, and infection. The use of a bioluminescent reporter strain served in a nondestructive way as a proxy for pathogen counts (18, 21) and allowed us to predict disease outcome at 6 days postinfection, before disease symptoms can be readily detected. In total, the modifications reduced the time needed for in planta screening for disease outcome to 3 weeks.
For Sphingomonas sp. Fr1, 3,539 genes are predicted according to the automatic annotation of the draft genome sequence. Considering this number and the completion of a screen using a total of 5,088 Tn5 mutants of Sphingomonas sp. Fr1 for loss of plant protection in the model system A. thaliana/P. syringae DC3000, we have not achieved a full statistical coverage screen of knocking down all nonessential genes. However, the fact that we identified a number of targets two or three times (independent insertions) suggests that a substantial coverage of mutants was achieved. This observation also indicates a certain robustness of the screening procedure, although variability of the disease severity from plant to plant within one experiment and between experiments using the same genotypes was observed. Such variability may be due to small differences in the environment, such as relative humidity and temperature, that may affect the plant-pathogen interaction (7). It may also be due to stochastic events introduced by the localization of the tested antagonist and/or the pathogen in heterogeneous microniches (40) or to effects occurring as a consequence of phenotypic variation of one of the three partners.
Although we have chosen the candidates tested in the microbox system based not only on the bioluminescence of the pathogen but also on the colonization ability of the candidate in the 24-well system, most candidates identified showed lower population densities than the wild type in the microbox system. It is known that preemptive colonization of target sites can be important for efficient biological control in some pathosystems (22, 52), whereas in others, a reduction in plant colonization by one order of magnitude does not affect plant protection (50). In our system, we cannot exclude that some of the biocontrol reduction may be accounted for by reduced population density. However, no correlation between reduced Sphingomonas and increased Pseudomonas population densities was observed (Table 2). Therefore, we do not expect that all phenotypes can be explained solely by the colonization abilities of the different Sphingomonas mutants. Clearly, colonization ability is not the determinant of loss of protection for mutants with insertions within the operon encoding the cytochrome o ubiquinol oxidase, as these population densities are comparable to that of the parental strain.
Plant protection may be multifactorial; i.e., several traits can contribute to the biocontrol activity of bacterial strains (28, 45). It is therefore not unexpected that none of the 10 candidates identified showed a complete plant protection-deficient phenotype but that they displayed intermediate phenotypes instead (Table 2). However, it is also possible that some mutations may not lead to a full loss of functionality of the gene product or may partially affect the expression of other genes or the expression of downstream genes (16). Such positional effects may also explain the differences in the severity of the phenotypes of mutants in the same genomic region (Table 2). Additionally, redundancy may be present in the system, thus rendering the identification of candidate genes difficult. The generation of “clean” knockouts and multiple knockout mutants of the most promising candidate genes will allow for future validation of the genes identified in this study and further examination of whether the effect of plant protection is additive.
Three mutants showed insertions within an operon encoding a cytochrome o ubiquinol oxidase, a terminal oxidase in the respiratory chain. Because these mutants show as high a level of plant colonization as the wild type, we do not expect that they are affected in growth per se. Interestingly, Pseudomonas putida cytochrome o ubiquinol oxidase was shown not only to act as an important terminal oxidase but also to contribute to global control of gene expression (33). The loss of a terminal oxidase can also influence the transcriptome comprehensively in other bacteria (43). It is possible that the mutations in the genes for cytochrome o ubiquinol oxidase identified in Sphingomonas sp. Fr1 are also of regulatory importance and thus have indirect effects on the trait(s) affecting plant protection. Another mutant impaired in plant protection was disrupted in SpoT, a GTP pyrophosphokinase. This enzyme catalyzes the reactions of GDP and GTP to ppGpp (guanosine 3′,5′-bipyrophosphate) and pppGpp (guanosine 3′-diphosphate 5′-triphosphate), respectively. These molecules are synthesized in response to different environmental stresses (e.g., starvation) and are involved in the regulation of various cellular processes, such as growth, survival, production of secondary metabolites, motility, biofilm formation, and others (reviewed in reference 38). In mutant 4JA3, the insertion mapped to a gene encoding a histidine kinase which shares homology with ChvG from Agrobacterium tumefaciens C58 (38% amino acid identity) and ExoS from Sinorhizobium meliloti 1021 (38% amino acid identity). ChvG is the sensor kinase of the ChvG/ChvI two-component system. This two-component system and its orthologues have been found to be important in the regulation of different host-microbe interactions (8, 13, 23, 39, 48), and mutations in this system can affect various cellular processes, such as metabolism, exopolysaccharide synthesis, motility, and biofilm formation (8, 48). For all of these regulatory mutants, pleiotropic phenotypes are expected, and it will be important to deduce which factors are most important for plant protection.
In mutant 16AB2, a transposon insertion was mapped to a gene encoding a multimodular transpeptidase-transglycosylase with homology to penicillin-binding proteins, the main peptidoglycan biosynthesis enzymes in E. coli (24). The putative membrane protein disrupted in the mutants 16EC12 and 4CA8 shares homology with RodZ, a protein required for proper actin cytoskeleton assembly and cell shape (2, 9, 44, 47). It is also possible that a regulatory function in these mutants was disrupted, as it was recently shown that in Shigella sonnei, RodZ is important not only for cell shape but also for posttranscriptional regulation of the type III secretion system (32).
Using a forward genetic screening procedure, we were able to identify a number of genes in Sphingomonas sp. Fr1 that affect the outcome of pathogen proliferation on A. thaliana. In-depth studies of these candidates will be the subject of future research.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Vontobel Foundation.
We thank Jun Fan (John Innes Centre, Norwich, United Kingdom) for providing lux-tagged Pseudomonas syringae DC3000.
Footnotes
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 2. Alyahya SA, et al. 2009. RodZ, a component of the bacterial core morphogenic apparatus. Proc. Natl. Acad. Sci. U. S. A. 106:1239–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andrews JH. 1992. Biological control in the phyllosphere. Annu. Rev. Phytopathol. 30:603–635 [DOI] [PubMed] [Google Scholar]
- 4. Andrews JH. 1985. Strategies for selecting antagonistic microorganisms from the phylloplane, p 31–44 In Windels CE, Lindow SE. (ed), Biological control on the phylloplane. The American Phytopathological Society, St. Paul, MN [Google Scholar]
- 5. Aziz RK, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75 doi:10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bashan Y, de Bashan LE. 2002. Protection of tomato seedlings against infection by Pseudomonas syringae pv. tomato by using the plant growth-promoting bacterium Azospirillum brasilense. Appl. Environ. Microbiol. 68:2637–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beattie GA. 2011. Water relations in the interaction of foliar bacterial pathogens with plants. Annu. Rev. Phytopathol. 49:533–555 [DOI] [PubMed] [Google Scholar]
- 8. Bélanger L, Dimmick KA, Fleming JS, Charles TC. 2009. Null mutations in Sinorhizobium meliloti exoS and chvI demonstrate the importance of this two-component regulatory system for symbiosis. Mol. Microbiol. 74:1223–1237 [DOI] [PubMed] [Google Scholar]
- 9. Bendezu FO, Hale CA, Bernhardt TG, de Boer PAJ. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 28:193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bertani G. 1951. Studies on lysogenesis. 1. The mode of phage liberation by lysogenic Escherichia-coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bonaldi K, et al. 2010. Large-scale transposon mutagenesis of photosynthetic Bradyrhizobium sp. strain ORS278 reveals new genetic loci putatively important for Nod-independent symbiosis with Aeschynomene indica. Mol. Plant Microbe Interact. 23:760–770 [DOI] [PubMed] [Google Scholar]
- 12. Braun SD, et al. 2010. In vitro antibiosis by Pseudomonas syringae Pss22d, acting against the bacterial blight pathogen of soybean plants, does not influence in planta biocontrol. J. Phytopathol. 158:288–295 [Google Scholar]
- 13. Charles TC, Nester EW. 1993. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 175:6614–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cuppels DA. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das S, Noe JC, Paik S, Kitten T. 2005. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J. Microbiol. Methods 63:89–94 [DOI] [PubMed] [Google Scholar]
- 16. de Bruijn FJ, Lupski JR. 1984. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids—a review. Gene 27:131–149 [DOI] [PubMed] [Google Scholar]
- 17. Delmotte N, et al. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:16428–16433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan J, Crooks C, Lamb C. 2008. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 53:393–399 [DOI] [PubMed] [Google Scholar]
- 19. Farias ME, et al. 2011. Genome sequence of Sphingomonas sp. strain S17, isolated from an alkaline, hyperarsenic, and hypersaline volcano-associated lake at high altitude in the Argentinean Puna. J. Bacteriol. 193:3686–3687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fravel DR. 1988. Role of antibiosis in the biocontrol of plant diseases. Annu. Rev. Phytopathol. 26:75–91 [Google Scholar]
- 21. Gau AE, Dietrich C, Kloppstech K. 2002. Non-invasive determination of plant-associated bacteria in the phyllosphere of plants. Environ. Microbiol. 4:744–752 [DOI] [PubMed] [Google Scholar]
- 22. Giddens SR, Houliston GJ, Mahanty HK. 2003. The influence of antibiotic production and pre-emptive colonization on the population dynamics of Pantoea agglomerans (Erwinia herbicola) Eh1087 and Erwinia amylovora in planta. Environ. Microbiol. 5:1016–1021 [DOI] [PubMed] [Google Scholar]
- 23. Guzman-Verri C, et al. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. U. S. A. 99:12375–12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Höltje JV. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Innerebner G, Knief C, Vorholt JA. 2011. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77:3202–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jacobsen BJ. 2006. Biological control of plant diseases by phyllosphere applied biological control agents, p 133–147 In Bailey MJ, Lilley AK, Timms-Wilson TM, Spencer-Phillips PTN. (ed), Microbial ecology of aerial plant surfaces. CAB International, Wallingford, United Kingdom [Google Scholar]
- 27. Katagiri F, Thilmony R, He SY. 2002. The Arabidopsis thaliana-Pseudomonas syringae interaction. In Somerville CR, Meyerowitz EM. (ed), The Arabidopsis book. American Society of Plant Biologists, Rockville, MD: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kempf HJ, Wolf G. 1989. Erwinia herbicola as a biocontrol agent of Fusarium culmorum and Puccinia recondita f. sp. tritici on wheat. Phytopathology 79:990–994 [Google Scholar]
- 29. Kim H, et al. 1998. High population of Sphingomonas species on plant surface. J. Appl. Microbiol. 85:731–736 [Google Scholar]
- 30. King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 31. Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitobe J, et al. 2011. RodZ regulates the post-transcriptional processing of the Shigella sonnei type III secretion system. EMBO Rep. 12:911–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morales G, Ugidos A, Rojo F. 2006. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 8:1764–1774 [DOI] [PubMed] [Google Scholar]
- 34. Morris CE, Kinkel LL. 2002. Fifty years of phyllosphere microbiology: significant contributions to research in related fields, p 365–375 In Lindow SE, Hecht-Poinar EI, Elliott VJ. (ed), Phyllosphere microbiology. APS Press, St. Paul, MN [Google Scholar]
- 35. Muller EEL, et al. 2011. Functional genomics of dichloromethane utilization in Methylobacterium extorquens DM4. Environ. Microbiol. 13:2518–2534 [DOI] [PubMed] [Google Scholar]
- 36. Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15:473 [Google Scholar]
- 37. Peyraud R, et al. 2009. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc. Natl. Acad. Sci. U. S. A. 106:4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51 [DOI] [PubMed] [Google Scholar]
- 39. Quebatte M, et al. 2010. The BatR/BatS two-component regulatory system controls the adaptive response of Bartonella henselae during human endothelial cell infection. J. Bacteriol. 192:3352–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Remus-Emsermann MN, Tecon R, Kowalchuk GA, Leveau JH. 2012. Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J. 6:756–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schlesier B, Breton F, Mock HP. 2003. A hydroponic culture system for growing Arabidopsis thaliana plantlets under sterile conditions. Plant Mol. Biol. Rep. 21:449–456 [Google Scholar]
- 42. Shaw JJ, Kado CI. 1986. Development of a Vibrio bioluminescence gene-set to monitor phytopathogenic bacteria during the ongoing disease process in a nondisruptive manner. Nat. Biotechnol. 4:560–564 [Google Scholar]
- 43. Shepherd M, Sanguinetti G, Cook GM, Poole RK. 2010. Compensations for diminished terminal oxidase activity in Escherichia coli: cytochrome bd-II-mediated respiration and glutamate metabolism. J. Biol. Chem. 285:18464–18472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shiomi D, Sakai M, Niki H. 2008. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 27:3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stockwell VO, Johnson KB, Sugar D, Loper JE. 2002. Antibiosis contributes to biological control of fire blight by Pantoea agglomerans strain Eh252 in orchards. Phytopathology 92:1202–1209 [DOI] [PubMed] [Google Scholar]
- 46. Suarez A, et al. 1997. Green fluorescent protein-based reporter systems for genetic analysis of bacteria including monocopy applications. Gene 196:69–74 [DOI] [PubMed] [Google Scholar]
- 47. van den Ent F, Johnson CM, Persons L, de Boer P, Lowe J. 2010. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 29:1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vanderlinde EM, Yost CK. 2012. Mutation of the sensor kinase chvG in Rhizobium leguminosarum negatively impacts cellular metabolism, outer membrane stability, and symbiosis. J. Bacteriol. 194:768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Welham PA, Stekel DJ. 2009. Mathematical model of the Lux luminescence system in the terrestrial bacterium Photorhabdus luminescens. Mol. Biosyst. 5:68–76 [DOI] [PubMed] [Google Scholar]
- 50. Wensing A, et al. 2010. Impact of siderophore production by Pseudomonas syringae pv. syringae 22d/93 on epiphytic fitness and biocontrol activity against Pseudomonas syringae pv. glycinea 1a/96. Appl. Environ. Microbiol. 76:2704–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Whalen MC, Innes RW, Bent AF, Staskawicz BJ. 1991. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3:49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilson M, Lindow SE. 1994. Ecological similarity and coexistence of epiphytic ice-nucleating (Ice+) Pseudomonas syringae strains and a non-ice-nucleating (Ice−) biological control agent. Appl. Environ. Microbiol. 60:3128–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.